Published online Mar 14, 2015. doi: 10.3748/wjg.v21.i10.2949
Peer-review started: July 17, 2014
First decision: August 6, 2014
Revised: August 29, 2014
Accepted: October 21, 2014
Article in press: October 21, 2014
Published online: March 14, 2015
Processing time: 242 Days and 2.2 Hours
AIM: To investigate the function and mechanism of miR-133a in gastric cancer (GC) and its relationship with clinicopathological characteristics of GC.
METHODS: A total of 105 GC patients who underwent surgical resection as primary treatment were selected for this study. Real-time quantitative reverse transcriptase polymerase chain (qRT-PCR) was used to examine the expression levels of miR-133a in human GC and adjacent non-tumor tissues, as well as in GC cell lines (SGC-7901, BGC-823, MGC-803, and AGS) and a human gastric mucosal epithelial cell line (GES-1). The biological role of miRNA (miR)-133a was assessed in the GC cell lines using MTT, apoptosis, migration and invasion, and colony formation assays, and xenograft tumorigenesis. qRT-PCR and western blot analyses were used to evaluate the potential target gene expression of miR-133a. Pearson’s correlation was calculated to evaluate the correlation between miR-133a and insulin-like growth factor 1 receptor (IGF1R) expression. The regulation of IGF1R by miR-133a was verified using the luciferase reporter assay.
RESULTS: In 80% of the 105 GC patients, the mean expression of miR-133a was significantly downregulated in tumor tissues compared with adjacent normal tissues (1.215 ± 0.1477 vs 3.093 ± 0.4104, P < 0.0001). Downregulation of miR-133a was significantly correlated with the degree of differentiation (P = 0.01), local invasion (P = 0.001) and TNM stage (P = 0.02) in GC patients. Compared with a control construct, forced expression of miR-133a in GC cell lines inhibited proliferation (0.4787 ± 0.0219 vs 0.7050 ± 0.0147, P = 0.0013 in SGC-7901 cells; and 0.5448 ± 0.0085 vs 0.7270 ± 0.0084, P = 0.001 in MGC-803 cells); migration (0.6333 ± 0.0233 vs 1.037 ± 0.0584, P = 0.003 in SGC-7901 cells; 0.6126 ± 0.0311 vs 1.024 ± 0.0456, P = 0.0017 in MGC-803 cells); and invasion (0.613 ± 0.0399 vs 1.033 ± 0.0278, P = 0.0013 in SGC-7901 cells; 0.7433 ± 0.0221 vs 1.017 ± 0.0311, P = 0.002 in MGC-803 cells). It also induced apoptosis (18.19% ± 0.2483% vs 5.887% ± 0.3837%, P < 0.0001 in SGC-7901 cells; 22.69% ± 0.7846% vs 9.347% ± 0.3012%, P < 0.0001 in MGC-803 cells). Furthermore, miR-133a inhibited tumor growth and xenograft tumorigenesis of SGC -7901 cells in vivo. In addition, we identified IGF1R as a regulatory target of miR-133a in GC.
CONCLUSION: This study suggests that miR-133a is downregulated in GC and functions as a tumor suppressor in vitro and in vivo partly by repressing IGF1R.
Core tip: miRNA (miR)-133a is a tumor suppressor in several cancers. It was recently shown that miR-133a inhibits proliferation, migration and invasion in gastric cancer (GC) by targeting transcription factor Sp1. Here, we further investigated the biological role of miR-133a in apoptosis and xenograft tumorigenesis, and its relationship with clinicopathological parameters. miR-133a downregulation was associated with degree of differentiation, local invasion and TNM stage, whereas forced miR-133a expression inhibited cell proliferation, migration and invasion, and xenograft tumorigenesis, and induced apoptosis. Furthermore, insulin-like growth factor 1 receptor was found to be another target gene of miR-133a in GC.
- Citation: Gong Y, Ren J, Liu K, Tang LM. Tumor suppressor role of miR-133a in gastric cancer by repressing IGF1R. World J Gastroenterol 2015; 21(10): 2949-2958
- URL: https://www.wjgnet.com/1007-9327/full/v21/i10/2949.htm
- DOI: https://dx.doi.org/10.3748/wjg.v21.i10.2949
Gastric cancer (GC) is the fourth most common cancer and the second leading cause of cancer-related death worldwide[1]. Surgical resection continues to be the main option for treating GC. Despite advances in surgery and other treatment modalities, the prognosis of GC patients remains poor because of the advanced stage of the disease at the time of diagnosis. The detection of early GC is < 15%[2]. Therefore, it is imperative to elucidate the regulatory network underlying gastric carcinogenesis to develop novel biomarkers for diagnosis and targeted therapy.
miRNAs are small noncoding RNAs that are 18-22 nucleotides in length. They negatively regulate gene expression through base pairing with the 3’ untranslated regions (UTRs) of target mRNAs, causing mRNA degradation and/or translational repression[3,4]. Abundant evidence suggests that miRNAs may function as oncogenes or tumor suppressors by regulating physiological processes[5,6]. In GC, ectopic expression of miRNAs is thought to play a crucial role in cancer initiation and progression[7,8]. Here, we focus on miR-133a, which has been validated as a tumor suppressor in ovarian cancer[9], colorectal cancer[10], bladder cancer[11,12], breast cancer[13] and prostate cancer[14]. Although a recent study has demonstrated that miR-133a suppresses the proliferation, migration, invasion and cell cycle progression of GC cells by decreasing the expression of transcription factor Sp1[15], the roles of miR-133a in apoptosis and xenograft tumorigenesis remain unclear, as does the relationship between miR-133a expression and clinicopathological parameters.
