Published online Oct 7, 2014. doi: 10.3748/wjg.v20.i37.13477
Revised: March 28, 2014
Accepted: June 2, 2014
Published online: October 7, 2014
Processing time: 265 Days and 8.8 Hours
MicroRNAs (miRNAs) are a class of small noncoding RNAs that post-transcriptionally regulate the expression of many target genes via mRNA degradation or translation inhibition. Many studies have shown that miRNAs are involved in the modulation of gene expression and replication of hepatitis B virus (HBV) and hepatitis C virus (HCV) and play a pivotal role in host-virus interactions. Increasing evidence also demonstrates that viral infection leads to alteration of the miRNA expression profile in hepatic tissues or circulation. The deregulated miRNAs participate in hepatocellular carcinoma (HCC) initiation and progression by functioning as oncogenes or tumor suppressor genes by targeting various genes involved in cancer-related signaling pathways. The distinct expression pattern of miRNAs may be a useful marker for the diagnosis and prognosis of virus-related diseases considering the limitation of currently used biomarkers. Moreover, the role of deregulated miRNA in host-virus interactions and HCC development suggested that miRNAs may serve as therapeutic targets or as tools. In this review, we summarize the recent findings about the deregulation and the role of miRNAs during HBV/HCV infection and HCC development, and we discuss the possible mechanism of action of miRNAs in the pathogenesis of virus-related diseases. Furthermore, we discuss the potential of using miRNAs as markers for diagnosis and prognosis as well as therapeutic targets and drugs.
Core tip: Chronic hepatitis B virus or hepatitis C virus infection changed miRNA expression profiles at the tissue or serum level, and the altered miRNAs play pivotal roles in gene expression and replication of the viruses and the development of virus-related diseases. These findings suggest that miRNAs have the potential as novel diagnostic and prognostic biomarkers and therapeutic targets in virus-related diseases.
- Citation: Fan HX, Tang H. Complex interactions between microRNAs and hepatitis B/C viruses. World J Gastroenterol 2014; 20(37): 13477-13492
- URL: https://www.wjgnet.com/1007-9327/full/v20/i37/13477.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i37.13477
Hepatocellular carcinoma (HCC) is the sixth most common cancer and the second leading cause of cancer-related death worldwide. Chronic hepatitis B virus (HBV) and/or hepatitis C virus (HCV) infections resulting in chronic liver diseases are the major causative factors for HCC[1]. Numerous studies have showed that HCC develops through aberrantly activation of various signaling pathways involved in cellular proliferation, differentiation, angiogenesis, and metastasis. But the precise molecular mechanisms underlying virus induced-HCC development and progression are still unclear.
MiRNAs are endogenous, small noncoding RNAs containing an average of 22 nucleotides that mainly regulate gene expression at the post-transcription levels through sequence-specific binding to the 3’-untranslated region (UTR), coding region or 5’-UTR of the target mRNA[2]. As most miRNAs guide the recognition of imperfect matches of target mRNAs, each miRNA can have tens to hundreds of different mRNAs targets and further one mRNA can be simultaneously regulated by multiple miRNAs. More than 60% of all human protein-coding genes were predicted to be under miRNA regulations, which enable miRNAs to have numerous regulatory roles in many physiological processes, such as cell proliferation, differentiation and apoptosis[3].
Due to their fundamental role in biological process, aberrant expression of miRNAs in the liver may be involved in pathological processes of various liver diseases. In fact, the specific deregulation of certain miRNAs, along with the roles of the miRNAs, was observed in liver diseases such as HBV/HCV infection, fibrosis, cirrhosis and HCC[4]. In this review, we will focus on the regulatory role of deregulated miRNA in HBV/HCV infection and HCC development. In addition, we briefly discuss miRNAs as potential diagnostic markers, prognostic markers and therapeutic targets for virus-related liver diseases.
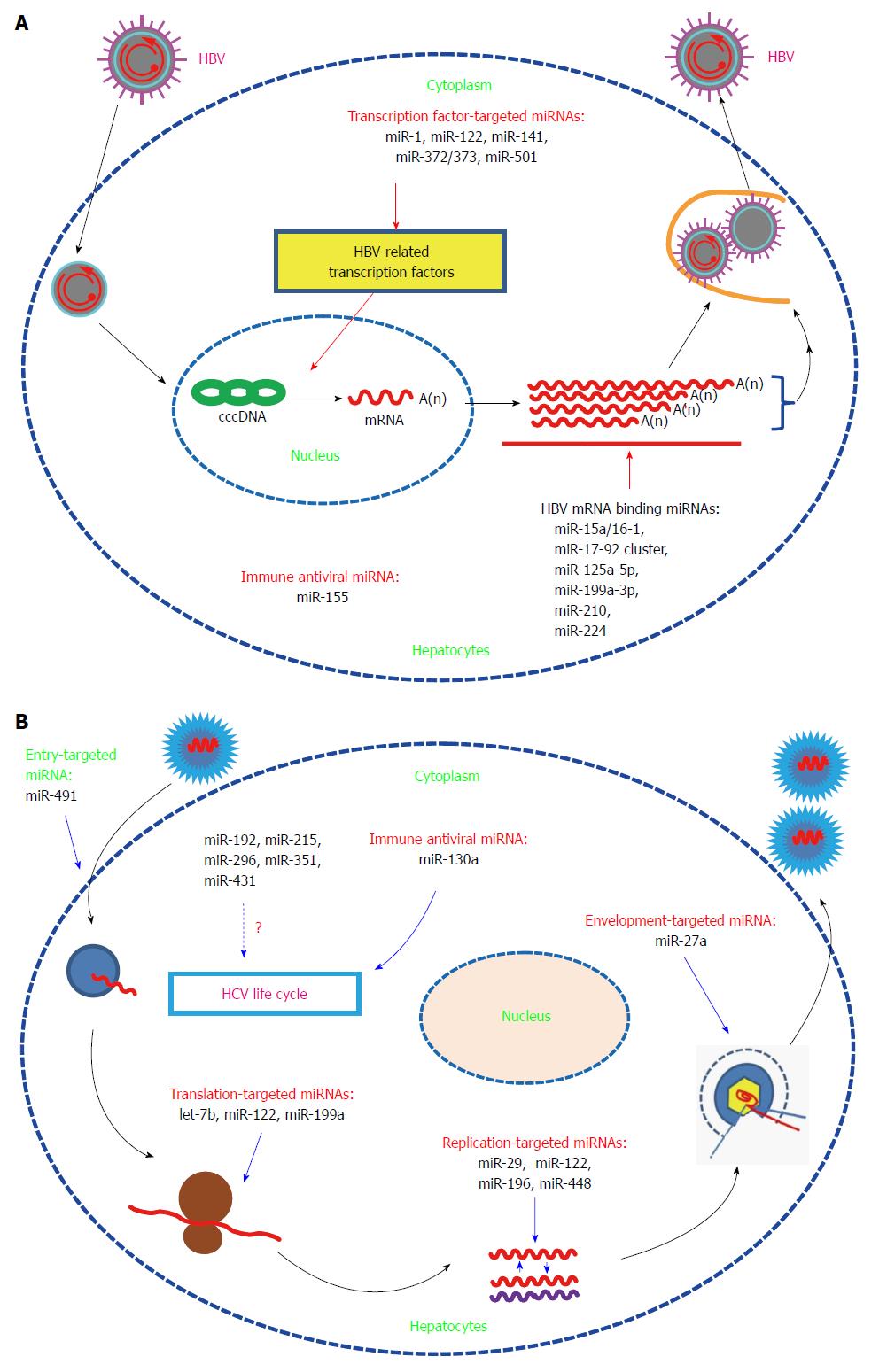
MiRNAs are reported to be involved in regulation of virus expression and replication mainly by binding directly to virus transcripts or targeting the related host genes[5,6]. Here, we will focus on a subset of miRNAs that are reported to affect gene expression and replication of HBV/HCV.
To systemically screen for host miRNAs that affected HBV replication, our lab employed a loss-of-function approach (transfecting chemically synthesized antisense oligonucleotides of 328 identified human miRNAs into HepG2.2.15 cells) and found that miR-199a-3p and miR-210 inhibitors could increase HBsAg expression and HBV replication without a significant effect on host cell proliferation. Further bioinformatics analysis and a GFP reporter assay validated that miR-199a-3p and miR-210 reduced HBV replication by binding to the HBV S protein coding region and pre-S1 region, respectively[7]. This is the first report that human miRNAs can directly target HBV genes. In addition, Potenza et al[8] reported that miR-125a-5p, a miRNA highly expressed in human liver tissue, was able to interfere with the translation of the HBV S gene. Wu et al[9] used four well-established target-prediction programs to predict the targets of human miRNAs in the HBV genome, and they found that let-7, miR-196b, miR-433, and miR-511 targeted the polymerase or S gene, miR-205 targeted the X gene, and miR-345 targeted the preC gene. Moreover, the target regions are conserved among different HBV clades, which implied that these miRNAs can be used in antiviral therapy. However, the anti-HBV activities of the miRNAs need further experimental validation. miR-15a/miR-16-1,MiR-20a and miR-92a-1 (two members of miR-17-92 cluster), and miR-224, were shown to suppress HBV replication in vitro possibly by directly binding to HBV genes[10-12].
MiRNAs can also modulate HBV replication by targeting HBV-associated host proteins. The study by Zhang et al[13] showed that miR-1 enhanced HBV replication by increasing HBV core promoter activity through augmenting farnesoid X receptor α expression. MiRs-372/373 stimulated the production of HBV proteins and HBV core-associated DNA in HepG2 cells by targeting nuclear factor I/B[14]. MiR-501 was recently reported to promote HBV replication in part by targeting HBXIP, an inhibitor of HBV replication in HepG2.215 cells[15]. MiR-141 suppressed HBV replication by down-regulating peroxisome proliferator-activated receptor alpha, a positive transcription factor of HBV[16]. MiR-122, an abundant liver-specific miRNA, could inhibit gene expression and replication of HBV in vitro via binding to highly conserved regions of the mRNA for the viral polymerase and the 3’-untranslated region of the mRNA for the core protein[17]. Further mechanism studies showed that miR-122 suppressed HBV replication partially by modulation of p53-mediated inhibition of HBV replication through down-regulation of cyclin G1[18,19]. MiR-155 mildly inhibited HBV infection in human hepatoma cells by suppressing suppressor of cytokine signaling 1 (SOCS1) expression and subsequently promoting JAK/STAT(signal transducer and activator of transcription) signaling pathway, which leads to enhanced innate antiviral immunity[20] (Figure 1A).
