Published online Jul 7, 2016. doi: 10.3748/wjg.v22.i25.5822
Peer-review started: December 31, 2015
First decision: March 22, 2016
Revised: April 8, 2016
Accepted: April 20, 2016
Article in press: April 20, 2016
Published online: July 7, 2016
Processing time: 186 Days and 2.7 Hours
AIM: To investigate clarithromycin resistance positions 2142, 2143 and 2144 of the 23SrRNA gene in Helicobacter pylori (H. pylori) by nested-allele specific primer-polymerase chain reaction (nested-ASP-PCR).
METHODS: The gastric tissue and saliva samples from 99 patients with positive results of the rapid urease test (RUT) were collected. The nested-ASP-PCR method was carried out with the external primers and inner allele-specific primers corresponding to the reference strain and clinical strains. Thirty gastric tissue and saliva samples were tested to determine the sensitivity of nested-ASP-PCR and ASP-PCR methods. Then, clarithromycin resistance was detected for 99 clinical samples by using different methods, including nested-ASP-PCR, bacterial culture and disk diffusion.
RESULTS: The nested-ASP-PCR method was successfully established to test the resistance mutation points 2142, 2143 and 2144 of the 23SrRNA gene of H. pylori. Among 30 samples of gastric tissue and saliva, the H. pylori detection rate of nested-ASP-PCR was 90% and 83.33%, while the detection rate of ASP-PCR was just 63% and 56.67%. Especially in the saliva samples, nested-ASP-PCR showed much higher sensitivity in H. pylori detection and resistance mutation rates than ASP-PCR. In the 99 RUT-positive gastric tissue and saliva samples, the H. pylori-positive detection rate by nested-ASP-PCR was 87 (87.88%) and 67 (67.68%), in which there were 30 wild-type and 57 mutated strains in gastric tissue and 22 wild-type and 45 mutated strains in saliva. Genotype analysis showed that three-points mixed mutations were quite common, but different resistant strains were present in gastric mucosa and saliva. Compared to the high sensitivity shown by nested-ASP-PCR, the positive detection of bacterial culture with gastric tissue samples was 50 cases, in which only 26 drug-resistant strains were found through analyzing minimum inhibitory zone of clarithromycin.
CONCLUSION: The nested-ASP-PCR assay showed higher detection sensitivity than ASP-PCR and drug sensitivity testing, which could be performed to evaluate clarithromycin resistance of H. pylori.
Core tip: In recent years, antibiotic resistance in Helicobacter pylori (H. pylori) has become a global problem, especially the resistances towards metronidazole, clarithromycin and amoxicillin. To combat this growing problem, it is important to determine antimicrobial resistance of a patient’s infection before treatment. Normally, the detection of clarithromycin resistance is based mainly on phenotypic methods performed after culturing. However, bacterial culture has many inherent disadvantages. In the present study, the nested-allele specific primer-polymerase chain reaction was established to detect the different mutations at positions 2142, 2143 and 2144 in the 23SrRNA gene of H. pylori, which can be completed within several hours. This method is expected to contribute towards improving the efficacy of H. pylori eradication therapy.
- Citation: Luo XF, Jiao JH, Zhang WY, Pu HM, Qu BJ, Yang BY, Hou M, Ji MJ. Establishment of a nested-ASP-PCR method to determine the clarithromycin resistance of Helicobacter pylori. World J Gastroenterol 2016; 22(25): 5822-5830
- URL: https://www.wjgnet.com/1007-9327/full/v22/i25/5822.htm
- DOI: https://dx.doi.org/10.3748/wjg.v22.i25.5822
Helicobacter pylori (H. pylori) is a microaerobic, Gram-negative, spiral bacterium that colonizes the human stomach in a sustainable manner and is found in more than half of the world’s population[1]. Eradication of H. pylori infection is required for the treatment of upper gastrointestinal disorders, peptic ulcer diseases, gastric mucosa-associated lymphoid tissue lymphoma and gastric cancer[2]. In most regimens, clarithromycin (CLA) is the key ingredient of strategies to eradicate H. pylori. Bacterial susceptibility to CLA is significantly related to eradication rates of H. pylori by CLA-based therapy. However, this therapy has gradually come under questioning because of increased eradication failure rates. Many factors contribute to the increased risk of failure, such as ineffective penetration of antibiotics into the gastric mucosa, antibiotic inactivation by low stomach pH, and lack of patient compliance[3-5]. Another important cause of failure of H. pylori eradication therapy is resistance to antibiotics. According to many reports[6-8], the incidence of resistance to CLA has increased rapidly in different geographical regions, such as from 29% in 2004 to 77% in 2007 in France[9], from 6% to 55% in Belgium during two decades of observation (1990-2009)[10,11] and from 15% to 65% in 10 years (2000-2009) in China[12,13].