In our study, we not only confirmed the findings of Qiu et al[15], but also observed an inhibitory role of miR-133a in xenograft tumorigenesis in vivo and a role in the promotion of apoptosis. Additionally, we demonstrated for the first time that miR-133a expression was inversely associated with degree of differentiation, local invasion and TNM stage of GC. Finally, we found another regulatory target gene of miR-133a: insulin-like growth factor 1 receptor (IGF1R), which plays a crucial role in cancer progression[16]. These data provide new insight into the tumor suppressor role of miR-133a in GC.
This was a retrospective study in which surgically resected human GC and adjacent non-tumor tissues were obtained from 105 patients who were admitted to the Department of Gastrointestinal Surgery, Nanjing Medical University Affiliated Changzhou No. 2 People’s Hospital, China from January 2012 to December 2013. The project was approved by the Ethics Committee of the hospital and written informed consent was obtained from each patient who enrolled in the study. All clinical specimens were histopathologically confirmed and the corresponding normal tissue samples were collected from > 5 cm away from the tumors. All clinical specimens were stored at -80 °C for further RNA isolation.
The SGC-7901, BGC-823, MGC-803 and AGS human GC cell lines; GES-1 human gastric mucosal epithelial cell line; and HEK293T cell line were purchased from the Cell Resource Center, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. All of the cell lines were maintained under the recommended culture conditions and incubated at 37 °C in a humidified environment with 5% CO2.
Total RNA was isolated from gastric tissues or cell lines using TRIzol reagent (Invitrogen, Carlsbad, CA, United States) following the manufacturer’s instructions. cDNA was synthesized using the PrimeScript RT Reagent Kit (TaKaRa). Real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) was performed using a SYBR Premix Ex Taq kit (TaKaRa) in a 7500 Real-Time PCR System (ABI). The PCR primers were obtained from Invitrogen, and the reaction conditions included the following steps: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. U6 small nuclear (sn)RNA and GAPDH mRNA were used as endogenous controls. The results were presented as fold changes relative to U6 or GAPDH and were calculated using the 2-△△CT method.
Pri-miR-133a was amplified from normal human genomic DNA using PCR with the following primers: 133a-Forward: 5’-ATAAGAATGCGGCCGCATTCCAAACTAGCAGCACTA-3’ and 133a-Reverse: 5’-AGCTTTGTTTAAACTTAACCATTCTAGCTTTTCC-3’. The PCR fragment was inserted into the MSCV-P2GM vector to generate P2GM-miR-133a. The empty P2GM vector was used as a negative control. The wild-type and mutant 3’-UTR sequences of IGF1R were inserted into the region where a cytomegalovirus promoter drove firefly luciferase cassette downstream directly in a pGL3-promoter vector. The following primers were used to clone the wild-type IGF1R 3’UTR, Forward: 5’-GCTCTAGAGCAGTGTAGTGCCCATCATAGC-3’ and Reverse: 5’-GCTCTAGAGCGACATCCCACGAAGGAGACC-3’. The mutant 3’UTR of IGF1R was constructed by overlapping PCR following two pairs of primers: Forward 1: 5’-GCTCTAGAGCAGTGTAGTGCCCATCATAGC-3’, Reverse 1: 5’-AAGGTGGGTGGGTGAACCCAGTAAA-3’; Forward 2: 5’-TGGGTTCACCCACCCACCTTGACTA-3’, Reverse 2: 5’-GCTCTAGAGCGACATCCCACGAAGGAGACC-3’. All constructs were verified by sequencing.
SGC-7901 and MGC-803 cells was transfected with plasmids using Lipofectamine 2000 (Invitrogen), and steady clones carrying P2GM-miR-133a or the empty vector were selected based on puromycin resistance (concentration: 10 g/mL). The miR-133a and negative control mimics were obtained from Ribobio (Guangzhou, China) and SGC-7901 and MGC-803 cells were transfected with these mimics using Lipofectamine 2000 (Invitrogen). SGC-7901 or MGC-803 cells plated the prior day were transfected with 100 nmol/L of miR-133a or negative control mimics when they reached 50% confluence. After 48 h, the cells were processed for further analysis.
Cell proliferation was measured using MTT assay. SGC-7901 or MGC-803 cells were harvested after 48 h transfection and reseeded onto 96-well plates at a density of 2500 cells per well. MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide] was added to the culture medium at specified intervals for 24 h, and the absorbance at 490 nm was measured using a spectrophotometer. Each assay was performed in triplicate and independently repeated three times.
Stable SGC-7901 or MGC-803 cells carrying P2GM-miR-133a or the empty vector control were seeded onto six-well plates at a density of 1000 cells per well. After 14 d culture, the cells were fixed with 3.7% methanol and stained with 0.1% crystal violet. Colonies containing at least 50 cells were recorded. Each assay was performed in triplicate and independently repeated three times.
SGC-7901 or MGC-803 cells transfected with miR-133a or negative control mimics were trypsinized and resuspended in 1× binding buffer at a density of 1 × 106 cells/mL. The cell suspension (100 μL) was incubated with 5 μL fluorescein isothiocyanate (FITC)-Annexin V and 5 μL propidium iodide (PI) for 15 min in the dark at room temperature. The reaction was terminated by adding 400 μL 1× binding buffer and examined using flow cytometry. The early apoptotic cells were identified as FITC-Annexin-V-positive and PI-negative cells, and the experiments were independently repeated three times.
Following 16 h transfection with miRNA mimics, 5 × 104 cells were added to the upper chamber of the insert (BD Science, Sparks, MD, United States), and 1 × 105 cells were used to carry out Matrigel invasion assays. The cells were trypsinized and resuspended in serum-free medium before being seeded into the upper chamber. The culture medium containing 10% fetal bovine serum was added to the lower chamber. The cells were incubated for an additional 48 h for the migration assay and 72 h for the invasion assay. At the end of the experiments, the cells on the lower surface were fixed and stained with 0.1% crystal violet. Three visual fields from each insert were randomly selected and the stained cells were counted under a light microscope. Each condition was assayed in triplicate and each experiment was independently performed three times.