In contrast to the inhibitory role on HBV replication, miR-122 is essential for HCV RNA replication. Jopling et al[21] found that sequestration of miR-122 led to a marked loss of HCV RNAs in vitro. Further study showed that the interaction between miR-122 and two binding sites in the 5’-noncoding region of the HCV genome were essential for HCV RNA maintenance. The binding of miR-122 to the HCV genome could also stimulate HCV translation by enhancing ribosome binding to the viral RNA[22]. However, results from the double-binding-site mutation and the IRES mutation assays showed that miR-122 may affect viral replication at other steps of the viral life cycle[23]. Expression of miR-122 in non-hepatic cells could facilitate efficient viral replication, which further supports the finding that miR-122 is essential for HCV replication[24,25]. Further structure and function analysis showed that nearly all nucleotides in miR-122 were involved in binding to the second binding site, and this additional interaction enhanced HCV replication[26]. MiR-122 could bind to HCV RNA in association with Ago2 to protect the viral genome from 5’-exonuclease activity and thus stabilize the HCV genome and slow its decay[27]. A recent study showed that the formation of a stable ternary complex between two copies of miR-122 and the HCV 5’-UTR could stabilize the HCV genome, which also implied that miR-122 had additional functions in the viral life cycle[28]. In addition to direct binding to the HCV genome, miR-122 may promote HCV replication through other pathways. Shan et al[29] found that suppression of miR-122 with an miR-122-specific antagomir decreased HCV replication and increased HO-1 expression, thus suggesting that miR-122 may promote HCV replication in part by decreasing heme oxygenase 1(HO-1) expression. HO-1, a key enzyme with anti-oxidant and anti-inflammatory activities, can suppress HCV RNA replication[30].
Although the specific mechanism of miR-122-supported HCV replication needs further exploration, the above studies indicated that miR-122-based therapy may be useful in limiting HCV infection. Silencing of miR-122 with a locked nucleic acid (LNA)-modified oligonucleotide (SPC3649) complementary to miR-122 resulted in universal antiviral activity against diverse HCV genotypes in vitro and led to long-lasting suppression of HCV viremia in chronically infected chimpanzees with no evidence of viral resistance or side effects[31,32]. In 2009, the miR-122 inhibitor miravirsen was applied to phase 1 clinical trials by Santaris Pharma to test its safety. The data demonstrated that miravirsen was well tolerated and has no dose-limiting toxicities[33]. In 2010, Santaris Pharma initiated phase 2a clinical trials and showed that miravirsen was efficient in reducing HCV RNA in patients with chronic HCV genotype 1 infection. Moreover, this antiviral effect was long-lasting, and no detectible adverse events or escape mutations were observed[34]. However, it has been reported that miR-122 could inhibit nuclear factor (NF)-κB activation and subsequently its downstream pro-inflammatory events[35]. So silencing of miR-122 may augment liver inflammation. One study showed that interferon (IFN)-β treatment leads to a significant reduction in the expression of miR-122[36]. Other studies showed that during chronic HCV infection, patients with decreased pretreatment liver miRNA-122 levels responded poorly to interferon therapy[37-39]. All these issues should be considered when using miravirsen alone or in combination with IFN therapy. Furthermore, the suppression effect of miR-122 on HBV should be taken into consideration when treating HCV infection with miravirsen in HBV and HCV co-infected patients.
In contrast to the inhibitory effect on HBV, HCV infection-induced miR-141 could promote efficient HCV replication in primary human hepatocytes, which was dependent on miR-141 mediated suppression of the tumor suppressor gene DLC-1[40]. MiR-192, miR-215, and miR-491, which were found to be altered after HCV infection, could enhance HCV replication. Among these miRNAs, miR-491 promoted HCV replication through the PI3 kinase/Akt pathway which may enhance entry of HCV into cells, while the mechanisms for the other two miRNAs were not mentioned[41]. In contrast to the enhancing capabilities of the above-mentioned miRNAs, miR-199a inhibited HCV replication in a cell culture system by directly targeting to the sequence in domain II of the IRES region in the HCV 5’-UTR[42]. MiR-196 inhibited HCV replication in vitro mainly through repressing the expression of Bach1, a basic leucine zipper mammalian transcriptional repressor of HO-1[43]. MiR-29 was down-regulated in both hepatocytes and hepatic stellate cells after HCV infection, and function analysis showed that miR-29 overexpression reduced HCV RNA abundance and inhibited collagen and extracellular matrix expression[44]. Therefore, miR-29 has potential as a therapy tool by inhibiting HCV replication and decreasing fibrosis. Expression of let-7b markedly suppressed HCV replication and down-regulated HCV accumulation and had a synergistic inhibitory effect on HCV infection with IFN-α-2a[45]. Although bioinformatics analysis identified conserved binding sites for let-7b among various HCV genotypes within the coding region of NS5B and the 5’-UTR of the HCV genome, mechanism studies showed that let-7b-mediated suppression of HCV RNA accumulation was independent of translation inhibition. Let-7b represents the first miRNA with a target site in the coding region of the HCV genome. Interestingly, a recent study showed that let-7b was regulated by IFNα and IL-28B and inhibited HCV replication and viral protein translation by targeting host factor insulin-like growth factor 2 mRNA-binding protein 1 (IGF2BP1)[46]. HCV-induced miR-130a inhibits HCV replication/production by restoring host innate immune responses and/or down-regulating pro-HCV miR-122[47,48]. MiR-27a induced by HCV infection and lipid overload could in turn repress HCV infection and lipid storage in vitro. This negative feedback loop between miR-27a and HCV may be beneficial for immune escape and establishment of persistent HCV infection[49]. Pedersen et al[36] found that IFNβ-induced miRNAs miR-196, miR-296, miR-351, miR-431 and miR-448 could substantially attenuate HCV replication. Among them, miR-196 and miR-448 may inhibit HCV replication by directly targeting the HCV genomic RNA (Figure 1B).
To date, no HCV-encoded miRNAs have been reported, which may be due to the nature of RNA viruses or to the limitations of current detection technology. By using computational approaches, Jin et al[50] found that HBV putatively encodes only one candidate pre-miRNA. The presence of the mature form of the predicted miRNA in HBV-infected patients was validated by Northern blot. Further target search results showed that there was only one potential viral mRNA target, which suggested that HBV has evolved to produce a miRNA to regulate its own replication. Studies investigating the specific function of this viral miRNA are in progress.
Increasing evidence showed that HBV or HCV has evolved various strategies to evade host immune surveillance and establish a persistent chronic infection, which includes modulating the expression of host miRNAs. The alteration of the miRNA expression profile by HBV or HCV infection, the possible mechanism responsible for miRNA deregulation, and miRNA roles in virus-related liver pathogenesis are reviewed.
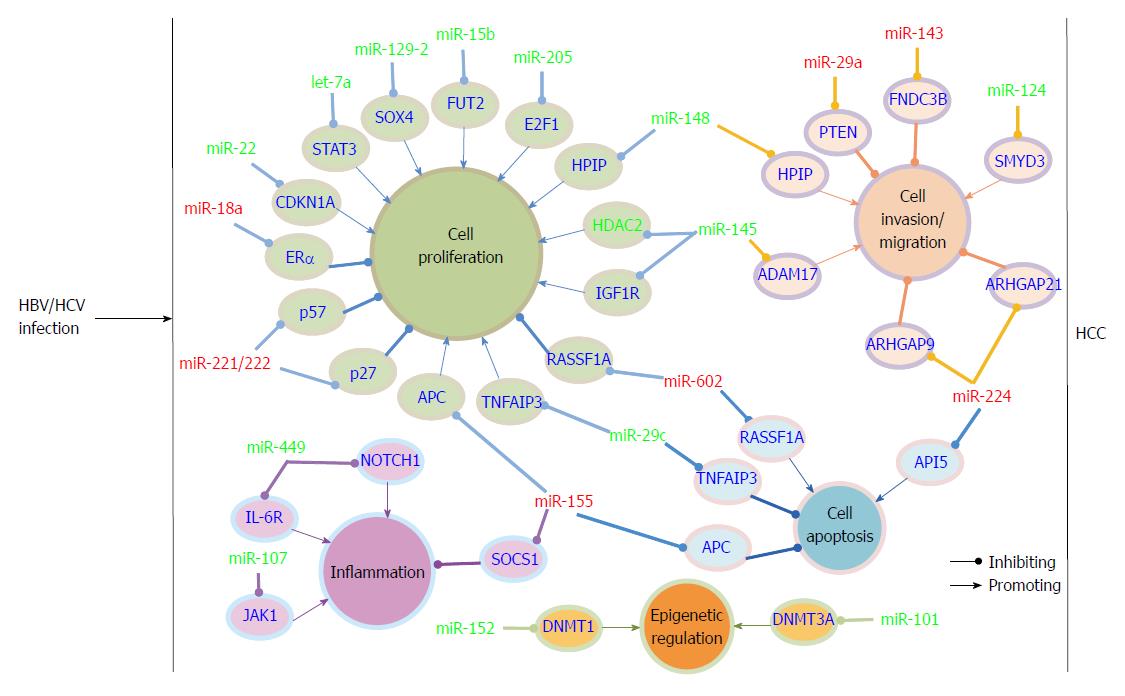
Many reports have examined miRNA expression alteration in cell culture, HCC liver tissue and serum samples after HBV infection by miRNA-based microarray or sequencing. The cell line HepG2.2.15, which stably expresses HBV, and the parental cell line HepG2 are often used to compare the effect of HBV on miRNA expression. MiRNA microarray analysis showed that 11 miRNAs were up-regulated and 7 miRNAs were down-regulated in HepG2.2.15 cells compared to HepG2 cells[51]. After this initial report, additional studies have compared the miRNA expression profile in these two cell lines by miRNA array[14,52,53]. Among the up-regulated miRNAs, miR-199a, miR-210, miR-371/372, miR-501, and the miR-17-92 cluster were involved in HBV replication[7,11,14,15]. The mechanism by which HBV regulates miRNAs is largely unknown. It has been reported that the viral protein HBx could transactivate a variety of viral and host genes, including miRNAs. To date, it has been reported that HBx modulated miRNA expression mainly through 3 pathways. First, HBx regulates miRNA expression at the transcription level via direct binding to the transcription factor essential for miRNA transcription or by modulating the transcription factor expression[54-56]. Second, HBx mRNA functions as a sponge that can specifically down-regulate miRNA via the miRNA target sequence in the viral RNA[10,57]. Third, HBx modulates pri-miRNA biogenesis by reducing the protein levels of the RNase III enzyme Drosha[58]. However, there is evidence in HBV-associated HCC that the key regulators of miRNA biosynthesis, including Drosha, DGCR8, Ago1, and Ago2, are frequently overexpressed[59]. Except for HBx, the other HBV proteins have not been reported to be directly involved in regulating miRNA expression. The specific mechanism will be discussed in the related reference. MiRNA array analysis of HBx-expressing versus control HepG2 cells showed that 7 miRNAs were significantly up-regulated and 11 were markedly down-regulated in HBx-expressing cells[60]. HBx-mediated down-regulation of let-7a promoted cell proliferation and hepatocarcinogenesis in part via up-regulation of STAT3. HBx could also reduce the levels of another let-7 family member, let-7i, which regulated CD59 expression and thus protected the HCC cells from complement-dependent cytotoxicity[61]. Tumor suppressor-like