Many studies have confirmed that bacterial resistance of H. pylori to CLA is associated with structural change of the 23SrRNA. This structural change is caused by the single nucleotide polymorphism (SNP) of 23SrRNA[14]. A-to-G point mutations at three positions of 2142, 2143 and 2144 within domain V (A2142G, A2143G and A2144G) have been found to be associated with CLA resistance[2,15-21]. Generally, the assessment of CLA resistance is mainly based on phenotypic methods that are performed by sensitivity testing after bacterial culturing, including the agar diffusion for the E-test or the agar dilution method. However, these methods are time-consuming and lack sensitivity. Moreover, the E-test is unable to provide any information regarding the genetic mutations involved in the resistance mechanism. In the last decade, novel culture-free polymerase chain reaction (PCR)-based techniques have been established to detect these mutations, and they include the techniques of PCR-restriction fragment length polymorphism (RFLP), PCR-DNA-enzyme immunoassay[22], nested PCR[23-26] and so on. In this study, the nested-allele specific primer-polymerase chain reaction (nested-ASP-PCR) and ASP-PCR methods were implemented to determine SNPs of 23SrRNA in H. pylori. Furthermore, the feasibility of saliva as a noninvasive material for testing CLA resistance of H. pylori was evaluated, as compared to gastric tissue.
The standard strain NCTC11637 of H. pylori was kept in our laboratory. H. pylori isolates were collected from 99 patients at the Nanjing Qixia District Hospital (Nanjing, Jiangsu Province, China) from March 1, 2014 to December 30, 2014. The age range of these patients was from 18-years old to 65-years-old. All patients underwent gastroscopy examination and tested positive for H. pylori by the rapid urease test (RUT), in which the biopsy specimen was inoculated into the rapid urease reagent and a positive result was indicated when the color changed from yellow to pink within 15 min. Then, we collected the gastric tissue and saliva samples to detect 23SrRNA mutation points of CLA. All patients provided a signed informed consent form for study participation.
PCR primers were designed by the Primer 5.0 software, according to 23S ribosomal gene sequence of H. pylori (GenBank Accession No. U27270). Primers External-F and External-R were used as the nested-PCR outer primers to amplify the 23SrRNA gene fragment (1962-2466 bp, 505 bp) of H. pylori. Allele-specific primers (WT2142-F, MUT2142-F, WT2143-F, MUT2143-F, WT2144-F, MUT2144-F and Inner-R) were used as the nested-PCR inner primers to amplify the resistance mutation gene (2122-2415 bp, 294 bp). The primers are listed in Table 1.
| Gene name | Primer sequence, 5’-3’ | Product length (bp) |
| External-F | GCGTTGAATTGAAGCCCGAGTAAAC | 505 |
| External-R | CCGACTTTCGTCTCTGCTTGA | |
| WT2142-F | TCCTACCCGCGGCAAGACGGA | 294 |
| MUT2142-F | TCCTACCCGCGGCAAGACGGG | |
| WT2143-F | CCTACCCGCGGCAAGACGGAA | |
| MUT2143-F | CCTACCCGCGGCAAGACGGAG | |
| WT2144-F | CCTACCCGCGGCAAGACGGAAA | |
| MUT2144-F | CCTACCCGCGGCAAGACGGAAG | |
| Inner-R | GCCATTACACTCAACTTGCGATTTC |
The extracted DNA from the standard strain NCTC11637 was used as the positive control, while DNA extracted from H. pylori-negative samples determined by RUT and PCR as the negative control. Genomic DNAs extracted from gastric mucosa samples and saliva of 30 H. pylori-positive patients by RUT were used to evaluate the detection sensitivity between ASP-PCR and nested-ASP-PCR methods.
The ASP-PCR amplification reaction mixture (50 μL) contained 5 μL 10 × buffer, 1 μL dNTPs, 1 μL allele-specific primers (WT2142-F, MUT2142-F, WT2143-F, MUT2143-F, WT2144-F, MUT2144-F respectively and Inner-R), 1 μL rTaq DNA polymerase, 1 μL genomic DNA template and 40 μL double-distilled H2O. The PCR cycling conditions were 95 °C for 5 min, hot start, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 30 s, with a final extension step at 72 °C for 5 min. Amplified fragments were visualized by 1.5% agarose gel electrophoresis and staining with ethidium bromide.
The nested-ASP-PCR technique is carried out as a combination of nested-PCR and ASP-PCR, which improves detection sensitivity and specificity by using two rounds of the PCR procedure. The outer primers (External-F and External-R) were utilized to amplify the genomic DNA in the first round of PCR. Then, 1 μL outer PCR product was applied as a template to amplify the resistance mutations fragment with allele-specific primers in the second round of PCR. The reaction conditions for the two rounds of PCR were the same as outlined for the ASP-PCR. Finally, the amplified 23SrRNA gene fragments obtained by nested-ASP-PCR were sequenced in order to confirm the corresponding position’s mutation.
The CLA resistance of 99 RUT-positive H. pylori gastric mucosa and saliva samples was evaluated by various methods. The resistance mutation points were detected by nested-ASP-PCR. Gastric specimens were subjected to bacterial culturing and antimicrobial sensitivity testing. The biopsy samples were incubated at 37 °C under microaerobic conditions (5% O2, 10% CO2 and 85% N2) for 3 d to 5 d. Then, colonies were sub-cultured to determine the minimum inhibitory zone of CLA by the disk diffusion method (TianHe Microbial Ltd, Hangzhou, China). The minimum inhibitory zone used to define resistance to CLA was ≤ 13 mm.