SGC-7901 cells (1.5 × 106) that stably carried either P2GM-miR-133a or the empty vector control were injected subcutaneously into the flanks of female BALB/c nude mice (aged 6 wk; 5 mice per group). The tumor size was measured using a caliper every 3 d from the sixth day after injection. The tumor volume was calculated using the following formula: volume = (length × width2)/2. The mice were sacrificed on day 24 and the tumors were separated for further analysis. Nude mice were manipulated and cared for at the Experiment Animal Center of the Nanjing Medical University (Nanjing, Jiangsu, China) according to the NIH Animal Care and Use Committee guidelines.
Total protein was extracted from SGC-7901 cells by lysis buffer, separated by 10% SDS-PAGE and transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA, United States). The membranes were blocked with 5% nonfat milk and then incubated with primary anti-IGF1R or anti-β-actin (Abcam, Cambridge, MA, United States) overnight at 4 °C, followed by incubation with horseradish-peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, United States) for 1 h at 37 °C. Protein bands were visualized using the ECL-kit according to the manufacturer’s instructions. β-Actin was used as a loading control.
HEK293T cells were seeded onto 24-well plates and allowed to settle for 24 h. The cells were cotransfected with 250 ng wild-type or mutant luciferase reporter constructs and 100 nM miR-133a mimics or negative control mimics plus 20 ng pRL-TK plasmid using Lipofectamine 2000 (Invitrogen). After 48 h, the firefly and Renilla luciferase activities were measured using Dual-Luciferase Reporter System (Promega, Madison, WI, United States). The Renilla luciferase activity was used as an internal control to confirm transfection efficiency. Each condition was performed in triplicate and independently repeated three times.
The data were expressed as the mean ± SEM. Paired-sample t tests were used to analyze the difference in miR-133a expression between the tumors and normal tissues. For in vitro and in vivo experiments, an independent-sample t test was used to evaluate differences between the treatment and control groups. Pearson’s correlation was used to analyze the correlation between miR-133a and IGF1R expression. All statistical analyses were conducted using SPSS version 16. (SPSS Inc, Chicago, IL, United States). P < 0.05 was considered to be statistically significant.
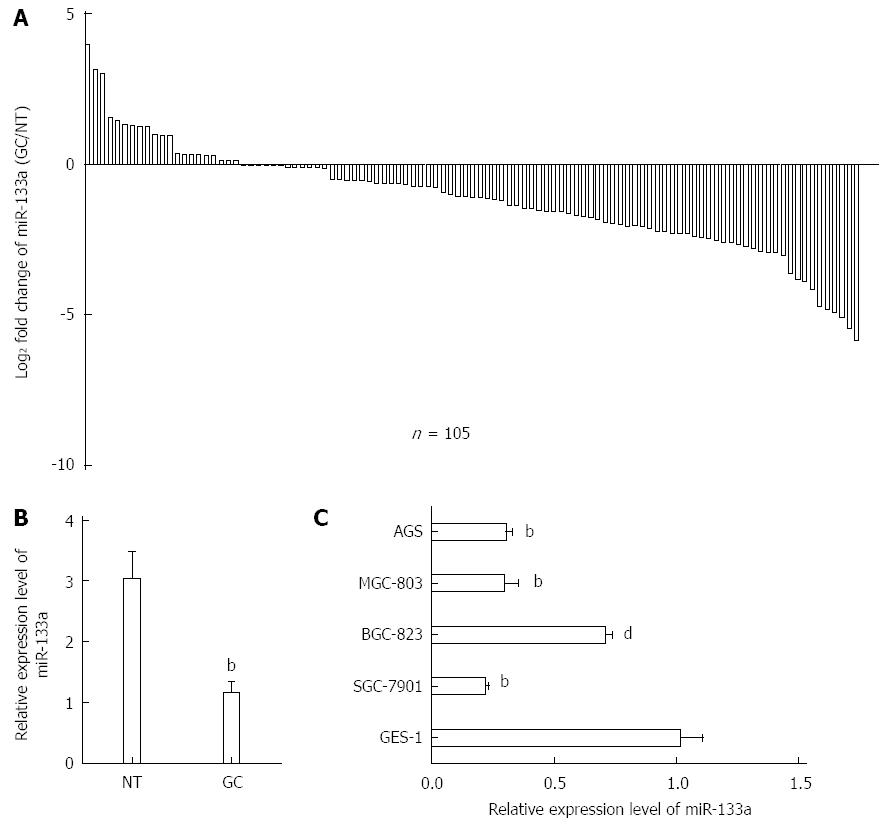
To determine the expression of miR-133a in GC, qRT-PCR was performed on 105 pairs of tissue samples and five human GC cell lines (SGC-7901, BGC-823, MGC-803, AGS and GES-1). As shown in Figure 1A, miR-133a generally exhibited lower expression in GC tissues than in normal tissues. The average expression levels of miR-133a were lower in GC tissues than in normal tissues (Figure 1B). This phenomenon was also observed in the SGC-7901, BGC-823, MGC-803 and AGS GC cell lines compared with GES-1 cells (Figure 1C). An analysis of the clinicopathological features of GC revealed that a reduction in miR-133a expression was significantly associated with the degree of differentiation, local invasion and TNM stage (Table 1). Overall, these results suggest that miR-133a may play a crucial role in the development of GC.
