INTRODUCTION
Colorectal cancer (CRC) is the third leading cause of cancer death in the United States and one of the most common types of cancer in the western societies[1]. Surgical intervention remains the mainstay of therapy for patients without metastatic disease while adjuvant or neo-adjuvant chemotherapy and radiation are considered to improve the survival of patients with stage 2 and 3 disease[2]. Despite the improvements of secondary prevention and surgical intervention, the prognosis for patients with metastatic disease remains poor. The role of adjuvant therapy for patients with stage 2 is still not clear suggesting that better stratification of these patients may be critical to improve the survival rates[2]. Finally, therapeutic approaches for patients with stage 4 are mainly palliative[3] which clearly implies that better understanding of the molecular biology of this disease may reveal new targets for the development of novel agents for CRC patients with poor response to the conventional chemotherapy.
Epidemiologic and genetic studies have shown that there is a clear link between the disruption of circadian rhythms and cancer development and progression in humans including breast, endometrial, prostate and colon cancer[4,5]. Interestingly, alterations of the circadian rhythm have been related to modulations of tumor growth in animal models[6], differences in recurrence rates, stage and prognosis in human cancers[7,8]. The master circadian clock generating and sustaining 24 h periodicity is located in the suprachiasmatic nucleus (SCN) in the anterior hypothalamus orchestrating peripheral clocks located in other organs and tissues[9]. The existence of a circadian clock in the cellular level generating and regulating multiple activities related to metabolism, cell cycle, DNA synthesis and repair has been recently identified. In particular, the molecular mechanisms underlying circadian clock involve transcriptional and translational positive and negative feedback loops[10].
Recent molecular and genetic data strongly suggest that among the most important targets downstream of circadian clock are molecules related to DNA damage response (DDR) such as ATM, CHK2 [11] and BRCA1[12], cell cycle progression such as c-Myc and p21[13,14] and Wnt/β-catenin pathway[15]. Given that all these pathways are involved in the molecular biology of CRC it is not surprising that numerous epidemiological, genetic and molecular studies highlight the implication of clock genes not only in the initiation and progression of CRC, but in the development of resistance to chemotherapeutic agents as well. In particular, Soták et al[16] using a model of chemically induced CRC, recently found that the circadian rhythmicity of critical mediators of the circadian system, namely PER1, PER2, REV-ERBA is significantly decreased in CRC tissues while the rhythmicity of BMAL1, another circadian rhythm component is completely abolished not only in the CRC tissues but in the surrounding healthy colon tissue as well in tumor bearing animals. These results clearly support that deregulation of the Circadian system is strongly implicated in the development of CRC.
The aim of this review is to summarize the involvement of clock genes in the molecular pathways related to the development and progression of CRC and the implication of clock genes’ genetic alterations in the aggressiveness, therapeutic response and prognosis of the disease.
MOLECULAR DETERMINANTS OF THE CIRCADIAN RHYTHM
Recent data from expression pattern analysis and generation of transgenic mice with hyperactive clock genes such as PER1, PER2 and BMAL1 have shown that the activity of the SCN is not essential for the peripheral oscillation but is critical for the synchronization of these “peripheral clocks”[17,18]. These results suggest that each individual cell exhibits an independent regulation of its own circadian system.
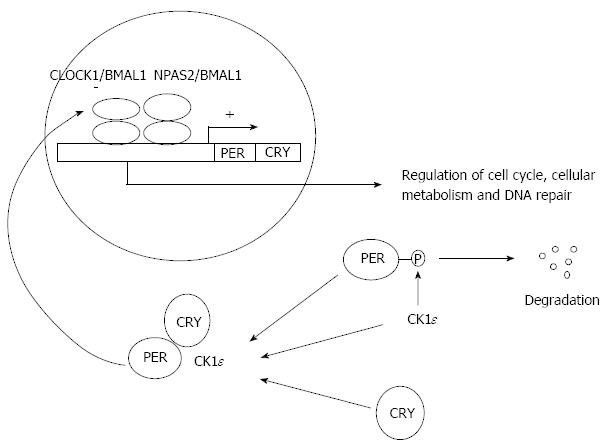
As mentioned above the circadian system and its downstream effects on various cellular activities are regulated by positive and negative feedback loops which are tightly connected. BMAL1, CLOCK1 and NPAS2 form heterodimers, which bind to the promoters of PER, CRY, REV-ERBa, RORα, DEC1 and DEC2 genes activating their transcription[19] (Figure 1). PER and CRY proteins are negatively regulate the BMAL1/CLOCK1 and BMAL1/NPAS2 heterodimers activity suppressing their own expression and inhibiting the circadian system[9] (Figure 1). Moreover, the DEC1 and DEC2 proteins compete with the above mentioned heterodimers for a common DNA binding site and supress the expression of clock genes[20]. It should be noted that PER proteins can also form heterodimers with the TIM protein maintaining their own integrity[21]. Finally, being involved in an additional pathway of the circadian system, REV-ERBα and RORα compete each other for binding to RORE elements inhibiting or activating the expression of the BMAL1 gene respectively[22].