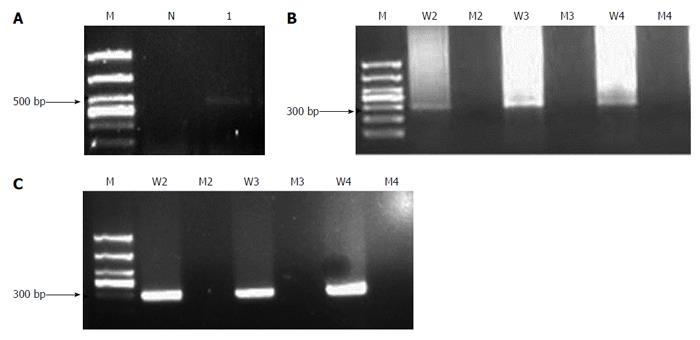
The H. pylori reference strain was used to establish the nested-ASP-PCR method. Then, PCR products were amplified from patient samples of gastric mucosa (Figure 1) and saliva (Supplementary Figure 1). The wild-type bacteria strain was amplified into PCR products with 2142A, 2143A and 2144A primers, while the mutated primers were unable to produce any amplification. As Figure 1A and B shows, the 505 bp outer gene fragment of 23SrRNA and the 294 bp inner PCR product were amplifiable by the outer primers and inner primers of the wild-type strain respectively. Furthermore, we found that the result of nested-PCR after the two-round PCR exhibited a stronger band at 294 bp (Figure 1C) than the one-round PCR procedure (Figure 1B). A similar finding was obtained in the detection of saliva samples. Because the DNA content was very low in saliva, the one-round PCR produced no bands (Supplementary Figure 1B), whereas the nested-PCR amplified the relative bands at 294 bp (Supplementary Figure 1C). Thus, the nested-PCR presented more detection sensitivity than the one-round PCR, especially for the saliva samples.
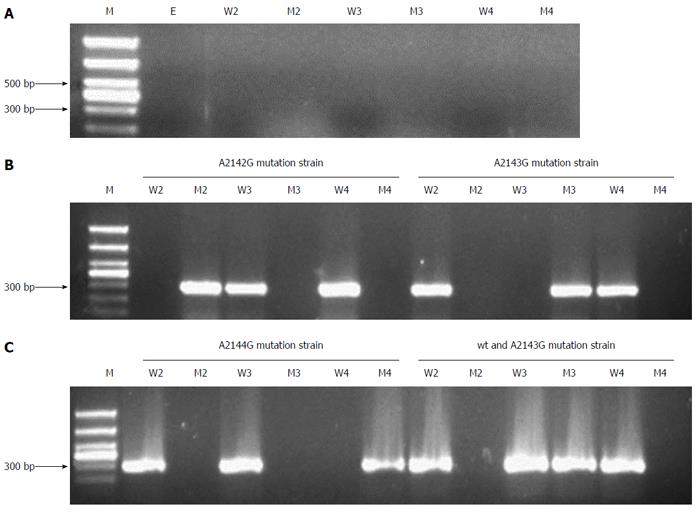
Next, we used the allele-specific primers to perform the second PCR in order to identify each gene mutation’s position. As Figure 2 shows, in gastric mucosa, H. pylori-negative samples yielded no PCR product (Figure 2A). The A2142G, A2143G and A2144G mutated H. pylori strains were identified according to the corresponding PCR products using different mutated primers, while other positions showed no mutation (Figure 2B and C). Figure 2C shows the mixed bacteria strains of wild-type and A2143G mutation. The saliva samples (Supplementary Figure 2) showed similar results as the gastric mucosa samples, but the bands amplified from saliva were weaker than those from gastric mucosa. The amplified PCR products were sent for sequencing, and the results were consistent with those previously reported in the NCBI GenBank database (Accession No. U27270). The sequences of fragments with 2142, 2143, 2144 position A-to-G mutation are shown in the Supplementary Text 1.
To ascertain that the nested-ASP-PCR method has higher sensitivity compared to ASP-PCR, we implemented a small-sample experiment. The gastric mucosa and saliva samples from 30 RUT-positive patients were detected to determine the resistance mutation position by the nested-ASP-PCR and ASP-PCR methods. The H. pylori-positive detection includes the amplification detection of wild-type and mutated fragments of H. pylori strains. As shown in Table 2, among the 30 gastric tissue samples, the H. pylori-positive rate detected by nested-ASP-PCR was 90%, but only 63% by ASP-PCR. In the 30 saliva samples, the H. pylori-positive detection rates of nested-ASP-PCR and ASP-PCR were 83.33% and 56.67% respectively. Thus, the detection rate of nested-ASP-PCR was obviously higher than that of ASP-PCR. Furthermore, H. pylori-resistance mutation detection of 23SrRNA in the 30 gastric tissue and saliva samples by nested-ASP-PCR was 11 and 16 respectively, while the number of total mutation was 11 and 10 detected by ASP-PCR. These results suggested that nested-ASP-PCR was more suitable for the detection of low levels of H. pylori in saliva samples.