| Categories | Patients, n | Mean expression of miR-133a (mean ± SEM) | P value |
| Sex | |||
| Male | 63 | 1.30 ± 0.21 | 0.47 |
| Female | 42 | 1.08 ± 0.19 | |
| Age (yr) | |||
| ≥ 60 | 57 | 1.30 ± 0.21 | 0.52 |
| < 60 | 48 | 1.11 ± 0.20 | |
| Diameter (cm) | |||
| ≥ 5 | 39 | 1.09 ± 0.30 | 0.52 |
| < 5 | 66 | 1.29 ± 0.16 | |
| Location | |||
| Middle and proximal third | 69 | 1.22 ± 0.20 | 0.95 |
| Distal third | 36 | 1.20 ± 0.21 | |
| Degree of differentiation | |||
| Well and moderately | 36 | 1.71 ± 0.29 | 0.01 |
| Poorly | 69 | 0.96 ± 0.14 | |
| Local invasion | |||
| T1+T2 | 30 | 1.89 ± 0.32 | 0.001 |
| T3+T4 | 75 | 0.95 ± 0.13 | |
| Lymph node metastasis | |||
| No | 42 | 1.53 ± 0.28 | 0.07 |
| Yes | 63 | 1.0 ± 0.15 | |
| TNM stage | |||
| I + II | 36 | 1.68 ± 0.30 | 0.02 |
| III + IV | 69 | 0.97 ± 0.14 |
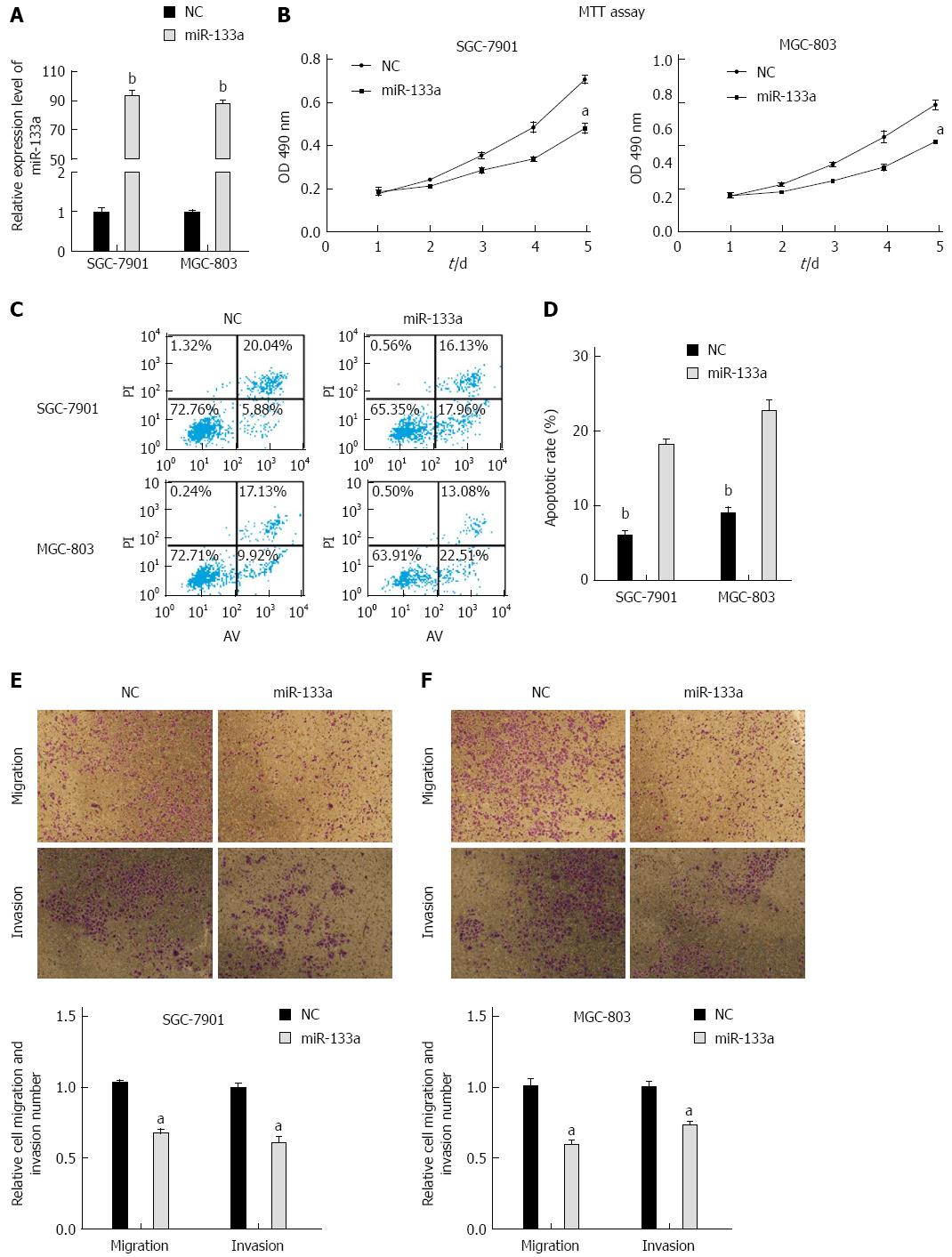
To investigate whether miR-133a inhibited GC proliferation, we performed MTT assays on the SGC-7901 and MGC-803 cell lines. Overexpression of miR-133a was confirmed using stem-loop PCR in SGC-7901 and MGC-803 cells after transfection (Figure 2A). We found that the proliferation rate of cells transfected with miR-133a mimics was reduced over a 5-day period compared with that of cells transfected with negative control mimics (Figure 2B). Apoptosis analysis revealed that artificially increasing the level of miR-133a via mimics also caused a significant increase in apoptosis after 48 h in the SGC-7901 and MGC-803 cell lines (Figure 2C and D). Furthermore, we performed migration and invasion assays in SGC-7901 and MGC-803 cells and determined that upregulation of miR-133a significantly decreased migration and invasion compared with those of a control construct (Figure 2E and F). Taken together, these results indicate that the restoration of miR-133a significantly inhibits cell proliferation, migration and invasion, and induces apoptosis in SGC-7901 and MGC-803 cells.