Figure 1 The molecular mechanism of the cellular circadian system.
CLOCK1/BMAL1 and NPAS2/BMAL1 complexes promote the expression of PER and CRY. PER is phosphorylated by CK1ε kinase resulting to its degradation while the accumulation of CRY leads to the formation of PER/CRY/CK1ε complex which inserts to the nucleus and down-regulates the CLOCK1 and BMAL1 activity. Among the transcriptional targets of CLOCK1 and BMAL1 are some cell cycle mediators, tumor suppressor genes and oncogenes regulating cell cycle, cellular metabolism and DNA repair.
Interestingly, various posttranscriptional and posttranslational modulations of the above mentioned proteins further confer to the complexity of this system. In particular, protein kinases CK1epsilon and CK1δ phosphorylate multiple molecules implicated in this signaling altering their nuclear translocation and subsequently their transcriptional activities[23] (Figure 1). Moreover, epigenetic modifications through acetylation, deacetylation and methylation of histones in the promoter of various clock genes are critical for the activation of this cellular clock. In particular, it is known that the BMAL1/CLOCK1 heterodimers promote the acetylation activating the expression of their downstream targets[24] while PER/CRY heterodimers induce deacetylation and methylation of histones downregulating the expression of the clock genes.
CIRCADIAN SYSTEM REGULATES CELL CYCLE THROUGH c-MYC, P21 AND WEE1
Recent studies support that the circadian system in the regulates the cell cycle progression through c-Myc/p21 signaling which has been implicated in the development of CRC since c-Myc is found to be overexpressed in 70% of colon cancers[25]. In particular, microarray data revealed that critical molecules involved in the cell cycle machinery such as p21, cyclin D1, cyclin B1, c-Myc, p53, Wee1 and Mdm2 are regulated by the circadian system[9]. BMAL1 deletion results in an imbalance in the expression of REV-ERBα and RORα which are positive and negative regulators of p21 leading to inhibition and promotion of the cell cycle respectively[14]. It has also been shown that Bmal1-/- transgenic mice exhibit dramatically increased expression of p21 which is no longer rhythmic in their liver cells suggesting that the peripheral oscillators control critical biological processes such as cell cycle progression[14]. On the other hand, Zeng et al[13] showed that downregulation of BMAL1 in colon cancer cells leads to increased cyclin B1, CDC2, cyclin D1 and E expression accelerating tumor growth. Finally, Alhopuro et al[12] using ChIP technology showed that one of the targets of CLOCK1 gene is p21, while its regulation is believed to be p53 independent.
c-Myc is a critical regulator of the cell cycle through downregulation of p21 and activation of cyclin D1. Its promoter contains multiple E-box sequences, which are controlled by molecular components of the circadian system including PER1 and PER2[26]. Of note, Per1-/- and Per2Brdm1 mutant mice exhibit increased expression of c-Myc leading to elevated cyclin D1 and induced cell cycle[27,28]. Moreover, overexpression of PER2 in K562 leukemic cells led to downregulation of c-Myc and cyclin B1 suppressing cell’s proliferation and inducing their apoptosis[29]. Finally, BMAL1/CLOCK1 and BMAL1/NPAS2 activate WEE1 expression activating the phosphorylation of CDK1/Cyclin B complex which leads to G2/M arrest and inhibition of cell proliferation[30]. Consistent with these data Clock1 deficient mice present significantly decreased levels of WEE1 mRNA[9] while CRY1 and CRY2 deletion leads to higher WEE1 levels inhibiting cell proliferation[30,31]. Notably, WEE1 has been found to be suppressed in colon cancer tissues and cell lines[32]. Collectively these data suggest that potential alterations of critical clock genes such as BMAL1 and CLOCK1 can modulate the G2-to-M transition with subsequent effects on cell cycle progression and cell proliferation through c-Myc, p21 and Wee1.