miR-15a/16 was first reported to be repressed by HBx-induced c-myc[62]. Two recent findings showed that HBx transcripts could directly lead to miR-15a/16 down-regulation via the so-called miRNA sponge effect[10,57]. Therefore, the virus gene could regulate one miRNA expression in different ways. Moreover, miR-15b, another member of miR-16 family, was also repressed by HBx and suppressed cell proliferation via directly targeting fucosyltransferase 2 (FUT2), which leads to decreased levels of Globo H[63]. A recent study showed that HBx down-regulated tumor suppressor miRNA miR-205 through inducing hypermethylation of miR-205 promoter, which may be another pathway by which HBx regulating miRNA expression[64]. Quantitative real-time PCR analysis showed that miR-101, one of the most abundantly expressed miRNAs in human normal liver, was reduced by HBx in vitro. Function analysis showed that down-regulated miR-101 promoted aberrant DNA hypermethylation, which contributes to hepatocarcinogenesis by allowing DNMT3A expression[65]. Another association study of genetic variations in miR-101-1 and miR-101-2 with HBV-related liver diseases showed that the single nucleotide polymorphism (SNP) in miR-101-1 was associated with the development of HCC and liver cirrhosis[66]. To date, it has been reported that SNPs in the 3’-UTR of three miRNA processing genes, miRNA encoding genes or binding sites are associated with miRNA deregulation in HCC[67-78]. MiR-29a was dramatically increased in p21-HBx transgenic mice, HBx-transfected HepG2-X (or H7402-X) cells and HepG2.2.15 cells. MiR-29a enhanced hepatoma cell migration by targeting phosphatase and tensin homolog (PTEN)[79]. HBx-induced miR-21 enhanced cell proliferation by targeting tumor-suppressor genes Programmed Cell Death Protein-4 (PDCD4) and PTEN[80]. One report showed that miR-148 was repressed by HBx to promote cancer growth and metastasis in a mouse model by activating hematopoietic pre-B cell leukemia transcription factor- interacting protein (HPIP)-mediated mTOR signaling[54]. The mechanism studies showed that HBx suppressed miR-148a expression by directly interacting with p53. However, another study by Yuan et al[81] reported that miR-148 is induced by HBx, and the up-regulated miR-148 stimulated hepatocellular growth and tumorigenesis by repressing PTEN and subsequent induction of β-catenin expression. The mechanism responsible for the HBx-mediated miR-148 induction is not mentioned in this study. The opposite effect of HBx on miR-148 in the latter report may be because the authors examined the miR-148 levels in stable HBx-expressing HepG2 cell lines and because the biological characterization of HepG2 cells have changed for HBx integration. The specific reason for the observation requires further exploration. Carboxyl-terminal truncated HBx (Ct-HBx) is frequently over-expressed in HBV-associated HCC and plays a critical role in hepatocarcinogenesis[55]. Ct-HBx down-regulated a set of growth-inhibitory miRNAs that were often induced by its full-length counterpart. Moreover, Ct-HBx could directly bind to some of these miRNA promoters to repress transcription. All of the above-mentioned miRNAs regulated by HBx play important roles in tumor formation. Therefore, HBx, which has been implicated in HBV-related HCC pathogenesis, may perform its pro-carcinogenic function partly through regulating host miRNA expression. Recently, one report showed that HBV up-regulated miR-181a expression by enhancing its promoter activity, HBV induced-miR-181a promoted cell growth in vitro and tumor formation in vivo possibly by targeting transcription factor E2F5[82].
Jiang et al[83] showed that a large number of mature and precursor miRNA expression was increased in hepatitis B or C virus-positive, cirrhotic tissues compared to normal liver. This is the first report that hepatitis infection altered miRNA expression in liver tissues. Later, Ladeiro et al[84] examined the expression of 250 miRNAs in 46 benign and malignant hepatocellular tumors with 4 normal livers as controls with real-time PCR and found that miR-96 was specifically overexpressed in HBV tumors. Connolly et al[85] found that two known oncomiRs, the miR-17-92 polycistron and miR-21, were significantly increased in human HBV-positive HCCs and woodchuck hepatics virus-positive HCCs. Ura et al[86] analyzed the expression of 188 miRNAs in liver tissues from 12 HBV-related HCCs and 14 HCV-related HCCs, and 6 miRNAs were found to be markedly decreased in the HBV group and 13 miRNAs were decreased in the HCV group. Guo et al[14] analyzed the miRNA expression profiles of 39 cases of HBsAg positive and 11 cases of HBsAg negative liver tissues using microarrays. They showed that 9 miRNAs were up-regulated, with miR-373 being the most significantly up-regulated, and the up-regulation was associated with copy number variation of the miRs-371-3 gene cluster. The copy number variation of the miRs-371-3 gene cluster may be due to gene instability resultant from HBV integration as the genomic locus for miRs-372/373 is near to the fragile genomic site FRA19A, a known hotspot for the integration of HBV DNA[87,88]. Mizuguchi et al[89] analyzed the miRNA transcriptome in HBV-related HCC using combined conventional cloning with GS 454 sequencing technology. They identified more than 314000 reliable reads from HCC and more than 268000 from corresponding liver tissues for human miRNAs registered in miRBase. MiR-122, miR-21, and miR-34a were found to be expressed aberrantly in liver cancer by clone count analysis. Therefore, the combination of sequencing and bioinformatics may benefit the discovery of novel markers in HBV-related liver disease.
The identification of altered miRNA profiles in HBV-associated HCC has led to miRNA target identification and has increased our understanding of the mechanisms underlying HCC development and progression. MiRNAs can be classified as tumor suppressor-like and oncogene-like based on their functions in HCC. Furthermore, tumor suppressor-like miRNAs are often down-regulated and oncogene-like miRNAs are often up-regulated in HCC. To date, most of the miRNAs discovered in HBV-associated HCC belong to the tumor suppressor-like class. Ji et al[90] examined miRNA expression patterns, survival and response to IFN-α in a panel of 455 patients with mainly HBV-associated HCC. They found that miR-26 expression was frequently decreased in HCC patients and determined that patients with low miR-26 expression in tumors had a better response to interferon therapy but shorter survivals than patients with high miRNA-26 expression. In a study by Huang et al[91], miR-152 was found to be frequently down-regulated in HBV-related HCC compared to the paired non-tumor hepatic tissues and was found to be involved in the regulation of abnormal DNA methylation status. Further study showed that down-regulated miR-152 induced global DNA hypermethylation and increased the methylation levels of two tumor suppressor genes, glutathione S-transferase pi 1 (GSTP1) and E-cadherin 1 (CDH1), by allowing expression of DNA methyltransferase 1(DNMT1). Chen et al[92] found that miR-129-2 expression was repressed in HBV-HCC tissues compared to the adjacent non-tumor tissues, and miR-129-2 methylation was significantly increased both in frequency and intensity in tumor tissues. Further investigation showed this methylation-mediated repression of miR-129-2 may be involved in HCC development through enhancing oncogenic SOX4 expression. Together with other reports, all of the findings suggested that epigenetic regulation, such as DNA methylation and histone modification, of miRNA expression play an important role in HCC development[93,94]. Wang et al[95] reported that miR-29c was significantly down-regulated in HBV-related HCC. They showed that miR-29c functioned as a tumor suppressor in HCC development by targeting tumor necrosis factor alpha-induced protein 3 (TNFAIP3), a key regulator in inflammation and immunity.
One study reported that miR-22 was overexpressed in male tumor adjacent tissues as determined by SOLiD sequencing[96]. Overexpressed miR-22 was correlated with low estrogen receptor α (ERα) and high IL-1a expression in HBV-associated patients. These results may explain the high incidence of HBV-associated HCC in the male population. Another miRNA associated with ERα is miR-18a. Elevated miR-18a was observed in female HCC tissues compared to male HCC tissues in HBV-related HCC. MiRNA-18a may promote HCC development in females by suppressing the expression of ERα[97]. In Zhang et al[56]’s study, they demonstrated that miR-143 was dramatically increased by HBx-mediated NF-κB activation in metastatic HBV-HCC in both p21-HBx transgenic mice and HCC patients. Additionally, miR-143 was found to promote cancer cell invasion/migration. MiR-143 behaved as an oncogene in HCC through repressing expression of fibronectin type III domain containing 3B (FNDC3B).
In a study by Yang et al[98], 14 miRNAs were found to be aberrantly expressed in HBV-related patients. MiR-602 showed a progressive increase trend from chronic hepatitis to HCC compared to normal livers and played a pro-carcinogenic role in HBV- associated hepatocarcinogenesis. Up-regulated miR-602 acted as an oncogene by repressing expression of tumor suppressive gene RAS association domain family 1A (RASSF1A). In a study by Gao et al[99], they investigated the expression of 7 cancer-related miRNAs during the early stages of HCC. Down-regulation of tumor suppressive-like miR-145 and miR-199b, and up-regulation of miR-224, was frequently observed from low grade dysplastic nodule to high grade dysplastic nodule and HCC. Moreover, in HBV-associated HCC patients, the expression of low-Atg5 (autophagy-related genes), high-miR-224 and low-Smad4 showed significant correlation with HBV infection and poor overall survival rate[100].