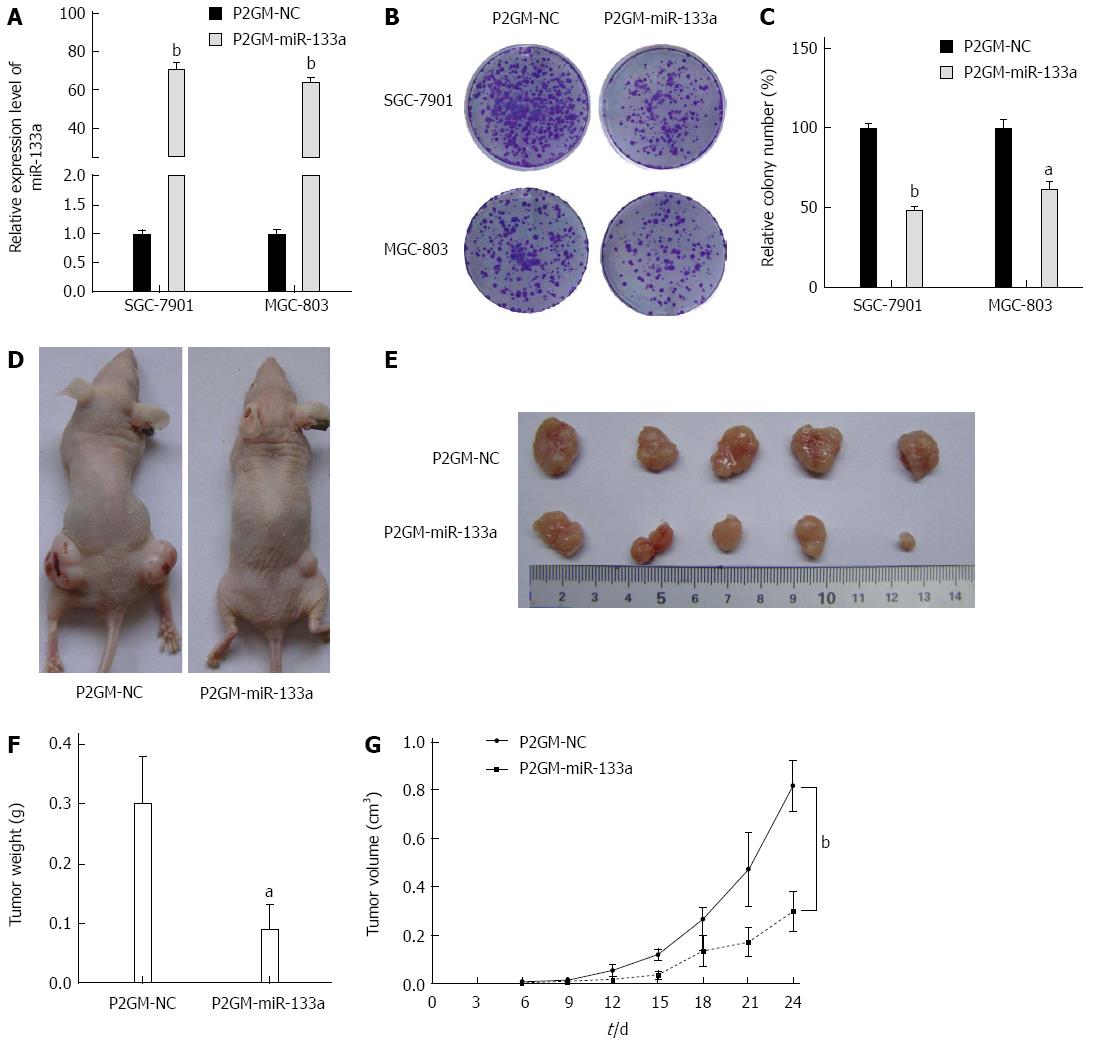
To examine the effects of miR-133a on cell growth and colony formation, we generated stable cell lines from SGC-7901 and MGC-803 cells that carried either a P2GM-miR-133a vector or an empty vector control. We first examined the expression levels of miR-133a in these stable cell lines and observed that miR-133a was dramatically elevated in the stable P2GM-miR-133a cell lines (Figure 3A). Then, 1 × 103 stable cells carrying P2GM-miR-133a or P2GM-NC were seeded directly onto Petri dishes. After 14 d culture, we visualized the cell colonies using crystal violet staining. The stable cells carrying an empty vector (P2GM-NC) formed significantly more colonies than did the stable cells carrying P2GM-miR-133a (P2GM-miR-133a) (Figure 3B and C). Furthermore, we injected stable SGC-7901 cell lines subcutaneously and bilaterally into the lower backs of nude mice and observed tumor growth every day. These nude mice were sacrificed after 24 d, and the tumor was excised and then weighed. The miR-133a-expressing SGC-7901 tumors were significantly smaller than were the control SGC-7901 tumors (Figure 3D and E). The average weight and volume of the miR-133a-expressing tumors were also markedly reduced (Figure 3F and G). These results indicate that miR-133a can repress the growth of GC cells.