WNT/β-CATENIN SIGNALING AND CIRCADIAN SYSTEM
Wnt/β-catenin singaling is frequently de-regulated in colorectal cancer and APC, a central component of this pathway is mutated in 50% of sporadic CRCs[33]. Wnt ligand binds the N terminal domain of a Frizzled family receptor, a G-protein coupled receptor. This interaction disrupts the function of the APC/Axin/GSK-3β destruction complex inhibiting the degradation of β-catenin. This results in increased nuclear accumulation of β-catenin inducing the expression of several mediators of cell proliferation such as c-Myc and cyclin D1[34], and activates cadherin cell adhesion complexes promoting migration and metastasis. Patients with Familial Adenomatous Polypodiasis harbor APC mutations resulting in dysfunctional destruction complex and sustained β-catenin signaling. Overexpression of BMAL1 in NIH-3T3 fibroblasts leads to increased β-catenin expression and Wnt activation contributing to induced cell proliferation[15]. Moreover, it was found that downregulation of PER2 in HCT116 and SW480 colon cell lines induces β-catenin expression and accelerates cell proliferation mediated by increased cyclin D levels[35]. Consistently, deletion of PER2 was related to increased colonic and small intestine polyps formation in mice with APC mutation[35]. Interestingly, the same group showed that activation of β-catenin signaling leads to destabilization of PER2 in the intestinal mucosa of mice with APC mutation altering the circadian rhythm and its downstream targets such as WEE1[36]. Finally, Sahar et al[37] showed that under Wnt signaling activation, the absence of GSK-3β mediated phosphorylation leads to increased BMAL1 stabilization and activity while active GSK-3β promotes BMAL1 ubiquitylation. These results suggest that Wnt/β-catenin interacts with clock system probably through a positive feedback mechanism maintaining and de-regulating colon cancer cell proliferation. Further studies are needed to validate the effect of circadian system on cell migration and metastasis through de-regulated β-catenin pathway.
DNA DAMAGE RESPONSE AND CLOCK GENES
It is known that 90% of hereditary non-polyposis colon cancers and 10%-15% of sporadic CRCs carry inactivating mutations in genes involved in the mismatch repair (MMR) system such as MLH1 and MSH2 leading to microsatellite instability (MSI) related to deficient DNA repair[38]. It should be mentioned that Alhopuro et al[12] in a recent study showed that CLOCK1 gene is somatically mutated in 53% of CRC characterized by MSI. According to this study, CLOCK1 promotes growth arrest, DNA repair and apoptosis upon genotoxic stress caused by UV radiation suggesting that this molecule may represent an important “caretaker” promoting cell cycle arrest upon DNA damage. Further studies are needed to establish the important effects of clock genes and circadian system on the cellular responses following DNA damaging events.
Apart from MMR other components of DDR such as repair of double strand breaks through ATM and CHK2 activation have been implicated in the development of CRC. In particular, reduced expression of BRCA1 and ATM, which are critical nodes in the double strand break repair system, is more frequent in CRC compared to normal colonic mucosa and related to decreased overall survival in patients with CRC[39,40]. Moreover, Takabayashi et al[41] showed that DNA damage response is significantly reduced during CRC progression. These results suggest that DNA repair and its defects are correlated to CRC development, progression and potentially clinical outcome since the deficient DNA damage response can be proved to be the Achilles’ heel of these cells.
Interestingly, Gery et al[11] showed that PER1, a critical component of the circadian system promotes the ATM mediated CHK2 activation upon exposure to radiation leading to increased G1/S arrest in colon cancer cell lines while PER1 levels are significantly reduced in human colorectal cancer samples. Moreover, it has been shown that TIM protein, another mediator of the circadian regulation at the molecular level, is also required for CHK2 activation promoting arrest of the cancer cell in the G2 phase upon DNA damage[42]. Collectively, these data support that circadian system regulates the ATM/CHK2 signaling which is critical for the repair of DNA.
More reports demonstrated that circadian system modulates other aspects of DNA repair upon genotoxic stimuli. In particular recent studies have also highlighted the role of PER1/TIM complex in the ATR mediated CHK1 activation which also leads to cell cycle arrest upon genotoxic events[43,44]. Hoffman et al[45] found that knockdown of the circadian gene NPAS2, which as described above creates heterodimers with BMAL1 and CLOCK1, leads to impaired DNA repair and inhibition of cell cycle delay upon mutagen treatment. Finally, loss of circadian CRY1 and CRY2 genes increases the sensitivity to DNA damage induced apoptosis in p53 deficient cancer cells through increased expression of the p53 related gene p73. These results suggest that impaired DNA damage response, which is partially related to altered peripheral circadian function, promotes genomic instability contributing to the development of CRC.
CLINICAL CORRELATIONS BETWEEN CLOCK GENES AND COLORECTAL CANCER
Multiple recent studies correlating the expression of clock genes with clinical outcomes highlight the role of Circadian system in the development and progression of CRC. In particular, Mazzoccoli et al[46] demonstrated that PER1, PER2, PER3 and CRY2 are significantly down-regulated in CRC tissues compared to healthy colonic mucosa while lower PER1 and PER3 expression was associated with poorer survival rates. These results support the hypothesis that these genes act mainly as tumor suppressors and their downregulation is implicated in CRC development and progression. Consistent with these data, Oshima et al[47] showed that PER1 and PER3 are downregulated in CRC tissue compared to adjacent normal mucosa while reduced expression of PER1 is related to increased incidence of liver metastasis highlighting the potential negative impact of clock genes in the aggressiveness of CRC. On the contrary the same group showed that CLOCK1 and CK1epsilon are upregulated in CRC compared to healthy mucosa, while increased expression of BMAL1 is related to decreased overall survival. These results were further supported by our group showing in a recent study that CLOCK1 and BMAL1 are upregulated while PER1 and PER3 are downregulated in CRC tissues compared to healthy mucosa[48]. These conclusions suggest that different components of the circadian system may have different effects on the development of this disease based on their implications in different oncogenic pathways.
It is known that the expression and activity of dihydropyrimidine dehydrogenase (DPD) which determine the efficacy and outcome of 5-fluorouracil (5-FU) treatment in CRC are regulated by a circadian rhythm. This conclusion led to the introduction of the “chronomodulated chemotherapy” with variable rate infusions of 5-FU for treatment of advanced CRC[49,50]. Interestingly, Krugluger et al[51] found that reduced PER1 mRNA levels are correlated with decreased DPD expression in undifferentiated CRC, a result which was more pronounced in female patients. This result suggest that in advanced CRC characterized by lower PER1 mRNA levels the circadian regulation of DPD is probably lost making cancer cells more susceptible to 5-FU treatment especially in female patients.
The role of PER2 as a negative regulator of circadian system and potential tumor suppressor for CRC has been supported by a study by Zeman et al[52] who found that tumor staging is negatively correlated with PER2 gene expression. Interestingly, a recent study demonstrated that immunohistochemical staining for PER2 is weaker in CRC cancer cells following a heterogenous pattern compared to normal colonic cells[53]. In the same study, the well differentiated cancer cells were found to have comparable PER2 levels with that in non-cancerous cells suggesting that loss of PER2 is related to increased aggressiveness of CRC[53]. Finally, the authors showed that decreased PER2 mRNA and protein levels are correlated with higher histological grade, deeper tumor invasion, lymph node metastasis, advanced TNM stage and higher Ki67 score, which suggests that reduction of PER2 may lead to attenuated cancer cell growth[53]. Collectively these data support that low expression of PER2 and potentially activated circadian system is implicated in the CRC development and progression.
On the contrary, according to a recent report by Yu et al[54], CRY was found to be upregulated in CRC cell lines and human CRC samples while higher CRY expression was associated with lymph node metastasis, increased TNM staging and poorer prognosis. At the molecular level the authors showed that upregulation of CRY increased CRC cell proliferation and migration while downregulation of CRY significantly decreased the colony formation and migration in a CRC cell line[54]. Further studies are needed to clarify the role of CRY since as mentioned above it negatively regulates the BMAL1 and CLOCK1 activities, which is more consistent with a role of tumor suppressor in CRC.