Recently, change in serum or plasma miRNA expression pattern was also discovered in HBV-associated patients. Li et al[101] examined the miRNA expression profiles in serum from 210 controls and 135 HBV-, 48 HCV-, and 120 HCC-related individuals by Solexa sequencing followed by TaqMan probe-based quantitative real-time PCR validation. They identified 13 miRNAs that were differentially expressed in HBV serum compared to control serum. Two of them, miR-375 and miR-92a, were also identified as HBV specific. This is the first report that serum miRNA profiles can serve as a biomarker for HBV infection. Later, additional miRNAs were found to be significantly deregulated in serum or plasma from patients with chronic HBV infection (CHB), HBV-associated HCC or occult HBV infection compared with healthy controls, which suggests that serum or plasma miRNAs have the potential to serve as biomarkers in detecting HBV-specific cases[102-111]. Interestingly, a recent study reported that serum HBsAg particles carried selective pools of hepatocellular miRNAs and may play a role in viral persistence, which enriched the knowledge of HBV-host interaction[112] (Figure 2).
Different HCV genotypes induced different miRNA expression changes in hepatoma cells. Liu et al[113] compared the miRNA expression profile between Huh7.5.1 hepatoma cells infected with HCV JFH-1 virus and the same cell type infected with UV-inactivated HCV virus with miRNA microarray analysis. One-hundred eight miRNAs were differentially expressed (greater than 2-fold change) at 4 d post infection. Most (60%) were up-regulated in HCV-infected cells, and some of them were identified to be involved in HCV entry, replication and propagation. In Huh7 cells, miR-192/miR-215 and miR-194 were up-regulated and miR-320 and miR-491 were down-regulated after HCV infection. Moreover, overexpression of miR-192/miR-215 and miR-491 in vitro significantly increased HCV replicon abundance[41]. In Huh7.5 cells, 7 miRNAs (miR-30b, miR-30c, miR-130a, miR-192, miR-301, miR-324-5p, and miR-565), which were linked to genes associated with HCV entry and replication, were down-regulated after infection with HCV genotype 2a[114]. The miR-192 expression is inconsistent in the two studies. This may be due to the difference in cell type, HCV genotype and sampling time after HCV infection. Steuerwald et al[115] found that 43 miRNAs were differently expressed between Con1 cells containing a full-length HCV genotype 1b replicon and Huh 7.5 parental hepatoma cells. In another hepatoma cell line, HepG2, stably transfected with full length HCV genotype 1b, 75 miRNAs were found to be aberrantly expressed. The miRNAs miR-193b, miR-768-3p, and miR-585 were the most significantly up-regulated. Up-regulated miR-193b made the malignant hepatocytes more sensitive to sorafenib by modulation of the anti-apoptotic protein Mcl-1. The miRNAs that had been reported to be regulated by the HCV core protein or non-structure proteins (NS3/4A, NS4B and NS5A) included miR-93, miR-345, miR-124, miR-21, and miR-27[116-120].
Varnholt et al[121] examined the miRNA expression profiles in HCV-infected patients with premalignant dysplastic liver nodules and hepatocellular carcinomas by real-time PCR. Ten miRNAs were more than 2-fold overexpressed, and 19 were less than 0.4-fold underexpressed. Among the overexpressed miRNAs, miR-122, miR-100, and miR-10a were consistently overexpressed across all liver nodules. MiR-198 and miR-145 were underexpressed progressively from low-grade dysplastic nodule to high-grade dysplastic nodule to HCC and further to kidney metastasis. Marquez et al[122] found that miR-122 and miR-21 expression were altered in HCV-infected liver. The miR-21 levels were correlated, while miR-122 levels were inversely correlated, with viral load, fibrosis and serum liver transaminase levels. Zhang et al[123] found that miR-155 was significantly increased by HCV-RNA-induced NF-κB activation in chronic HCV-infected patients and HCV-HCC compared to healthy controls. Up-regulated miR-155 repressed Adenomatous polyposis coli (APC) and subsequently activated Wnt signaling, which resulted in hepatocyte proliferation and tumorigenesis. P65/NF-κB activated miR-224 expression in both peri-tumoral cirrhotic livers and HCC samples from HCV-infected patients as determined by real-time PCR. MiR-224 promoted HCC cell migration and invasion, which is a key determinant in HCC development and progression[124]. A genome-wide microarray analysis performed by Sarma et al[125] showed that a distinct miRNA expression profile existed in HCV-infected liver tissue compared to the normal liver. Particularly, miR-449a was specifically down-regulated in HCV-infected patients compared to the other three groups, and down-regulated miR-449a promoted inflammatory marker YKL40 expression by modulating the NOTCH signaling pathway. Diaz et al[126] examined the expression of 2226 miRNAs in patients with HCV-associated HCC, HCC-associated non-tumorous cirrhosis, HCV-associated cirrhosis without HCC, HBV-associated ALF and normal liver tissue using microarrays. They found 18 miRNAs that were specifically expressed in HCV-associated HCC and were connected in a regulatory network, including p53 tumor suppressor, the PTEN phosphatase and retinoic acid signaling.
Ogawa et al[127] found that the miR-221/222 levels increased with the progression of liver fibrosis in HCV-infected patients and significantly correlated with the expression of a1 (I) collagen (Col1A1) and α-smooth muscle actin (αSMA). MiR-221/222 was the first miRNA that was reported to be related to liver fibrosis and may be useful in the prediction of liver fibrosis progression. Ramachandran et al[128] found that several miRNAs expression were altered by chronic HCV infection by using miRNA expression profiling. HCV-induced-miR-200c promotes hepatic fibrosis by repressing the expression of FAS associated phosphatase 1 (FAP-1), a critical regulator of Src and MAP kinase pathway. It was reported that miRNA-107 and miRNA-449a were decreased following HCV infection in patients with HCV-mediated liver diseases. Further function analysis showed that they regulate CCL2 expression by activation of the IL-6-mediated signaling cascade, which may be involved in HCV-mediated induction of inflammatory responses and fibrosis[129]. A report showed that 5 miRNAs were significantly overexpressed and 7 miRNAs were markedly reduced in splenic marginal zone lymphoma (SMZL) from HCV-positive patients[130]. MiR-26b, a known tumor suppressor, was found to be significantly reduced in SMZL. This result suggested that HCV may contribute to lymphoma development. With the hypothesis that miRNA expression profiles from liver grafts could distinguish the severity of HCV recurrence and discriminate this from acute cellular rejection (ACR), Joshi et al[131] found that miR-146a, miR-19a, miR-20a, and let-7e were overexpressed in patients with slow HCV fibrosis progression compared to patients with fast HCV fibrosis progression after liver transplantation. Distinct miRNA profiles exist between patients with HCV recurrence and ACR. Moreover, altered hepatic expression of miRNA is associated with HCV recurrence and antiviral therapy in liver transplant recipients for hepatitis C virus cirrhosis[132].
In chronic HCV-infected patients, serum miR-122 was increased, and the levels of miR-122 were correlated with serum ALT but not with HCV RNA[133]. However, a later study showed that miR-122 may just mirror acute liver injury with the finding that the levels of miR-122 that were high in the early stage of fibrosis were reduced in advanced fibrosis stages during chronic HCV infection[134]. However, Cermelli et al[135] found that serum levels of miR-122, miR-34a, and miR-16 in chronic HCV-infected patients were steadily increased during the course of HCV infection, and miR-122 and miR-34a levels were positively correlated with disease severity. The discordant change trend of miR-122 may be due to the lack of a consistent internal control for circular miRNA measurements or due to the small patient population. Moreover, the serum miR-122 kinetics could predict the treatment results, as patients with higher serum miR-122 showed early and sustained virologic response to pegylated IFN plus ribavirin therapy[38]. In addition to miRNA-122, other miRNAs were also found to be significantly deregulated in serum from HCV-related patients and were associated with either HCV infection or HCV-mediated liver disease progress[136-140]. MiRNA deregulation was also found in peripheral blood mononuclear cells (PBMCs) of HCV-infected patients. Deregulation of miRNAs may play a key role in HCV-related pathogenesis[141,142]. One study showed that 27 miRNAs and 476 mRNAs were differentially expressed in PBMCs from HCV/HIV co-infected patients when compared to controls by performing genome-wide mRNA and miRNA analysis, this is the first report of miRNAs specific for HCV/HIV co-infection[143]. Abdalla et al[144] found that miR-323, miR-449, miR-502d, miR-92b, miR-516-5p, and miR-650 were down-regulated while miR-335, miR-618, miR-625, miR-532, and miR-7 were up-regulated in urine from both the HCC-post HCV-positive group and the HCV-positive group compared to the control group. Further study showed that the aberrant expression of miR-618 and miR-650 can be used as a marker for the early diagnosis of HCC from HCV-positive patients (Figure 2).
It has been shown that miRNAs play important roles in HBV/HCV infection and the associated liver disease. Moreover, differential expression patterns of miRNAs have been observed in the livers or serum of HBV- and HCV-infected patients. The unique miRNA expression profile in HBV- or HCV-related diseases suggested that they may represent novel biomarkers for diagnosis or prognosis or can be exploited as therapeutic agents or targets in the treatment and prevention of these diseases.
In HBV-related liver diseases, some miRNAs were aberrantly expressed in liver tissues, such as miR-148a, miR-96, miR-29c, and miR-602, were related to virus infection and disease progression[81,84,95,98]. Therefore, miRNAs may serve as diagnostic markers. For example, miR-602 was overexpressed from the very early stage of chronic hepatitis to HBV-positive cirrhosis and HCC[98]. MiR-602 played an important regulatory role in HBV-mediated hepatocarcinogenesis by inhibiting the tumor suppressive gene RASSF1A. Therefore, this miRNA can serve as an early diagnostic marker for HBV-mediated HCC. In HCV-related liver diseases, some deregulated miRNAs in liver tissues also have diagnostic potential. HCV-induced miR-155 could promote hepatocyte proliferation and tumorigenesis by activating Wnt signaling and thus may be a useful biomarker for early diagnostics of HCV-HCC[123]. With high specificity and selectivity, 18 miRNAs exclusively expressed in HCV-HCC were identified by Diaz et al[126] and displayed particular clinical value in diagnosing HCV-HCC. Up-regulated miR-199a, miR-199a*, miR-200a, miR-200b, and miR-221/222 were significantly correlated to the grade of liver fibrosis and may serve as biomarkers for stellate cell activation and liver fibrosis progression[127].