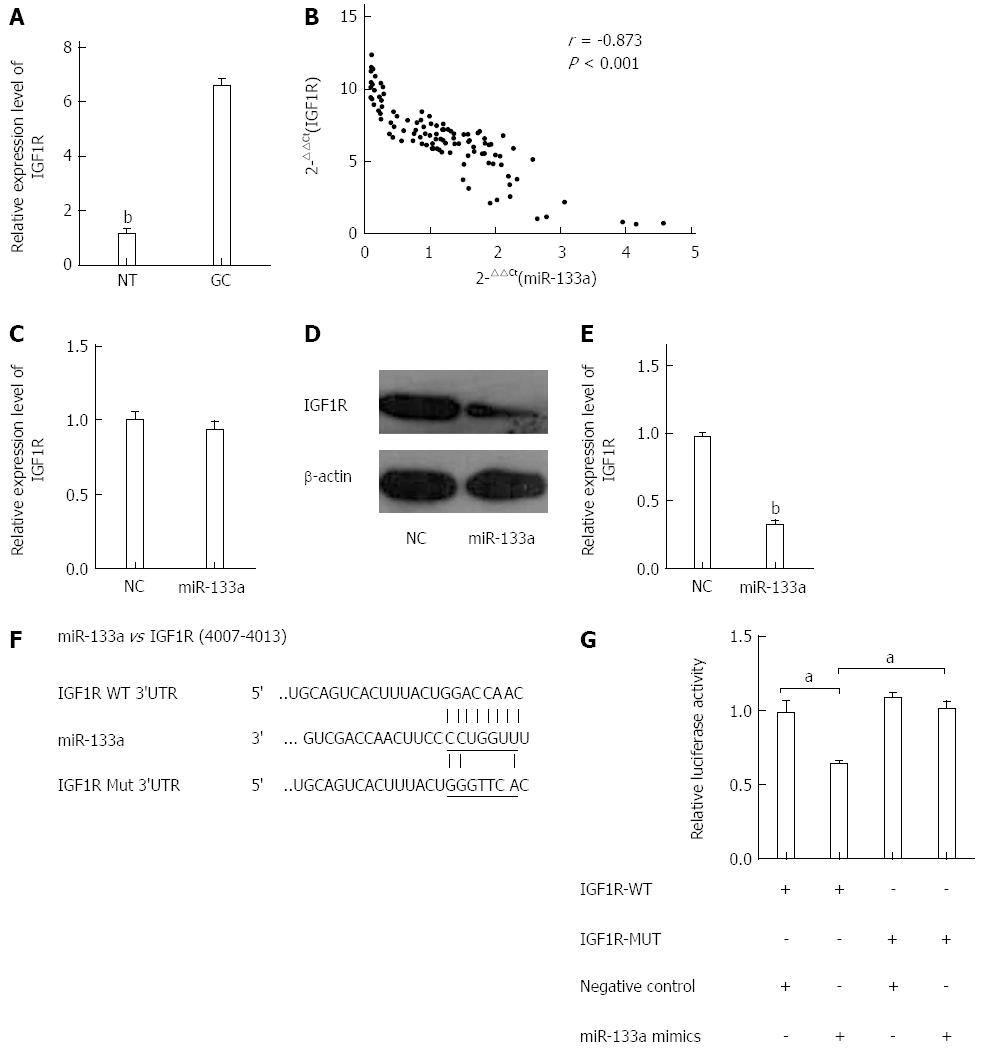
To elucidate the mechanism of miR-133a function in GC, we performed miRNA target gene prediction using the TargetScan and Miranda databases. Among several candidate target genes, IGF1R attracted our attention. We first analyzed the expression levels of IGF1R in the 105 pairs of tissues using qRT-PCR. The expression level of IGF1R was significantly higher in the GC tissues than in the normal gastric tissues (Figure 4A). We then assessed the effect of miR-133a on IGF1R using data obtained from the qRT-PCR. A significant inverse correlation was observed between IGF1R mRNA and miR-133a (Figure 4B). These data suggested that miR-133a downregulation plays a causal role in IGF1R overexpression in GC. To determine further whether miR-133a could regulate the expression of IGF1R, we analyzed the mRNA and protein levels of IGF1R in SGC-7901 cells that had been transfected with miR-133a or negative control mimics. The mRNA level of IGF1R was not affected by transfection with miR-133a, but the protein level exhibited a significant reduction (Figure 4C-E). To confirm that the IGF1R gene is a target of miR-133a, we cloned the predicted IGF1R 3’-UTR binding site and a mutant form downstream of a firefly luciferase reporter gene (Figure 4F). HEK-293T cells were cotransfected with the wild-type or mutant vector and miR-133a or negative control mimics. Analysis of a dual-luciferase reporter gene assay revealed that overexpression of miR-133a dramatically decreased the relative luciferase activity of constructs containing the wild-type IGF1R 3’ UTR, but the luciferase activity of the reporter containing the mutant binding site did not drop as sharply as that of the wild-type construct (Figure 4G). These findings indicate that miR-133a directly regulates IGF1R expression at the post-transcriptional level.
Abundant evidence has indicated that miRNAs play a crucial role in carcinogenesis and cancer progression[5,6,17,18]. It is therefore important to identify cancer-related miRNAs and their target genes to understand their roles in tumorigenesis. Several miRNAs have been reported to be dysregulated in GC[19-21]. If we identify specific miRNAs involved in GC, we may improve our understanding of GC and develop new targets for diagnosis and therapy. Here, we focused on the role of miR-133a in GC.