| H. pylori-positive detection rate | WT | Total mutation, A-to-G | 2142 G | 2143 G | 2144 G | |
| Gastric mucosa | ||||||
| ASP-PCR | 19 | 8 | 11 | 10 | 10 | 10 |
| Ratio | 63% | 42.11% | 57.89 | 52.63% | 52.63% | 52.63% |
| (19/30) | (8/30) | (11/19) | (10/19) | (10/19) | (10/19) | |
| Nested-ASP-PCR | 27 | 16 | 11 | 11 | 10 | 11 |
| Ratio | 90% | 59.26% | 40.74% | 40.74% | 37.04% | 40.74% |
| (27/30) | (16/30) | (11/19) | (11/19) | (10/19) | (10/19) | |
| Saliva | ||||||
| ASP-PCR | 17 | 7 | 10 | 10 | 4 | 0 |
| Ratio | 56.67% | 41.18% | 58.82% | 58.82% | 23.53% | 0 |
| (17/30) | (7/30) | (10/17) | (10/17) | (4/17) | ||
| Nested-ASP-PCR | 25 | 9 | 16 | 16 | 13 | 9 |
| Ratio | 83.33% | 36% | 64% | 64% | 52% | 36% |
| (25/30) | (9/30) | (16/25) | (16/25) | (13/25) | (9/25) | |
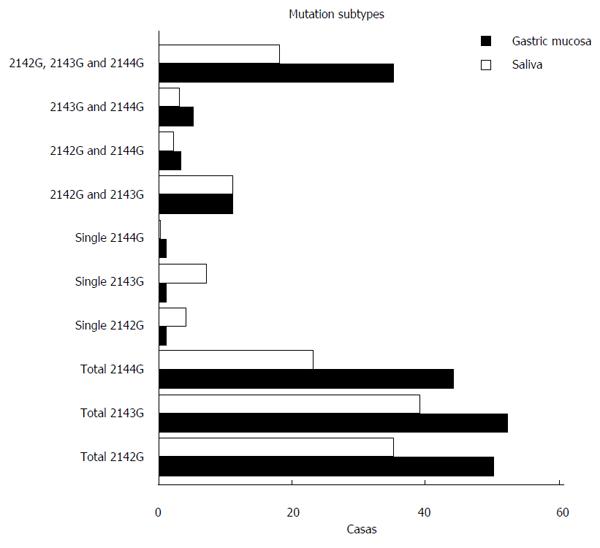
Furthermore, 99 RUT-positive patients were analyzed by nested ASP-PCR. Among these patients, 87 (87/99, 87.88%) cases were detected as H. pylori-positive by testing gastric mucosa by nested-ASP-PCR, and included 30 patients (30/87, 34.48%) with wild-type strains and 57 patients (57/87, 65.52%) with 23SrRNA point mutations associated with CLA resistance. Fifty patients (50/87, 57.47%) were infected with strains with A2142G mutations, 52 patients (52/87, 59.77%) with A2143G mutations and 44 patients (44/87, 50.57%) with A2144G mutations. Among the 99 RUT-positive saliva samples, 67 (67/99, 67.68%) were detected as H. pylori-positive by nested-ASP-PCR, and included 22 patients (22/67, 32.84%) with wild-type strains and 45 patients (45/67, 67.16%) with 23SrRNA point mutations. The numbers of patients with A2142G, A2143G and A2144G mutations were 35 (35/67, 52.24%), 39 (39/67, 58.21%) and 23 (23/67, 34.34%) respectively (Table 3). We observed all the mutation sub-types in these patients (Figure 3). The results showed that the mixed mutations rate with 2142, 2143 and 2144 positions was relatively high, while patients with a single mutation were rare, which suggested that the mixture strains existed commonly in gastric mucosa and saliva. We further analyzed the mutation genotypes in the gastric mucosa and saliva samples from each individual (Supplementary Table 1) and found that the rate of simultaneous mutation in gastric mucosa and saliva was 33. The mutation consistency rate was just 33.33% (11/33). In short, gastric mucosa and saliva samples exhibited different mutation genotypes.
| RUT-positive | H. pylori-positive detection rate | WT | Total mutation, A-to-G | 2142 G | 2143 G | 2144 G | |
| Gastric mucosa | 99 | 87 | 30 | 57 | 50 | 52 | 44 |
| ratio | 87.88% | 34.48% | 65.52% | 57.47% | 59.77% | 50.57% | |
| (87/99) | (30/87) | (57/87) | (50/87) | (52/87) | (44/87) | ||
| Saliva | 99 | 67 | 22 | 45 | 35 | 39 | 23 |
| ratio | 67.68% | 32.84% | 67.16% | 52.24% | 58.21% | 34.34% | |
| (67/99) | (22/67) | (45/67) | (35/67) | (39/67) | (22/67) |
In the 99 RUT-positive patients, the H. pylori detection rate in gastric mucosa by the nested-ASP-PCR was 87.88% (87/99), for which the resistance site mutation rate was 65.52% (57/87). This finding suggested that RUT can give some false-positive results.