Although the miRNA expression profiles in liver tissues were helpful in discriminating HCC from non-tumor tissues, they are not ideal as markers in the early diagnosis of HCC for their invasiveness. Circulating miRNAs were specific and stable in body fluids such as blood or urine and can be used as potential diagnostic markers not only for HBV or HCV infection but also for the development of HCC. In separating the control and HBV groups, serum miR-375 or miR-10a individually had a 97.5% area under curve (AUC) of receiver operating characteristic (ROC) while the combination of miR-375, miR-10a, miR-223, and miR-423 resulted in 100% AUC. Serum miRNAs including miR-23b, miR-423, miR-375, miR-23a, and miR-342-3p could clearly separate the HBV-positive HCC group from the control with an AUC of 99.9%[101]. A plasma miRNA panel (miR-122, miR-192, miR-21, miR-223, miR-26a, miR-27a, and miR-801) had a high diagnostic accuracy of HBV-positive HCC regardless of disease status (Barcelona Clinic Liver Cancer stages 0, A, B, and C). This miRNA panel could also discriminate HCC from healthy liver, chronic hepatitis B and cirrhosis[108]. The expression profiles of miR-21, miR-122, and miR-223 could serve as a novel biomarker for liver injury but not specifically for detection of HCC in chronic HBV-infected patients[105]. The serum miR-210 level may serve as a molecular biomarker for the severity of hepatitis in patients with hepatitis B[110]. The serum miR-125b-5p and miR223-3p are potentially very early diagnostic biomarkers of HBV-related HCC even at chronic hepatitis B stage of liver disease[111]. Four serum miRNA signatures (let-7c, miR-23b, miR-122, and miR-150) can clearly separate occult HBV-infected patients from the control group with an AUC of 99.9%, although larger population studies are required to validate these findings[107]. Plasma miR-20a and miR-92a were increased in acute and chronic HCV-infected patients and HCV-infected fibrosis patients and thus may serve as biomarkers for the early detection of HCV infection[137]. The urinary miRNAs signatures found by Abdalla et al[144] is of great value for the early diagnosis of HCC, before the onset of disease in HCV-positive patients (Table 1).
| miRNA | Expression(vs control) | HCC etiology | Clinical relevance | Ref. |
| Liver miRNAs | ||||
| miR-29c | Down | HBV | Diagnostic marker therapeutic agent | [95] |
| miR-96 | Up | HBV | Diagnostic marker | [84] |
| miR-148 | Up | HBV | Early diagnostic marker and/or therapeutic target for HBx-mediated HCC | [81] |
| miR-155 | Up | HCV | Diagnostic marker therapeutic target | [123] |
| miR-221/222 | Up | HCV | New markers for stellate cell activation and liver fibrosis progression | [127] |
| miR-602 | Up | HBV | Early diagnostic marker | [98] |
| Serum miRNAs | ||||
| miR-18b | Up | Mainly HCV | Prognostic marker | [146] |
| miR-20a | Up | HCV | Sensitive biomarker for early detection of HCV infection | [137] |
| miR-26 | Down | mainly HBV | Prognostic marker | [90] |
| miR-29a-5p | Up | HBV | Novel predictor for early recurrence of HBV-HCC after surgical resection | [145] |
| miR-92a | Up | HBV | Novel biomarker for HBV infection and HBV-HCC | [101] |
| miR-122 | Up | HBV | Novel biomarker for liver injury but not specifically for HCC | [102,105] |
| miR-125b-5p | Up | HBV | Novel biomarkers of HCC in very early, even at CHB stage of liver disease | [111] |
| miR-210 | Up | HBV | A molecular biomarker for the severity of hepatitis | [110] |
| miR-223 | Down | HBV | Novel biomarkers of HCC in very early, even at CHB stage of liver disease | [111] |
| miR-224 | Up | HBV | Autophagy-miR-224-Smad4 in combination could be used prognostic marker | [100] |
| miR-375 | Up | HBV | Novel biomarker for HBV infection and HBV-HCC | [101] |
| miR-572 | Up | HBV | Biomarker for liver injury | [102] |
| miR-575 | Up | HBV | Biomarker for liver injury | [102] |
| miR-638 | Up | HBV | Biomarker for liver injury | [102] |
| miR-744 | Down | HBV | Biomarker for liver injury | [102] |
| Urinary miRNAs | ||||
| miR-618 | Up | HCV | Early diagnostic marker of HCC among high-risk HCV patients | [144] |
| miR-650 | Down | HCV | Early diagnostic marker of HCC among high-risk HCV patients | [144] |
MiRNAs were also suitable for predicting the HCC clinical outcomes. A set of 19 miRNAs were significantly altered in cirrhotic and hepatitis B or C virus-positive livers, and this miRNA expression signature significantly correlated with disease outcome[83]. The expression levels of miR-26 were reduced in HCC compared with adjacent non-tumor tissues. Moreover, tumor tissue with low miRNA-26 expression was associated with shorter overall survival but better response to IFN therapy than tumor tissue with high miR-26. It hence appears to be a predictor of survival[90]. MiR-29a-5p can be a novel predictor for early recurrence of HBV-related HCC after HCC resection, especially in BCLC 0/A stage HCCs[145]. In HBV-associated HCC patients, the expression of low-Atg5 (autophagy-related genes), high-miR-224 and low-Smad4 shows significant correlation with HBV infection and poor overall survival rate[100]. ROC curve analysis revealed that the levels of miR-122, miR-572, miR-575, miR-638, and miR-744 in serum can serve as molecular markers to predict liver injury resulted from chronic hepatitis B[102]. Murakami et al[146] found that HCC patients (mainly HCV-HCC) with high miR-18b expression had a significantly shorter relapse-free period than those with low expression after surgical resection. Joshi et al[131] identified a panel of miRNAs associated with the severity of HCV recurrence which can be used as biomarkers that are predictive of aggressive recurrence after liver transplantation. Similarly, Gehrau et al[147] identified a 9 miRNA signature that can predict the severity of hepatitis C virus recurrence after liver transplantation (Table 1).
Many in vitro studies have indicated that miRNA-based therapy could efficiently inhibit HBV/HCV production and impair proliferation and invasion of the HCC cells. With the advantage that one miRNA can simultaneously target multiple genes and pathways, and one gene or pathway can be targeted by multiple miRNAs, targeting miRNA may be an efficient antiviral or anti-HCC strategy and be helpful in reducing the incidence of therapy resistance. The application of miRNA often involves two strategies: restoration of miRNA with antiviral or tumor suppressor functions by reintroducing miRNA mimics or expression from transfected/transduced vectors and the inhibition of miRNAs with virus promoting or oncogene-like functions by using miRNA inhibitors, such as antagomiRs and antimiRs. At present, only one miRNA-based drug has entered and passed through human clinical 1 and clinical 2a trials, the miRNA-122 inhibitor miravirsen. Miravirsen showed a promising antiviral activity with no obvious evidence of virus escape in treatment-naive HCV patients and thus may be useful in preventing HCV-related HCC[33,34].
Some studies also showed that miRNAs were involved in the response/resistance of HCC cells to chemotherapy[38,39,116,148-153]. The classical miRNA is miRNA-122, which accounts for more than 70% of all miRNAs present in hepatic cells. During chronic HCV infection, patients with decreased pretreatment liver miRNA-122 levels responded poorly to IFN therapy[128]. Similarly, patients with higher pretreatment serum miRNA-122 levels showed early and sustained virologic response to pegylated IFN plus ribavirin therapy[38]. Restoration of miRNA-122-sensitized HCC cells to doxorubicin, sorafenib, adriamycin and vincristine occurs through inactivation of the CDK4-PSMD10-UPR pathway or down-regulation of multidrug resistance-related genes[149-152]. Comprehensive expression profiling of miRNAs in DOX-resistant and parental HCC HepG2 cells were recently analyzed by deep sequencing[153]. The authors identified a panel of differentially expressed known and novel miRNAs, and their target genes were mainly involved in the MAPK signaling pathway by function annotation analysis, which may help us to overcome DOX resistance in future HCC chemotherapy.
The complex interactions between miRNA and HBV/HCV were discussed in this review. HBV/HCV infection elicited changes in the host miRNA expression profile, and the altered miRNAs could in turn facilitate the virus life cycle and the development of virus-associated diseases, which suggested that the deregulated miRNA profiles may serve as biomarkers and therapy targets for virus-mediated diseases. For their stability and specificity of expression in tissue and circulation, miRNAs are good biomarkers for early diagnosis and prognostic prediction. But the patient population, the sampling and testing technologies are often different in different studies, and the present testing technologies (including microarray, real time-PCR, and next generation sequencing) are either high cost or low sensitivity, all these factors limit their clinical application. With the broad regulatory roles of miRNAs, and for their small size and less antigenicity, miRNAs are good candidates for therapeutic targeting. However, in addition to the side effects and high cost, the stability of miRNA modulators and the specific delivery system are the major challenges to overcome in advancing miRNA-based strategies from bench to bedside. In view of the advantages and limitations of application of miRNA in clinic, further research into the precise molecular mechanism by which miRNAs regulate the pathogenesis of virus-associated diseases will be helpful in early diagnosis, prognostic prediction and effective treatment of these diseases.