It has been suggested that miR-133a functions as a tumor suppressor in various cancers[9-14]. Recently, Qiu et al[15] found that miR-133a was downregulated in GC tissues and cell lines, and identified an inhibitory role of miR-133a in proliferation, migration, invasion and cell cycle progression in vitro, and our data resemble theirs. Our results also indicate that miR-133a inhibits xenograft tumorigenesis in vivo and induces cell apoptosis, which are novel findings. Meanwhile, we observed that a reduction in miR-133a is significantly associated with the degree of differentiation, local invasion and TNM stage. These results indicate that miR-133a downregulation may be a potential diagnostic biomarker and that it may predict cancer progression in GC patients. These findings also strongly demonstrate that decreased miR-133a expression in GC might be a factor that contributes to the development of GC rather than a consequence of GC. However, the mechanism by which miR-133a is downregulated remains unknown. It has been suggested that ectopic DNA methylation within the promoter region of miRNA genes may play an important role in their downregulation. Chen et al[22] reported that DNA-methylation-mediated silencing of the miR-133a gene caused a downregulation of miR-133a during colorectal cancer. This mechanism may explain the downregulation of miR-133a during GC. However, further studies are needed to elucidate the underlying mechanisms of miR-133a downregulation.
Identifying miRNA targets that are essential for cancer development may clarify their mechanisms of action and the pathways that miRNAs modulate[23]. To determine how miR-133a functions as a tumor suppressor, we searched for physiological targets using bioinformatics analyses. Several oncogenes have been reported to be the targets of miR-133a in various cancers, including epidermal growth factor receptor (EGFR)[13], purine nucleoside phosphorylase[14] and fascin homolog 1[12]. These findings suggest that miR-133a targets multiple genes in various cancer types. In this study, we verified that the IGF1R gene is a direct target of miR-133a using luciferase reporter assays. miR-133a directly suppresses the expression of IGF1R through translational repression rather than mRNA degradation. IGF1R, a member of the insulin receptor family of receptor tyrosine kinases, is overexpressed in many cancers and acts as an oncogene[16]. Recently, several lines of evidence have supported the role of IGF1R in promoting carcinogenesis. The upregulation of IGF1R/insulin receptor substrate (IRS)1 pathway proteins, such as IGF1R and mammalian target of rapamycin (mTOR), is closely associated with a poor response to chemotherapy and poor prognosis during GC, and abnormalities in the IGF1R signaling pathway are closely associated with drug resistance and a poor chemotherapy response during GC[24,25]. Additionally, several studies have noted the role of miRNAs in the regulation of IGF1R. Yen et al[26] reported that miR-99a functions as a tumor metastasis suppressor by targeting IGF1R during oral squamous cell carcinoma. Qian et al[27] revealed that miR-143 suppressed growth and angiogenesis and sensitized colorectal cancers to oxaliplatin by inhibiting IGF1R. Together, these studies suggest that the miRNAs-IGF1R/IRS1 regulatory network plays an important role in carcinogenesis and cancer progression. Moreover, Qiu et al[15] reported that miR-133a can inhibit GC cell proliferation, migration and invasion in vitro by directly targeting Sp1. It seems that miR-133a has multiple targets and functions during GC as a novel tumor suppressor. Therefore, further studies are essential to clarify the role of miR-133a in GC.
In conclusion, our data provide new evidence to support the tumor suppressor function of miR-133a in GC. Meanwhile, we have determined that IGF1R is a novel target of miR-133a in GC. In the future, miR-133a may be used as a novel diagnostic biomarker and therapeutic target in the treatment of GC.
Gastric cancer (GC) is the fourth most common cancer and the second leading cause of cancer-related death worldwide. Despite advances in surgery and other treatment modalities, the prognosis of GC patients remains poor because diagnosis frequently occurs at an advanced stage of the disease. The detection percentage for early GC is < 15%. Further investigation of the underlying molecular mechanisms of GC development is important.
miRNAs have been reported to be involved in carcinogenesis and cancer progression. In GC, studies have shown that miRNAs play crucial roles in regulating tumor proliferation, migration and invasion. A recent study has demonstrated that miR-133a suppresses the proliferation, migration, invasion and cell cycle progression of GC cells by decreasing the expression of transcription factor Sp1. However, the role of miR-133a in apoptosis and xenograft tumorigenesis remains unclear. In addition, the relationship between miR-133a expression levels and clinicopathological parameters is still unknown.
The authors investigated the biological role of miR-133 in apoptosis and xenograft tumorigenesis and its relationship with clinicopathological parameters. Downregulation of miR-133a was associated with degree of differentiation, local invasion and TNM stage, whereas forced expression of miR-133a was able to inhibit cell proliferation, migration, invasion, and xenograft tumorigenesis, as well as induce apoptosis. Further experiments indicated that insulin-like growth factor 1 receptor (IGF1R) was a direct downstream target of miR-133a in GC. These results collectively demonstrated that miR-133a acts as a tumor suppressor in GC by repressing IGF1R, providing a valuable target for cancer therapy.
The findings of this study indicated that miR-133a was significantly downregulated during GC and that this downregulation was correlated with the degree of differentiation, local invasion and TNM stage. Moreover, the experiments in vivo and in vitro indicated that miR-133a acted as a tumor suppressor by repressing IGF1R. This study suggests a novel strategy for treating patients with GC.
miRNAs are small (18-22 nucleotides) noncoding RNAs. They negatively regulate gene expression through base pairing with the 3’ untranslated regions of target mRNAs, resulting in mRNA degradation and/or translational repression. Abundant evidence suggests that miRNAs may function as oncogenes or tumor suppressors by regulating physiological processes.
Emerging evidence has shown that microRNAs are involved in development and dissemination of gastric cancer. miR-133a could serve as a potential tumour suppressor in several distinct cancer types that include esophageal squamous cell carcinoma, bladder cancer, prostate cancer, colorectal cancer, osteosarcoma and breast cancer. However, the specific role of miR-133a in gastric cancer is largely unknown. The work documents a very interesting aspect and data looks convincing.