Next, H. pylori isolates were cultured from the gastric biopsy samples of 87 patients who had been identified as having H. pylori infection by nested-ASP-PCR, and only 50 H. pylori-positive strains were obtained by primary culture. The bacterial culture positive rate was 57.47% (50/87). Furthermore, we implemented the drug sensitivity test (DST) by disk diffusion method. Among these 50 cases, 26 (26/50, 52%) showing resistant strains of H. pylori, with the remaining 24 (24/50, 48%) showing wild-type strains (Table 4).
| RUT-positive | Nested-ASP-PCR detection | Nested-ASP-PCR mutation | Bacterial culture H. pylori-positive | Drug-resistant strains | |
| Cases | 99 | 87 | 57 | 50 | 26 |
| Ratio | 87.88% (87/99) | 65.52% (57/87) | 57.47% (50/87) | 52% (26/50) |
In recent years, antibiotic resistance in H. pylori infection has become a global problem, especially for resistance to amoxicillin, CLA and metronidazole. Some elements, including age, sex, in-house infection, colonization density, gastric pH and pre-exposure to antibiotics, are thought to be responsible for resistance development, which subsequently leads to failure of H. pylori eradication. It has been reported that CLA resistance doubled within a 5-year period, probably because of the wide usage of this antibiotic as treatment for community-based pneumonia[27]. To combat the growing problem of antibiotic resistance, it is important to choose the most appropriate first-line treatment regimen. This choice should be made on the basis of knowledge of the status of antimicrobial resistance before treatment[28].
A variety of diagnostic methods have been developed for antibiotic resistance to H. pylori, including invasive tests (gastric biopsy for histology and culture) and noninvasive tests (urea breath and stool antigen tests). Normally, the detection of CLA resistance is based mainly on phenotypic methods that are performed after culturing, such as agar diffusion for the E-test or the agar dilution method. Bacterial culturing is a widely available method that allows for antimicrobial susceptibility testing, and it can require tissues obtained through endoscopy. However, bacterial culture has many inherent disadvantages, such as special transportation and culture conditions, special media and environments, restrictive timelines and lengthy incubation time. These factors directly influence the detection of H. pylori. Detection of point mutations conferring resistance to CLA by molecular methods may constitute a more reliable approach. Some PCR-based techniques have been developed to detect these mutations, such as PCR-RFLP, PCR-DNA-enzyme immunoassay, real-time PCR and reverse hybridization line probe assay[29]. In the present study, the nested-ASP-PCR was established to detect the different mutations at positions 2142, 2143 and 2144 in the 23SrRNA gene of H. pylori. This method can determine whether H. pylori strains are sensitive or resistant to CLA within several hours, while traditional culture testing for bacterial susceptibility to antibiotics is expensive and requires 7-10 d[30]. Thus, nested-ASP-PCR molecular testing is a relatively rapid, sensitive and reliable technique to detect CLA resistance[31], which might help make an individualized treatment strategy feasible in daily clinical practice and contribute to H. pylori eradication therapy[27]. In our study, among 30 gastric tissue samples, the H. pylori-positive rates detected by nested-ASP-PCR and ASP-PCR were 90% and 63% respectively. Simultaneously, the H. pylori-positive detection rates for saliva samples by the two methods were 83.33% and 56.67%. Not only in H. pylori detection but also in resistance site mutation detection, the nested-ASP-PCR method exhibited much higher sensitivity than ASP-PCR, especially in saliva samples with low levels of H. pylori.
PCR detection of H. pylori has been reported using a variety of clinical samples, including gastric biopsy, gastric juice, saliva, dental plaque and stools. It has been reported that the oral cavity may be a reservoir for H. pylori, responsible for oral-to-oral transmission of this bacterium. Other studies have also shown that H. pylori infection in the oral cavity is closely related to that in the stomach and that the mouth is the first extra-gastric reservoir[32,33]. More recently, DNA present in saliva has been employed in PCR-based assays designed to detect mutations associated with fragile X syndrome[34,35]. DNA fingerprinting studies have suggested that the same H. pylori strain can colonize both the oral cavity and the stomach[36].
We sought to determine if genomic DNA recovered from whole saliva constitutes a reliable alternative for DNA isolated from gastric mucosa and can reflect the resistance site polymorphisms[37] of H. pylori. In our study, the CLA resistance rate in gastric mucosa and saliva was 65.52% and 67.16% respectively. On the one hand, the resistance to CLA in gastric mucosa and saliva is quite common now. Some studies in the literature have reported 72.44% CLA resistance in Italy[7], 65%-75% in Taiwan[8], and even as high as 84.9% in the children in Beijing, China[13]. On the other hand, we found that gastric mucosa and saliva samples can exhibit different mutation genotypes. Our results are similar to Wang and Song’s studies[38], which showed that DNA sequences of PCR products from saliva differed from those obtained from the gastric biopsy of the same individual, suggesting that different strains are present in the mouth and the stomach. However, more investigations with a larger number of patients are required to confirm this.
In conclusion, in the present study we established the nested-ASP-PCR method to increase the detection sensitivity of resistance site mutations of the H. pylori 23SrRNA gene[39,40]. This study provides an inexpensive, reliable method for the evaluation of CLA resistance, which will likely contribute to improving the efficacy of H. pylori eradication therapy.