P- Reviewer: Komatsu S, Silva ACS, Taylor JR, Tetsuya T S- Editor: Ma YJ L- Editor: A E- Editor: Wang CH
| 1. | Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1097] [Cited by in RCA: 1102] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 2. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16068] [Article Influence: 1004.3] [Reference Citation Analysis (2)] |
| 3. | Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5833] [Cited by in RCA: 6539] [Article Influence: 384.6] [Reference Citation Analysis (0)] |
| 4. | Wang XW, Heegaard NH, Orum H. MicroRNAs in liver disease. Gastroenterology. 2012;142:1431-1443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 228] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 5. | Lin Z, Flemington EK. miRNAs in the pathogenesis of oncogenic human viruses. Cancer Lett. 2011;305:186-199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 6. | Cullen BR. Viral and cellular messenger RNA targets of viral microRNAs. Nature. 2009;457:421-425. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 219] [Cited by in RCA: 216] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 7. | Zhang GL, Li YX, Zheng SQ, Liu M, Li X, Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antiviral Res. 2010;88:169-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 182] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 8. | Potenza N, Papa U, Mosca N, Zerbini F, Nobile V, Russo A. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 2011;39:5157-5163. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 136] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 9. | Wu FL, Jin WB, Li JH, Guo AG. Targets for human encoded microRNAs in HBV genes. Virus Genes. 2011;42:157-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 31] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 10. | Wang Y, Jiang L, Ji X, Yang B, Zhang Y, Fu XD. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J Biol Chem. 2013;288:18484-18493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 11. | Jung YJ, Kim JW, Park SJ, Min BY, Jang ES, Kim NY, Jeong SH, Shin CM, Lee SH, Park YS. c-Myc-mediated overexpression of miR-17-92 suppresses replication of hepatitis B virus in human hepatoma cells. J Med Virol. 2013;85:969-978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Scisciani C, Belloni L, Guerrieri F, Levrero M, Pediconi N. mir-224 is a direct target of hbx and modulates hbv replication. J Hepatol. 2011;54:S444. [DOI] [Full Text] |
| 13. | Zhang X, Zhang E, Ma Z, Pei R, Jiang M, Schlaak JF, Roggendorf M, Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology. 2011;53:1476-1485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 161] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 14. | Guo H, Liu H, Mitchelson K, Rao H, Luo M, Xie L, Sun Y, Zhang L, Lu Y, Liu R. MicroRNAs-372/373 promote the expression of hepatitis B virus through the targeting of nuclear factor I/B. Hepatology. 2011;54:808-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 15. | Jin J, Tang S, Xia L, Du R, Xie H, Song J, Fan R, Bi Q, Chen Z, Yang G. MicroRNA-501 promotes HBV replication by targeting HBXIP. Biochem Biophys Res Commun. 2013;430:1228-1233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Hu W, Wang X, Ding X, Li Y, Zhang X, Xie P, Yang J, Wang S. MicroRNA-141 represses HBV replication by targeting PPARA. PLoS One. 2012;7:e34165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 17. | Chen Y, Shen A, Rider PJ, Yu Y, Wu K, Mu Y, Hao Q, Liu Y, Gong H, Zhu Y. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. FASEB J. 2011;25:4511-4521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 18. | Wang S, Qiu L, Yan X, Jin W, Wang Y, Chen L, Wu E, Ye X, Gao GF, Wang F. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G(1) -modulated P53 activity. Hepatology. 2012;55:730-741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 209] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 19. | Ori A, Zauberman A, Doitsh G, Paran N, Oren M, Shaul Y. p53 binds and represses the HBV enhancer: an adjacent enhancer element can reverse the transcription effect of p53. EMBO J. 1998;17:544-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 81] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 20. | Su C, Hou Z, Zhang C, Tian Z, Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol J. 2011;8:354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 109] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 21. | Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577-1581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1993] [Cited by in RCA: 1986] [Article Influence: 99.3] [Reference Citation Analysis (0)] |
| 22. | Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Fehr C, Jünemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300-3310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 492] [Cited by in RCA: 527] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 23. | Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J Virol. 2010;84:6615-6625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 256] [Cited by in RCA: 247] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 24. | Chang J, Guo JT, Jiang D, Guo H, Taylor JM, Block TM. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J Virol. 2008;82:8215-8223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 168] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 25. | Fukuhara T, Kambara H, Shiokawa M, Ono C, Katoh H, Morita E, Okuzaki D, Maehara Y, Koike K, Matsuura Y. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with hepatitis C virus. J Virol. 2012;86:7918-7933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 92] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 26. | Pang PS, Pham EA, Elazar M, Patel SG, Eckart MR, Glenn JS. Structural map of a microRNA-122: hepatitis C virus complex. J Virol. 2012;86:1250-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 21] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 27. | Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc Natl Acad Sci USA. 2012;109:941-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 303] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 28. | Mortimer SA, Doudna JA. Unconventional miR-122 binding stabilizes the HCV genome by forming a trimolecular RNA structure. Nucleic Acids Res. 2013;41:4230-4240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 73] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 29. | Shan Y, Zheng J, Lambrecht RW, Bonkovsky HL. Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology. 2007;133:1166-1174. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 141] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 30. | Zhu Z, Wilson AT, Mathahs MM, Wen F, Brown KE, Luxon BA, Schmidt WN. Heme oxygenase-1 suppresses hepatitis C virus replication and increases resistance of hepatocytes to oxidant injury. Hepatology. 2008;48:1430-1439. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 109] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 31. | Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5’ UTR. Proc Natl Acad Sci USA. 2011;108:4991-4996. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 148] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 32. | Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1291] [Article Influence: 86.1] [Reference Citation Analysis (0)] |
| 33. | Hildebrandt-Eriksen ES, Bagger YZ, Knudsen TB, Petri A, Persson R, Boergesen HM, McHulchison JG, Levin AA. A unique therapy for HCV inhibits microRNA-122 in humans and results in HCV RNA suppression in chronically infected chimpanzees: results from primate and first-in-human studies. Hepatology. 2009;50:12A-12A. |
| 34. | Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685-1694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1665] [Cited by in RCA: 1699] [Article Influence: 141.6] [Reference Citation Analysis (0)] |
| 35. | Chen Y, Wang C, Liu Y, Tang L, Zheng M, Xu C, Song J, Meng X. miR-122 targets NOD2 to decrease intestinal epithelial cell injury in Crohn’s disease. Biochem Biophys Res Commun. 2013;438:133-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 36. | Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919-922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 722] [Cited by in RCA: 703] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 37. | Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15:31-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 252] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 38. | Su TH, Liu CH, Liu CJ, Chen CL, Ting TT, Tseng TC, Chen PJ, Kao JH, Chen DS. Serum microRNA-122 level correlates with virologic responses to pegylated interferon therapy in chronic hepatitis C. Proc Natl Acad Sci USA. 2013;110:7844-7849. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 39. | Köberle V, Waidmann O, Kronenberger B, Andrei A, Susser S, Füller C, Perner D, Zeuzem S, Sarrazin C, Piiper A. Serum microRNA-122 kinetics in patients with chronic hepatitis C virus infection during antiviral therapy. J Viral Hepat. 2013;20:530-535. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 40. | Banaudha K, Kaliszewski M, Korolnek T, Florea L, Yeung ML, Jeang KT, Kumar A. MicroRNA silencing of tumor suppressor DLC-1 promotes efficient hepatitis C virus replication in primary human hepatocytes. Hepatology. 2011;53:53-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 64] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 41. | Ishida H, Tatsumi T, Hosui A, Nawa T, Kodama T, Shimizu S, Hikita H, Hiramatsu N, Kanto T, Hayashi N. Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem Biophys Res Commun. 2011;412:92-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 42. | Murakami Y, Aly HH, Tajima A, Inoue I, Shimotohno K. Regulation of the hepatitis C virus genome replication by miR-199a. J Hepatol. 2009;50:453-460. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 169] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 43. | Hou W, Tian Q, Zheng J, Bonkovsky HL. MicroRNA-196 represses Bach1 protein and hepatitis C virus gene expression in human hepatoma cells expressing hepatitis C viral proteins. Hepatology. 2010;51:1494-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 44. | Bandyopadhyay S, Friedman RC, Marquez RT, Keck K, Kong B, Icardi MS, Brown KE, Burge CB, Schmidt WN, Wang Y. Hepatitis C virus infection and hepatic stellate cell activation downregulate miR-29: miR-29 overexpression reduces hepatitis C viral abundance in culture. J Infect Dis. 2011;203:1753-1762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 131] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 45. | Cheng JC, Yeh YJ, Tseng CP, Hsu SD, Chang YL, Sakamoto N, Huang HD. Let-7b is a novel regulator of hepatitis C virus replication. Cell Mol Life Sci. 2012;69:2621-2633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 46. | Cheng M, Si Y, Niu Y, Liu X, Li X, Zhao J, Jin Q, Yang W. High-throughput profiling of alpha interferon- and interleukin-28B-regulated microRNAs and identification of let-7s with anti-hepatitis C virus activity by targeting IGF2BP1. J Virol. 2013;87:9707-9718. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 47. | Bhanja Chowdhury J, Shrivastava S, Steele R, Di Bisceglie AM, Ray R, Ray RB. Hepatitis C virus infection modulates expression of interferon stimulatory gene IFITM1 by upregulating miR-130A. J Virol. 2012;86:10221-10225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 48. | Li S, Duan X, Li Y, Liu B, McGilvray I, Chen L. MicroRNA-130a inhibits HCV replication by restoring the innate immune response. J Viral Hepat. 2014;21:121-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 55] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 49. | Shirasaki T, Honda M, Shimakami T, Horii R, Yamashita T, Sakai Y, Sakai A, Okada H, Watanabe R, Murakami S. MicroRNA-27a regulates lipid metabolism and inhibits hepatitis C virus replication in human hepatoma cells. J Virol. 2013;87:5270-5286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 157] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 50. | Jin WB, Wu FL, Kong D, Guo AG. HBV-encoded microRNA candidate and its target. Comput Biol Chem. 2007;31:124-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 49] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 51. | Liu Y, Zhao JJ, Wang CM, Li MY, Han P, Wang L, Cheng YQ, Zoulim F, Ma X, Xu DP. Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin Med J (Engl). 2009;122:10-14. [PubMed] |