P- Reviewer: Mishra PK, Shehata MMM, Yao YS S- Editor: Yu J L- Editor: Kerr C E- Editor: Liu XM
| 1. | Danaei G, Vander Hoorn S, Lopez AD, Murray CJ, Ezzati M. Causes of cancer in the world: comparative risk assessment of nine behavioural and environmental risk factors. Lancet. 2005;366:1784-1793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 831] [Cited by in RCA: 783] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 2. | Varadhachary G, Ajani JA. Gastric cancer. Clin Adv Hematol Oncol. 2005;3:118-124. [PubMed] |
| 3. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [PubMed] |
| 4. | Macfarlane LA, Murphy PR. MicroRNA: Biogenesis, Function and Role in Cancer. Curr Genomics. 2010;11:537-561. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1008] [Cited by in RCA: 1336] [Article Influence: 95.4] [Reference Citation Analysis (0)] |
| 5. | Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259-269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5384] [Cited by in RCA: 5607] [Article Influence: 295.1] [Reference Citation Analysis (0)] |
| 6. | Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 524] [Cited by in RCA: 556] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 7. | Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J, Sung JJ. MicroRNA dysregulation in gastric cancer: a new player enters the game. Oncogene. 2010;29:5761-5771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 238] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 8. | Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni L, Zhang WG, Nan KJ, Song TS, Huang C. MicroRNA profiling of human gastric cancer. Mol Med Rep. 2009;2:963-970. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 147] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 9. | Guo J, Xia B, Meng F, Lou G. miR-133a suppresses ovarian cancer cell proliferation by directly targeting insulin-like growth factor 1 receptor. Tumour Biol. 2014;35:1557-1564. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 10. | Dong Y, Zhao J, Wu CW, Zhang L, Liu X, Kang W, Leung WW, Zhang N, Chan FK, Sung JJ. Tumor suppressor functions of miR-133a in colorectal cancer. Mol Cancer Res. 2013;11:1051-1060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 11. | Yoshino H, Chiyomaru T, Enokida H, Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N, Nakagawa M. The tumour-suppressive function of miR-1 and miR-133a targeting TAGLN2 in bladder cancer. Br J Cancer. 2011;104:808-818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 204] [Cited by in RCA: 233] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 12. | Chiyomaru T, Enokida H, Tatarano S, Kawahara K, Uchida Y, Nishiyama K, Fujimura L, Kikkawa N, Seki N, Nakagawa M. miR-145 and miR-133a function as tumour suppressors and directly regulate FSCN1 expression in bladder cancer. Br J Cancer. 2010;102:883-891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 256] [Cited by in RCA: 288] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 13. | Cui W, Zhang S, Shan C, Zhou L, Zhou Z. microRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J. 2013;280:3962-3974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 75] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 14. | Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M. Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer. 2012;106:405-413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 169] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 15. | Qiu T, Zhou X, Wang J, Du Y, Xu J, Huang Z, Zhu W, Shu Y, Liu P. MiR-145, miR-133a and miR-133b inhibit proliferation, migration, invasion and cell cycle progression via targeting transcription factor Sp1 in gastric cancer. FEBS Lett. 2014;588:1168-1177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 147] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 16. | Hartog H, Wesseling J, Boezen HM, van der Graaf WT. The insulin-like growth factor 1 receptor in cancer: old focus, new future. Eur J Cancer. 2007;43:1895-1904. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 128] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 17. | Othman N, Nagoor NH. The role of microRNAs in the regulation of apoptosis in lung cancer and its application in cancer treatment. Biomed Res Int. 2014;2014:318030. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 18. | Xia H, Sun S, Wang B, Wang T, Liang C, Li G, Huang C, Qi D, Chu X. miR-143 inhibits NSCLC cell growth and metastasis by targeting Limk1. Int J Mol Sci. 2014;15:11973-11983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 19. | Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, Wang Y. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 343] [Cited by in RCA: 370] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 20. | Xu YJ, Fan Y. MiR-215/192 participates in gastric cancer progression. Clin Transl Oncol. 2015;17:34-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 21. | Tsai MM, Wang CS, Tsai CY, Chen CY, Chi HC, Tseng YH, Chung PJ, Lin YH, Chung IH, Chen CY. MicroRNA-196a/-196b promote cell metastasis via negative regulation of radixin in human gastric cancer. Cancer Lett. 2014;351:222-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 22. | Chen WS, Leung CM, Pan HW, Hu LY, Li SC, Ho MR, Tsai KW. Silencing of miR-1-1 and miR-133a-2 cluster expression by DNA hypermethylation in colorectal cancer. Oncol Rep. 2012;28:1069-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 23. | Cho WC. MicroRNAs in cancer - from research to therapy. Biochim Biophys Acta. 2010;1805:209-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 95] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 24. | Ge J, Chen Z, Wu S, Chen J, Li X, Li J, Yin J, Chen Z. Expression levels of insulin-like growth factor-1 and multidrug resistance-associated protein-1 indicate poor prognosis in patients with gastric cancer. Digestion. 2009;80:148-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 25. | Murayama T, Inokuchi M, Takagi Y, Yamada H, Kojima K, Kumagai J, Kawano T, Sugihara K. Relation between outcomes and localisation of p-mTOR expression in gastric cancer. Br J Cancer. 2009;100:782-788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 26. | Yen YC, Shiah SG, Chu HC, Hsu YM, Hsiao JR, Chang JY, Hung WC, Liao CT, Cheng AJ, Lu YC. Reciprocal regulation of microRNA-99a and insulin-like growth factor I receptor signaling in oral squamous cell carcinoma cells. Mol Cancer. 2014;13:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 27. | Qian X, Yu J, Yin Y, He J, Wang L, Li Q, Zhang LQ, Li CY, Shi ZM, Xu Q. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle. 2013;12:1385-1394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 134] [Article Influence: 11.2] [Reference Citation Analysis (0)] |