We thank the Hospital of Qixia in Nanjing for providing sufficient financial support for this study.
In recent years, antibiotic resistance in Helicobacter pylori infection has become a global problem, especially of resistance to amoxicillin, clarithromycin (CLA) and metronidazole. According to many reports, CLA resistance increased from 29% in 2004 to 77% in 2007 in France and from 15% to 65% over a 10-year period (2000-2009) in China. Generally, the assessment of CLA resistance is based mainly on phenotypic methods performed by sensitivity testing after bacterial culturing, such as agar diffusion for the E-test or the agar dilution method. Furthermore, CLA resistance of H. pylori has been directly linked to the single nucleotide polymorphisms at positions 2142, 2143 and 2144 of 23SrRNA gene. Thus, in the last decade, novel culture-free polymerase chain reaction (PCR)-based techniques have been established to detect these mutations, and these include the PCR-restriction fragment length polymorphism, PCR-DNA-enzyme immunoassay and so on.
A variety of diagnostic methods have been developed for antibiotic resistance to H. pylori, including invasive tests (gastric biopsy for histology and culture) and noninvasive tests (urea breath and stool antigen tests). Bacterial culture is a widely available method that allows for antimicrobial susceptibility testing, but it is expensive and requires 7-10 d for results. In the present study, the nested-ASP-PCR technique was established to detect the CLA resistance mutations at positions 2142, 2143 and 2144 in the 23SrRNA gene of H. pylori. This method can determine whether H. pylori strains are sensitive or resistant to CLA within several hours.
Traditional culture testing for bacterial susceptibility to antibiotics has many inherent disadvantages, such as special transportation and culture conditions, special media and environments, restrictive timelines and lengthy incubation time. These factors directly influence the detection of H. pylori. Detection of point mutations conferring resistance to CLA by molecular methods may constitute a more reliable approach. It has been reported that the oral cavity may be a reservoir for H. pylori, responsible for oral-to-oral transmission of this bacterium. Other studies have also showed that H. pylori infection in the oral cavity is closely related to that in the stomach and that the mouth is the first extra-gastric reservoir. In the present study, the feasibility of saliva as a noninvasive material for use in detecting CLA resistance of H. pylori was evaluated compared to gastric tissue. The nested-ASP-PCR molecular test is a relatively rapid, sensitive and reliable method to detect CLA resistance.
The data in this study suggested that the nested-ASP-PCR method can increase the detection sensitivity of resistance site mutations of the 23SrRNA gene in H. pylori. The outcomes from this study are expected to contribute to improving the efficacy of H. pylori eradication therapy and to guide clinical treatment. Furthermore, this study also provided readers with some important information regarding CLA resistance of H. pylori in the populations of China.
Nested-ASP-PCR is nested-PCR combined with allele-specific primer-PCR.
Available papers concerning CLA resistance of H. pylori in mainland China are common. However, articles describing the detection of CLA resistant strains using gastric and saliva samples simultaneously are rare. The authors in this study analyzed the CLA resistance rates for each site and the mutations consistency between the saliva and gastric samples. This study showed that gastric mucosa and saliva samples can exhibit different mutation genotypes. The results are interesting and provide important information concerning the background and trends of H. pylori treatments for people in China.
Manuscript Source: Unsolicited manuscript
Specialty Type: Gastroenterology and Hepatology
Country of Origin: China
Peer-Review Report Classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: El-Zahaby SA, Pierzchalski P, Zalazar F S- Editor: Yu J L- Editor: Filipodia E- Editor: Ma S
| 1. | Agudo S, Pérez-Pérez G, Alarcón T, López-Brea M. High prevalence of clarithromycin-resistant Helicobacter pylori strains and risk factors associated with resistance in Madrid, Spain. J Clin Microbiol. 2010;48:3703-3707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 98] [Article Influence: 6.5] [Reference Citation Analysis (1)] |