| 52. | Zhang ZZ, Liu X, Wang DQ, Teng MK, Niu LW, Huang AL, Liang Z. Hepatitis B virus and hepatocellular carcinoma at the miRNA level. World J Gastroenterol. 2011;17:3353-3358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 50] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 53. | Pan XB, Ma H, Jin Q, Wei L. Characterization of microRNA expression profiles associated with hepatitis B virus replication and clearance in vivo and in vitro. J Gastroenterol Hepatol. 2012;27:805-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 54. | Xu X, Fan Z, Kang L, Han J, Jiang C, Zheng X, Zhu Z, Jiao H, Lin J, Jiang K. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J Clin Invest. 2013;123:630-645. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 140] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 55. | Yip WK, Cheng AS, Zhu R, Lung RW, Tsang DP, Lau SS, Chen Y, Sung JG, Lai PB, Ng EK. Carboxyl-terminal truncated HBx regulates a distinct microRNA transcription program in hepatocellular carcinoma development. PLoS One. 2011;6:e22888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 56. | Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 286] [Article Influence: 17.9] [Reference Citation Analysis (1)] |
| 57. | Liu N, Zhang J, Jiao T, Li Z, Peng J, Cui Z, Ye X. Hepatitis B virus inhibits apoptosis of hepatoma cells by sponging the MicroRNA 15a/16 cluster. J Virol. 2013;87:13370-13378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 58. | Ren M, Qin D, Li K, Qu J, Wang L, Wang Z, Huang A, Tang H. Correlation between hepatitis B virus protein and microRNA processor Drosha in cells expressing HBV. Antiviral Res. 2012;94:225-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 59. | Liu AM, Zhang C, Burchard J, Fan ST, Wong KF, Dai H, Poon RT, Luk JM. Global regulation on microRNA in hepatitis B virus-associated hepatocellular carcinoma. OMICS. 2011;15:187-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 60. | Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow P, Chung AY, Jooi LL, Lee CG. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J Hepatol. 2010;53:57-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 184] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 61. | Shan C, Zhang S, Cui W, You X, Kong G, Du Y, Qiu L, Ye L, Zhang X. Hepatitis B virus X protein activates CD59 involving DNA binding and let-7i in protection of hepatoma and hepatic cells from complement attack. Carcinogenesis. 2011;32:1190-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 62. | Wu G, Yu F, Xiao Z, Xu K, Xu J, Tang W, Wang J, Song E. Hepatitis B virus X protein downregulates expression of the miR-16 family in malignant hepatocytes in vitro. Br J Cancer. 2011;105:146-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 63. | Wu CS, Yen CJ, Chou RH, Chen JN, Huang WC, Wu CY, Yu YL. Downregulation of microRNA-15b by hepatitis B virus X enhances hepatocellular carcinoma proliferation via fucosyltransferase 2-induced Globo H expression. Int J Cancer. 2014;134:1638-1647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 64. | Zhang T, Zhang J, Cui M, Liu F, You X, Du Y, Gao Y, Zhang S, Lu Z, Ye L. Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia. 2013;15:1282-1291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 65. | Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X, Wu Z. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal. 2013;25:439-446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 123] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 66. | Bae JS, Kim JH, Pasaje CF, Cheong HS, Lee TH, Koh IS, Lee HS, Kim YJ, Shin HD. Association study of genetic variations in microRNAs with the risk of hepatitis B-related liver diseases. Dig Liver Dis. 2012;44:849-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 67. | Liu L, An J, Liu J, Wen J, Zhai X, Liu Y, Pan S, Jiang J, Wen Y, Liu Z. Potentially functional genetic variants in microRNA processing genes and risk of HBV-related hepatocellular carcinoma. Mol Carcinog. 2013;52 Suppl 1:E148-E154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 68. | Xu T, Zhu Y, Wei QK, Yuan Y, Zhou F, Ge YY, Yang JR, Su H, Zhuang SM. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126-2131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 278] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 69. | Qi P, Dou TH, Geng L, Zhou FG, Gu X, Wang H, Gao CF. Association of a variant in MIR 196A2 with susceptibility to hepatocellular carcinoma in male Chinese patients with chronic hepatitis B virus infection. Hum Immunol. 2010;71:621-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 96] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 70. | Akkız H, Bayram S, Bekar A, Akgöllü E, Ulger Y. A functional polymorphism in pre-microRNA-196a-2 contributes to the susceptibility of hepatocellular carcinoma in a Turkish population: a case-control study. J Viral Hepat. 2011;18:e399-e407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 71. | Kim HY, Yoon JH, Lee HS, Cheong JY, Cho SW, Shin HD, Kim YJ. MicroRNA-196A-2 polymorphisms and hepatocellular carcinoma in patients with chronic hepatitis B. J Med Virol. 2014;86:446-453. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 72. | Xiang Y, Fan S, Cao J, Huang S, Zhang LP. Association of the microRNA-499 variants with susceptibility to hepatocellular carcinoma in a Chinese population. Mol Biol Rep. 2012;39:7019-7023. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 101] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 73. | Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH, Park PW, Hong SP, Rim KS, Kwon SW, Hwang SG. Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene. 2012;504:92-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 74. | Liu Y, Zhang Y, Wen J, Liu L, Zhai X, Liu J, Pan S, Chen J, Shen H, Hu Z. A genetic variant in the promoter region of miR-106b-25 cluster and risk of HBV infection and hepatocellular carcinoma. PLoS One. 2012;7:e32230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 75. | Kwak MS, Lee DH, Cho Y, Cho EJ, Lee JH, Yu SJ, Yoon JH, Lee HS, Kim CY, Cheong JY. Association of polymorphism in pri-microRNAs-371-372-373 with the occurrence of hepatocellular carcinoma in hepatitis B virus infected patients. PLoS One. 2012;7:e41983. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 76. | Cheong JY, Shin HD, Kim YJ, Cho SW. Association of polymorphism in MicroRNA 219-1 with clearance of hepatitis B virus infection. J Med Virol. 2013;85:808-814. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 77. | Guo Z, Wu C, Wang X, Wang C, Zhang R, Shan B. A polymorphism at the miR-502 binding site in the 3’-untranslated region of the histone methyltransferase SET8 is associated with hepatocellular carcinoma outcome. Int J Cancer. 2012;131:1318-1322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 78. | Han Y, Pu R, Han X, Zhao J, Li W, Yin J, Zhang Y, Shen Q, Xie J, Zhang Q. Association of a potential functional pre-miR-218 polymorphism and its interaction with hepatitis B virus mutations with hepatocellular carcinoma risk. Liver Int. 2014;34:728-736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 79. | Kong G, Zhang J, Zhang S, Shan C, Ye L, Zhang X. Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS One. 2011;6:e19518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 149] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 80. | Damania P, Sen B, Dar SB, Kumar S, Kumari A, Gupta E, Sarin SK, Venugopal SK. Hepatitis B virus induces cell proliferation via HBx-induced microRNA-21 in hepatocellular carcinoma by targeting programmed cell death protein4 (PDCD4) and phosphatase and tensin homologue (PTEN). PLoS One. 2014;9:e91745. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 81. | Yuan K, Lian Z, Sun B, Clayton MM, Ng IO, Feitelson MA. Role of miR-148a in hepatitis B associated hepatocellular carcinoma. PLoS One. 2012;7:e35331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 82. | Zou C, Li Y, Cao Y, Zhang J, Jiang J, Sheng Y, Wang S, Huang A, Tang H. Up-regulated MicroRNA-181a induces carcinogenesis in Hepatitis B virus-related hepatocellular carcinoma by targeting E2F5. BMC Cancer. 2014;14:97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 83. | Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419-427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 402] [Cited by in RCA: 422] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 84. | Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955-1963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 527] [Cited by in RCA: 545] [Article Influence: 32.1] [Reference Citation Analysis (0)] |
| 85. | Connolly E, Melegari M, Landgraf P, Tchaikovskaya T, Tennant BC, Slagle BL, Rogler LE, Zavolan M, Tuschl T, Rogler CE. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 207] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 86. | Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology. 2009;49:1098-1112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 306] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 87. | Feitelson MA, Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252:157-170. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 179] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 88. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 89. | Mizuguchi Y, Mishima T, Yokomuro S, Arima Y, Kawahigashi Y, Shigehara K, Kanda T, Yoshida H, Uchida E, Tajiri T. Sequencing and bioinformatics-based analyses of the microRNA transcriptome in hepatitis B-related hepatocellular carcinoma. PLoS One. 2011;6:e15304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 90. | Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437-1447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 662] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 91. | Huang J, Wang Y, Guo Y, Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus-related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 274] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 92. | Chen X, Zhang L, Zhang T, Hao M, Zhang X, Zhang J, Xie Q, Wang Y, Guo M, Zhuang H. Methylation-mediated repression of microRNA 129-2 enhances oncogenic SOX4 expression in HCC. Liver Int. 2013;33:476-486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 93. | Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 462] [Cited by in RCA: 464] [Article Influence: 30.9] [Reference Citation Analysis (0)] |
| 94. | Au SL, Wong CC, Lee JM, Fan DN, Tsang FH, Ng IO, Wong CM. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56:622-631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 240] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 95. | Wang CM, Wang Y, Fan CG, Xu FF, Sun WS, Liu YG, Jia JH. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem Biophys Res Commun. 2011;411:586-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 115] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 96. | Jiang R, Deng L, Zhao L, Li X, Zhang F, Xia Y, Gao Y, Wang X, Sun B. miR-22 promotes HBV-related hepatocellular carcinoma development in males. Clin Cancer Res. 2011;17:5593-5603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 97. | Liu WH, Yeh SH, Lu CC, Yu SL, Chen HY, Lin CY, Chen DS, Chen PJ. MicroRNA-18a prevents estrogen receptor-alpha expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology. 2009;136:683-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 195] [Cited by in RCA: 212] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 98. | Yang L, Ma Z, Wang D, Zhao W, Chen L, Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is overexpressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol Ther. 2010;9:803-808. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 99. | Gao P, Wong CC, Tung EK, Lee JM, Wong CM, Ng IO. Deregulation of microRNA expression occurs early and accumulates in early stages of HBV-associated multistep hepatocarcinogenesis. J Hepatol. 2011;54:1177-1184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 100. | Lan SH, Wu SY, Zuchini R, Lin XZ, Su IJ, Tsai TF, Lin YJ, Wu CT, Liu HS. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59:505-517. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 163] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 101. | Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, Zhang JF, Shen HB, Zhang CY, Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010;70:9798-9807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 378] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 102. | Zhang H, Li QY, Guo ZZ, Guan Y, Du J, Lu YY, Hu YY, Liu P, Huang S, Su SB. Serum levels of microRNAs can specifically predict liver injury of chronic hepatitis B. World J Gastroenterol. 2012;18:5188-5196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 103. | Hayes CN, Akamatsu S, Tsuge M, Miki D, Akiyama R, Abe H, Ochi H, Hiraga N, Imamura M, Takahashi S. Hepatitis B virus-specific miRNAs and Argonaute2 play a role in the viral life cycle. PLoS One. 2012;7:e47490. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 104. | Waidmann O, Bihrer V, Pleli T, Farnik H, Berger A, Zeuzem S, Kronenberger B, Piiper A. Serum microRNA-122 levels in different groups of patients with chronic hepatitis B virus infection. J Viral Hepat. 2012;19:e58-e65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 106] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 105. | Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. PLoS One. 2011;6:e28486. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 212] [Cited by in RCA: 241] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 106. | Li L, Guo Z, Wang J, Mao Y, Gao Q. Serum miR-18a: a potential marker for hepatitis B virus-related hepatocellular carcinoma screening. Dig Dis Sci. 2012;57:2910-2916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 107. | Chen Y, Li L, Zhou Z, Wang N, Zhang CY, Zen K. A pilot study of serum microRNA signatures as a novel biomarker for occult hepatitis B virus infection. Med Microbiol Immunol. 2012;201:389-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 108. | Zhou J, Yu L, Gao X, Hu J, Wang J, Dai Z, Wang JF, Zhang Z, Lu S, Huang X. Plasma microRNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma. J Clin Oncol. 2011;29:4781-4788. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 497] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 109. | Winther TN, Bang-Berthelsen CH, Heiberg IL, Pociot F, Hogh B. Differential plasma microRNA profiles in HBeAg positive and HBeAg negative children with chronic hepatitis B. PLoS One. 2013;8:e58236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 43] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 110. | Song G, Jia H, Xu H, Liu W, Zhu H, Li S, Shi J, Li Z, He J, Chen Z. Studying the association of microRNA-210 level with chronic hepatitis B progression. J Viral Hepat. 2014;21:272-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 111. | Giray BG, Emekdas G, Tezcan S, Ulger M, Serin MS, Sezgin O, Altintas E, Tiftik EN. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol Biol Rep. 2014;41:4513-4519. [PubMed] |