In contrast to PER proteins which are important negative regulators of the circadian system, CLOCK1, NPAS2 and BMAL1 molecules are forming heterodimers controlling the transcription of about 10% of genes implicated in cell proliferation, apoptosis and cell cycle such as c-MYC, p21 and WEE1[55]. As mentioned above, CLOCK1 regulates a complicated response to DNA damage caused by UV radiation protecting cells from acquiring additional DNA alterations, which can promote the development of a cancerous phenotype. Finally, it has been shown that 2 single nucleotide polymorphisms (rs3749474 and rs1801260) located in the 3’UTR of the CLOCK1 gene decreasing its mRNA levels are related to decreased overall survival of CRC patients[56]. In a recent study evaluating the association between clock genes polymorphisms and CRC susceptibility we showed that the rs1801260 polymorphism in the 3’UTR of the CLOCK1 gene significantly increases the risk for CRC development but it does not alter the clinical outcome in CRC patients[57]. It should be noted that BMAL1 is the Clock gene most strongly related to poor prognosis in CRC patients. According to a recent report by Tan et al the micro RNA (miRNA) mir-142-3p directly targets the 3’UTR of BMAL1 while its expression is controlled by the CLOCK1/BMAL1 heterodimers[58]. miRNAs are believed to be a novel reasonable therapeutic approach in numerous cancerous diseases[59]. Based on the above mentioned results showing that high BMAL1 is associated with poor prognosis in CRC the introduction of mir-142-3p for high grade metastatic CRC could be considered as a novel therapeutic approach in the future.
CONCLUSION
The aim of this review was to highlight the implications of the circadian system to various intracellular events related to CRC development and progression explaining multiple epidemiological findings correlating clock genes’ expression with CRC progression. According to the literature the oscillating circadian clock with its components regulates numerous cellular activities such as cell cycle, cellular metabolism and DNA damage response with known implications in carcinogenesis. In particular, clock genes have been related to p21, c-Myc and Wee1 regulation explaining their effect on cell cycle progression and proliferation while recent evidence suggest that the circadian system influences the Wnt/β-catenin signaling which is a critical pathway for the development and progression of CRC. Moreover, it has been shown that clock genes have important implications in the regulation of DNA damage response. A defective circadian system may be related to impaired DNA repair which related to the initiation and development of CRC but at the same time may make these cells susceptible to various DNA damaging agents. Finally, we can conclude that clock genes are implicated in the pathogenesis of CRC with important correlations with prognosis but may constitute an important pathway for the identification of novel agents for modern therapeutic approaches in this type of cancer.
In general, PER proteins are considered to be inhibitors of cell cycle progression and β-catenin activation and mediators of DDR maintaining genomic integrity explaining the clinical data suggesting that lower levels of PER2 are associated with poorer prognosis and metastatic disease. BMAL1 on the other hand activates cell proliferation, β-catenin pathway and is strongly associated with CRC initiation and poor clinical outcome. Targeting BMAL1 with miRNA may be a reasonable approach for patients with metastatic disease. CLOCK1 is more complicated since molecular data suggest that it inhibits cell cycle progression and promotes DNA repair upon genotoxic stress but clinical correlations show that CLOCK1 expression is higher in CRC tissues. It should be noted though that polymosphisms related to dysfunctional CLOCK1 have been shown to increase the risk for CRC and are associated with poorer prognosis. Further studies are needed to clarify the role of this gene in CRC development but it could be hypothesized that the role of CLOCK1 as an inhibitor of cell proliferation upon genotoxic stimuli can be beneficial for cancer cells’ survival after the development of the disease. Finally, the discovery of the biological roles of these genes in disease’s initiation and progression may provide valuable prognostic biomarkers which can be particularly useful for patients with stage 2 disease regarding the addition of chemotherapy for their management.
P- Reviewers: Aytac E, Botaitis SC, Chen JZ, Massimo A, Maesawa C, Shi C, Sulkowski S S- Editor: Wen LL L- Editor: A E- Editor: Wang CH