| 2. | Nakamura A, Furuta T, Shirai N, Sugimoto M, Kajimura M, Soya Y, Hishida A. Determination of mutations of the 23S rRNA gene of Helicobacter pylori by allele specific primer-polymerase chain reaction method. J Gastroenterol Hepatol. 2007;22:1057-1063. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | Schmitt BH, Regner M, Mangold KA, Thomson RB, Kaul KL. PCR detection of clarithromycin-susceptible and -resistant Helicobacter pylori from formalin-fixed, paraffin-embedded gastric biopsies. Mod Pathol. 2013;26:1222-1227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Furuta T, Shirai N, Takashima M, Xiao F, Hanai H, Sugimura H, Ohashi K, Ishizaki T, Kaneko E. Effect of genotypic differences in CYP2C19 on cure rates for Helicobacter pylori infection by triple therapy with a proton pump inhibitor, amoxicillin, and clarithromycin. Clin Pharmacol Ther. 2001;69:158-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 197] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 5. | Maeda S, Yoshida H, Matsunaga H, Ogura K, Kawamata O, Shiratori Y, Omata M. Detection of clarithromycin-resistant helicobacter pylori strains by a preferential homoduplex formation assay. J Clin Microbiol. 2000;38:210-214. [PubMed] |
| 6. | Mégraud F. H pylori antibiotic resistance: prevalence, importance, and advances in testing. Gut. 2004;53:1374-1384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 607] [Cited by in RCA: 681] [Article Influence: 32.4] [Reference Citation Analysis (2)] |
| 7. | Di Giulio M, Di Campli E, Di Bartolomeo S, Cataldi V, Marzio L, Grossi L, Ciccaglione AF, Nostro A, Cellini L. In vitro antimicrobial susceptibility of Helicobacter pylori to nine antibiotics currently used in Central Italy. Scand J Gastroenterol. 2016;51:263-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 8. | Wu IT, Chuah SK, Lee CH, Liang CM, Lu LS, Kuo YH, Yen YH, Hu ML, Chou YP, Yang SC. Five-year sequential changes in secondary antibiotic resistance of Helicobacter pylori in Taiwan. World J Gastroenterol. 2015;21:10669-10674. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 25] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 9. | Raymond J, Lamarque D, Kalach N, Chaussade S, Burucoa C. High level of antimicrobial resistance in French Helicobacter pylori isolates. Helicobacter. 2010;15:21-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 75] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 10. | Boltin D, Ben-Zvi H, Perets TT, Kamenetsky Z, Samra Z, Dickman R, Niv Y. Trends in secondary antibiotic resistance of Helicobacter pylori from 2007 to 2014: has the tide turned? J Clin Microbiol. 2015;53:522-527. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Miendje Deyi VY, Bontems P, Vanderpas J, De Koster E, Ntounda R, Van den Borre C, Cadranel S, Burette A. Multicenter survey of routine determinations of resistance of Helicobacter pylori to antimicrobials over the last 20 years (1990 to 2009) in Belgium. J Clin Microbiol. 2011;49:2200-2209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Gao W, Cheng H, Hu F, Li J, Wang L, Yang G, Xu L, Zheng X. The evolution of Helicobacter pylori antibiotics resistance over 10 years in Beijing, China. Helicobacter. 2010;15:460-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 13. | Zhang YX, Zhou LY, Song ZQ, Zhang JZ, He LH, Ding Y. Primary antibiotic resistance of Helicobacter pylori strains isolated from patients with dyspeptic symptoms in Beijing: a prospective serial study. World J Gastroenterol. 2015;21:2786-2792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 88] [Cited by in RCA: 104] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 14. | Ioannidou S, Tassios PT, Kotsovili-Tseleni A, Foustoukou M, Legakis NJ, Vatopoulos A. Antibiotic resistance rates and macrolide resistance phenotypes of viridans group streptococci from the oropharynx of healthy Greek children. Int J Antimicrob Agents. 2001;17:195-201. [PubMed] |
| 15. | Shen J, Zhang JZ, Ke Y, Deng D. Formation of A2143G mutation of 23S rRNA in progression of clarithromycin resistance in Helicobacter pylori 26695. Microb Drug Resist. 2005;11:100-106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 16. | Pan ZJ, Su WW, Tytgat GN, Dankert J, van der Ende A. Assessment of clarithromycin-resistant Helicobacter pylori among patients in Shanghai and Guangzhou, China, by primer-mismatch PCR. J Clin Microbiol. 2002;40:259-261. [PubMed] |
| 17. | Slinger R, Yan L, Chan F, Forward K, Cooper-Lesins G, Best L, Haldane D, Veldhuyzen van Zanten S. Pyrosequencing assay to rapidly detect clarithromycin resistance mutations in Canadian Helicobacter pylori isolates. Can J Gastroenterol. 2009;23:609-612. [PubMed] |
| 18. | Liu G, Xu X, He L, Ding Z, Gu Y, Zhang J, Zhou L. Primary antibiotic resistance of Helicobacter pylori isolated from Beijing children. Helicobacter. 2011;16:356-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 19. | Versalovic J, Osato MS, Spakovsky K, Dore MP, Reddy R, Stone GG, Shortridge D, Flamm RK, Tanaka SK, Graham DY. Point mutations in the 23S rRNA gene of Helicobacter pylori associated with different levels of clarithromycin resistance. J Antimicrob Chemother. 1997;40:283-286. [PubMed] |
| 20. | de Francesco V, Margiotta M, Zullo A, Hassan C, Valle ND, Burattini O, Cea U, Stoppino G, Amoruso A, Stella F. Primary clarithromycin resistance in Italy assessed on Helicobacter pylori DNA sequences by TaqMan real-time polymerase chain reaction. Aliment Pharmacol Ther. 2006;23:429-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 21. | Kim JM, Kim JS, Kim N, Kim YJ, Kim IY, Chee YJ, Lee CH, Jung HC. Gene mutations of 23S rRNA associated with clarithromycin resistance in Helicobacter pylori strains isolated from Korean patients. J Microbiol Biotechnol. 2008;18:1584-1589. [PubMed] |
| 22. | Elviss NC, Lawson AJ, Owen RJ. Application of 3’-mismatched reverse primer PCR compared with real-time PCR and PCR-RFLP for the rapid detection of 23S rDNA mutations associated with clarithromycin resistance in Helicobacter pylori. Int J Antimicrob Agents. 2004;23:349-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Noguchi N, Rimbara E, Kato A, Tanaka A, Tokunaga K, Kawai T, Takahashi S, Sasatsu M. Detection of mixed clarithromycin-resistant and -susceptible Helicobacter pylori using nested PCR and direct sequencing of DNA extracted from faeces. J Med Microbiol. 2007;56:1174-1180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 24. | Maeda S, Yoshida H, Ogura K, Kanai F, Shiratori Y, Omata M. Helicobacter pylori specific nested PCR assay for the detection of 23S rRNA mutation associated with clarithromycin resistance. Gut. 1998;43:317-321. [PubMed] |
| 25. | Patel SK, Mishra GN, Pratap CB, Jain AK, Nath G. Helicobacter pylori is not eradicated after triple therapy: a nested PCR based study. Biomed Res Int. 2014;2014:483136. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.7] [Reference Citation Analysis (1)] |
| 26. | Mishra S, Singh V, Rao GR, Dixit VK, Gulati AK, Nath G. Prevalence of Helicobacter pylori in asymptomatic subjects--a nested PCR based study. Infect Genet Evol. 2008;8:815-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 27. | Karabiber H, Selimoglu MA, Otlu B, Yildirim O, Ozer A. Virulence factors and antibiotic resistance in children with Helicobacter pylori gastritis. J Pediatr Gastroenterol Nutr. 2014;58:608-612. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 34] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 28. | Oleastro M, Ménard A, Santos A, Lamouliatte H, Monteiro L, Barthélémy P, Mégraud F. Real-time PCR assay for rapid and accurate detection of point mutations conferring resistance to clarithromycin in Helicobacter pylori. J Clin Microbiol. 2003;41:397-402. [PubMed] |
| 29. | Pina M, Occhialini A, Monteiro L, Doermann HP, Mégraud F. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J Clin Microbiol. 1998;36:3285-3290. [PubMed] |
| 30. | Calvet X, Ducons J, Guardiola J, Tito L, Andreu V, Bory F, Guirao R. One-week triple vs. quadruple therapy for Helicobacter pylori infection - a randomized trial. Aliment Pharmacol Ther. 2002;16:1261-1267. [PubMed] |
| 31. | Mahachai V, Sirimontaporn N, Tumwasorn S, Thong-Ngam D, Vilaichone RK. Sequential therapy in clarithromycin-sensitive and -resistant Helicobacter pylori based on polymerase chain reaction molecular test. J Gastroenterol Hepatol. 2011;26:825-828. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 32. | Rimbara E, Sasatsu M, Graham DY. PCR detection of Helicobacter pylori in clinical samples. Methods Mol Biol. 2013;943:279-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 33. | Sugimoto M, Wu JY, Abudayyeh S, Hoffman J, Brahem H, Al-Khatib K, Yamaoka Y, Graham DY. Unreliability of results of PCR detection of Helicobacter pylori in clinical or environmental samples. J Clin Microbiol. 2009;47:738-742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 56] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 34. | van Schie RC, Wilson ME. Saliva: a convenient source of DNA for analysis of bi-allelic polymorphisms of Fc gamma receptor IIA (CD32) and Fc gamma receptor IIIB (CD16). J Immunol Methods. 1997;208:91-101. [PubMed] |
| 35. | Hagerman RJ, Wilson P, Staley LW, Lang KA, Fan T, Uhlhorn C, Jewell-Smart S, Hull C, Drisko J, Flom K. Evaluation of school children at high risk for fragile X syndrome utilizing buccal cell FMR-1 testing. Am J Med Genet. 1994;51:474-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 36. | Parsonnet J, Shmuely H, Haggerty T. Fecal and oral shedding of Helicobacter pylori from healthy infected adults. JAMA. 1999;282:2240-2245. [PubMed] |
| 37. | Adler I, Muiño A, Aguas S, Harada L, Diaz M, Lence A, Labbrozzi M, Muiño JM, Elsner B, Avagnina A. Helicobacter pylori and oral pathology: relationship with the gastric infection. World J Gastroenterol. 2014;20:9922-9935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 61] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (1)] |
| 38. | Song Q, Spahr A, Schmid RM, Adler G, Bode G. Helicobacter pylori in the oral cavity: high prevalence and great DNA diversity. Dig Dis Sci. 2000;45:2162-2167. [PubMed] |
| 39. | Zhen-Hua Z, De-Qiang H, Yong X, Lin-Lin L, Nong-Hua L. Characterization of 23S rRNA gene mutation in primary and secondary clarithromycin-resistant Helicobacter pylori strains from East China. Turk J Gastroenterol. 2013;24:5-9. [PubMed] |
| 40. | Juvonen R, Koivula T, Haikara A. Group-specific PCR-RFLP and real-time PCR methods for detection and tentative discrimination of strictly anaerobic beer-spoilage bacteria of the class Clostridia. Int J Food Microbiol. 2008;125:162-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 42] [Article Influence: 2.5] [Reference Citation Analysis (1)] |