| 112. | Novellino L, Rossi RL, Bonino F, Cavallone D, Abrignani S, Pagani M, Brunetto MR. Circulating hepatitis B surface antigen particles carry hepatocellular microRNAs. PLoS One. 2012;7:e31952. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 113. | Liu X, Wang T, Wakita T, Yang W. Systematic identification of microRNA and messenger RNA profiles in hepatitis C virus-infected human hepatoma cells. Virology. 2010;398:57-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 114. | Zhang X, Daucher M, Armistead D, Russell R, Kottilil S. MicroRNA expression profiling in HCV-infected human hepatoma cells identifies potential anti-viral targets induced by interferon-α. PLoS One. 2013;8:e55733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 115. | Steuerwald NM, Parsons JC, Bennett K, Bates TC, Bonkovsky HL. Parallel microRNA and mRNA expression profiling of (genotype 1b) human hepatoma cells expressing hepatitis C virus. Liver Int. 2010;30:1490-1504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 116. | Braconi C, Valeri N, Gasparini P, Huang N, Taccioli C, Nuovo G, Suzuki T, Croce CM, Patel T. Hepatitis C virus proteins modulate microRNA expression and chemosensitivity in malignant hepatocytes. Clin Cancer Res. 2010;16:957-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 117. | Shiu TY, Huang SM, Shih YL, Chu HC, Chang WK, Hsieh TY. Hepatitis C virus core protein down-regulates p21(Waf1/Cip1) and inhibits curcumin-induced apoptosis through microRNA-345 targeting in human hepatoma cells. PLoS One. 2013;8:e61089. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 118. | Huang S, Xie Y, Yang P, Chen P, Zhang L. HCV core protein-induced down-regulation of microRNA-152 promoted aberrant proliferation by regulating Wnt1 in HepG2 cells. PLoS One. 2014;9:e81730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 119. | Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, Liu Y, Wu J. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. 2013;9:e1003248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 200] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 120. | Singaravelu R, Chen R, Lyn RK, Jones DM, O’Hara S, Rouleau Y, Cheng J, Srinivasan P, Nasheri N, Russell RS. Hepatitis C virus induced up-regulation of microRNA-27: a novel mechanism for hepatic steatosis. Hepatology. 2014;59:98-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 88] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 121. | Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 347] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 122. | Marquez RT, Bandyopadhyay S, Wendlandt EB, Keck K, Hoffer BA, Icardi MS, Christensen RN, Schmidt WN, McCaffrey AP. Correlation between microRNA expression levels and clinical parameters associated with chronic hepatitis C viral infection in humans. Lab Invest. 2010;90:1727-1736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 181] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 123. | Zhang Y, Wei W, Cheng N, Wang K, Li B, Jiang X, Sun S. Hepatitis C virus-induced up-regulation of microRNA-155 promotes hepatocarcinogenesis by activating Wnt signaling. Hepatology. 2012;56:1631-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 256] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 124. | Scisciani C, Vossio S, Guerrieri F, Schinzari V, De Iaco R, D’Onorio de Meo P, Cervello M, Montalto G, Pollicino T, Raimondo G. Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J Hepatol. 2012;56:855-861. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 112] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 125. | Sarma NJ, Tiriveedhi V, Subramanian V, Shenoy S, Crippin JS, Chapman WC, Mohanakumar T. Hepatitis C virus mediated changes in miRNA-449a modulates inflammatory biomarker YKL40 through components of the NOTCH signaling pathway. PLoS One. 2012;7:e50826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 126. | Diaz G, Melis M, Tice A, Kleiner DE, Mishra L, Zamboni F, Farci P. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer. 2013;133:816-824. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 127. | Ogawa T, Enomoto M, Fujii H, Sekiya Y, Yoshizato K, Ikeda K, Kawada N. MicroRNA-221/222 upregulation indicates the activation of stellate cells and the progression of liver fibrosis. Gut. 2012;61:1600-1609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 191] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 128. | Ramachandran S, Ilias Basha H, Sarma NJ, Lin Y, Crippin JS, Chapman WC, Mohanakumar T. Hepatitis C virus induced miR200c down modulates FAP-1, a negative regulator of Src signaling and promotes hepatic fibrosis. PLoS One. 2013;8:e70744. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (1)] |
| 129. | Sarma NJ, Tiriveedhi V, Crippin JS, Chapman WC, Mohanakumar T. Hepatitis C virus-induced changes in microRNA 107 (miRNA-107) and miRNA-449a modulate CCL2 by targeting the interleukin-6 receptor complex in hepatitis. J Virol. 2014;88:3733-3743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 130. | Peveling-Oberhag J, Crisman G, Schmidt A, Döring C, Lucioni M, Arcaini L, Rattotti S, Hartmann S, Piiper A, Hofmann WP. Dysregulation of global microRNA expression in splenic marginal zone lymphoma and influence of chronic hepatitis C virus infection. Leukemia. 2012;26:1654-1662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 131. | Joshi D, Salehi S, Brereton H, Arno M, Quaglia A, Heaton N, O’Grady J, Agarwal K, Aluvihare V. Distinct microRNA profiles are associated with the severity of hepatitis C virus recurrence and acute cellular rejection after liver transplantation. Liver Transpl. 2013;19:383-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 132. | Gelley F, Zadori G, Nemes B, Fassan M, Lendvai G, Sarvary E, Doros A, Gerlei Z, Nagy P, Schaff Z. MicroRNA profile before and after antiviral therapy in liver transplant recipients for hepatitis C virus cirrhosis. J Gastroenterol Hepatol. 2014;29:121-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 133. | Bihrer V, Friedrich-Rust M, Kronenberger B, Forestier N, Haupenthal J, Shi Y, Peveling-Oberhag J, Radeke HH, Sarrazin C, Herrmann E. Serum miR-122 as a biomarker of necroinflammation in patients with chronic hepatitis C virus infection. Am J Gastroenterol. 2011;106:1663-1669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 134. | Trebicka J, Anadol E, Elfimova N, Strack I, Roggendorf M, Viazov S, Wedemeyer I, Drebber U, Rockstroh J, Sauerbruch T. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J Hepatol. 2013;58:234-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 138] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 135. | Cermelli S, Ruggieri A, Marrero JA, Ioannou GN, Beretta L. Circulating microRNAs in patients with chronic hepatitis C and non-alcoholic fatty liver disease. PLoS One. 2011;6:e23937. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 423] [Cited by in RCA: 466] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 136. | Bihrer V, Waidmann O, Friedrich-Rust M, Forestier N, Susser S, Haupenthal J, Welker M, Shi Y, Peveling-Oberhag J, Polta A. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One. 2011;6:e26971. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 137. | Shrivastava S, Petrone J, Steele R, Lauer GM, Di Bisceglie AM, Ray RB. Up-regulation of circulating miR-20a is correlated with hepatitis C virus-mediated liver disease progression. Hepatology. 2013;58:863-871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 89] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 138. | Shwetha S, Gouthamchandra K, Chandra M, Ravishankar B, Khaja MN, Das S. Circulating miRNA profile in HCV infected serum: novel insight into pathogenesis. Sci Rep. 2013;3:1555. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 139. | Murakami Y, Toyoda H, Tanahashi T, Tanaka J, Kumada T, Yoshioka Y, Kosaka N, Ochiya T, Taguchi YH. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS One. 2012;7:e48366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 140. | Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, Szabo G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 130] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 141. | Scagnolari C, Zingariello P, Vecchiet J, Selvaggi C, Racciatti D, Taliani G, Riva E, Pizzigallo E, Antonelli G. Differential expression of interferon-induced microRNAs in patients with chronic hepatitis C virus infection treated with pegylated interferon alpha. Virol J. 2010;7:311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 142. | Fognani E, Giannini C, Piluso A, Gragnani L, Monti M, Caini P, Ranieri J, Urraro T, Triboli E, Laffi G. Role of microRNA profile modifications in hepatitis C virus-related mixed cryoglobulinemia. PLoS One. 2013;8:e62965. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 143. | Gupta P, Liu B, Wu JQ, Soriano V, Vispo E, Carroll AP, Goldie BJ, Cairns MJ, Saksena NK. Genome-wide mRNA and miRNA analysis of peripheral blood mononuclear cells (PBMC) reveals different miRNAs regulating HIV/HCV co-infection. Virology. 2014;450-451:336-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 144. | Abdalla MA, Haj-Ahmad Y. Promising Candidate Urinary MicroRNA Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients. J Cancer. 2012;3:19-31. [PubMed] |
| 145. | Zhu HT, Dong QZ, Sheng YY, Wei JW, Wang G, Zhou HJ, Ren N, Jia HL, Ye QH, Qin LX. MicroRNA-29a-5p is a novel predictor for early recurrence of hepatitis B virus-related hepatocellular carcinoma after surgical resection. PLoS One. 2012;7:e52393. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 146. | Murakami Y, Tamori A, Itami S, Tanahashi T, Toyoda H, Tanaka M, Wu W, Brojigin N, Kaneoka Y, Maeda A. The expression level of miR-18b in hepatocellular carcinoma is associated with the grade of malignancy and prognosis. BMC Cancer. 2013;13:99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 147. | Gehrau RC, Mas VR, Villamil FG, Dumur CI, Mehta NK, Suh JL, Maluf DG. MicroRNA signature at the time of clinical HCV recurrence associates with aggressive fibrosis progression post-liver transplantation. Am J Transplant. 2013;13:729-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 148. | Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135-1142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 470] [Cited by in RCA: 509] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 149. | Fornari F, Gramantieri L, Giovannini C, Veronese A, Ferracin M, Sabbioni S, Calin GA, Grazi GL, Croce CM, Tavolari S. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761-5767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 321] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 150. | Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015-32027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 413] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 151. | Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, Liu J, Cui Y, Bian X, Bie P. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011;310:160-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 104] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 152. | Yang F, Zhang L, Wang F, Wang Y, Huo XS, Yin YX, Wang YQ, Zhang L, Sun SH. Modulation of the unfolded protein response is the core of microRNA-122-involved sensitivity to chemotherapy in hepatocellular carcinoma. Neoplasia. 2011;13:590-600. [PubMed] |
| 153. | Zhang J, Wang Y, Zhen P, Luo X, Zhang C, Zhou L, Lu Y, Yang Y, Zhang W, Wan J. Genome-wide analysis of miRNA signature differentially expressed in doxorubicin-resistant and parental human hepatocellular carcinoma cell lines. PLoS One. 2013;8:e54111. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |