Published online Jul 7, 2014. doi: 10.3748/wjg.v20.i25.8229
Revised: March 9, 2014
Accepted: April 15, 2014
Published online: July 7, 2014
Processing time: 290 Days and 4 Hours
AIM: To investigate the effect of Girdin knockdown on the chemosensitivity of colorectal cancer cells to oxaliplatin and the possible mechanisms involved.
METHODS: Four siRNAs targeting Girdin were transfected into the chemoresistant colorectal cancer cell line DLD1. Real-time polymerase chain reaction (PCR) was employed to assess Girdin mRNA expression and the most effective siRNA was chosen for conversion into shRNA. Then, DLD1 cells were infected with lentiviruses expressing the Girdin shRNA and a scramble control, respectively, and Girdin mRNA and protein expression levels were assessed by real-time PCR and Western blotting. Furthermore, microarray experiments were used to assess global gene expression profile after Girdin suppression in DLD1 cells. Finally, the cytotoxic effect of simultaneous treatment with oxaliplatin and adriamycin (an inhibitor of a significantly downregulated gene after Girdin suppression in DLD1 cells) was examined by MTT assay.
RESULTS: The most effective siRNA suppressed Girdin expression with an inhibition efficiency of 57%. Compared with the scramble control, DLD1 cells infected with the Girdin shRNA displayed decreased Girdin mRNA and protein levels (P < 0.05), and Girdin knockdown significantly enhanced chemosensitivity to oxaliplatin in colorectal cancer cells (P < 0.05). Microarray data revealed that 381 and 162 genes were upregulated and downregulated in response to Girdin reduction, respectively, with ratios > 1.2 or < 0.8 (P < 0.01). Interestingly, TOP2B (DNA topoisomerase 2-β) was downregulated (ratio = 0.78, P = 0.0001) and oxaliplatin/adriamycin combination resulted in increased cell death compared with treatments with individual agents (P < 0.05).
CONCLUSION: Girdin knockdown enhances chemosensitivity of colorectal cancer cells to oxaliplatin via TOP2B down-regulation. These findings provide a promising approach to overcome the chemoresistance of colorectal cancer cells.
Core tip: The chemoresistance to oxaliplatin is a major problem in the treatment of colorectal cancer (CRC). In this study, a lentivirus-mediated shRNA was constructed to investigate the role of Girdin, a potential regulator of chemotherapy sensitivity. We found that Girdin knockdown enhanced chemosensitivity of colorectal cells to oxaliplatin via reduction of TOP2B. This suggested that Girdin is a modulator of CRC chemoresistance and a potential therapeutic target.
- Citation: Zhang YJ, Li AJ, Han Y, Yin L, Lin MB. Inhibition of Girdin enhances chemosensitivity of colorectal cancer cells to oxaliplatin. World J Gastroenterol 2014; 20(25): 8229-8236
- URL: https://www.wjgnet.com/1007-9327/full/v20/i25/8229.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i25.8229
Colorectal cancer (CRC) is currently the third most common cancer worldwide and the fourth cause of cancer-related death[1]. Administration of oxaliplatin-based regimens is considered the first-line chemotherapy in the adjuvant and palliative settings for CRC patients[2-4]. However, treatment efficacy has reached a plateau with a response rate of 40%[5], which highlights the need for novel strategies to enhance the chemosensitivity to oxaliplatin in CRC.
Accumulating studies have demonstrated that the phosphatidylinositol 3-kinase (PI3K)/Akt pathway plays a crucial role in chemotherapy resistance[6]. This pathway affects chemotherapy-induced apoptosis in various cancers, including lung, ovarian, breast, liver and pancreatic tumors[7-11]. In CRC, activation of the PI3K/Akt pathway has been shown to potently affect oxaliplatin resistance[12]. Girdin (also called coiled-coil domain-containing protein 88A), a multidomain molecule, plays important roles in diverse biological processes, e.g., wound healing, tumor cell motility, and angiogenesis[13]. Recent data described Girdin as a novel nonreceptor guanine nucleotide exchange factor (GEF) for Gai proteins that enhances PI3K-Akt signals by activating Gαi1, 2, 3[14]. Moreover, Matsushita proposed that Girdin family proteins play an essential role in apoptosis via activation of the IRE1-JNK pathway[15]. Based on these findings, it is conceivable that Girdin may constitute a potential regulator of chemotherapy sensitivity, which prompted us to investigate the effect and possible mechanisms of Girdin knockdown on the chemosensitivity of CRC cells to oxaliplatin.
To date, previous studies have focused on the role of Girdin in the progression and metastasis of CRC cells[16,17]; however, studies assessing the contribution of Girdin to chemoresistance in CRC are inexistent. Herein, we report for the first time that Girdin knockdown enhances chemosensitivity of CRC cells to oxaliplatin via reduction of TOP2B.
The 17 cell lines (CACO-2, D2, DLD1, HCT15, HCT116, HUTU80, SW48, SW480, SW620, SW837, CX-1, COLO205, GP2D, GP5D, HCT15, LS174T and LS180) were obtained from American Type Culture Collection (ATCC, United States) and Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences. All cell lines were maintained according to ATCC protocols. 293FT cells were purchased from Invitrogen (United States) and maintained according to the manufacturer’s specifications in DMEM supplemented with 10% FBS, 0.1 mmol/L minimum essential medium (MEM) nonessential amino acids, 2 mmol L-glutamine, 1% penicillin/streptomycin and 500 μg/mL geneticin (Invitrogen). Sodium butyrate and polyethylene glycol (PEG)-8000 were obtained from Sigma-Aldrich (United States).
Four target sequences were chosen (Table 1) from the human Girdin gene (GenBank, Gene ID: 55704) to design siRNA online (WI siRNA selection program, http://sirna.wi.mit.edu/), and a scramble siRNA that does not match any known mammalian GenBank sequence was designed using the Invivogen scramble siRNA online program (Invivogen, United States). All oligonucleotides were synthesized by Shanghai GenePharma Co., Ltd (China) (Table 1).
| siRNA | Synthetic siRNA sequence |
| siRNA1 | Sense: CCAGAAUGUACCGAGAUGAUU |
| Antisense: UCAUCUCGGUACAUUCUGGUU | |
| siRNA2 | Sense: CUUCAUUAGUUCUGCGGGAUU |
| Antisense: UCCCGCAGAACUAAUGAAGUU | |
| siRNA3 | Sense: GGACCAACCUUGAUGAAUAUU |
| Antisense: UAUUCAUCAAGGUUGGUCCUU | |
| siRNA4 | Sense: CAAGAGUUGAGGAAUUAAAUU |
| Antisense: UUUAAUUCCUCAACUCUUGUU | |
| scramble siRNA | Sense: GAGGCGAAACGCTAAATATCT |
| Antisense: AGATATTTAGCGTTTCGCCTC |
Oxaliplatin was purchased from Sigma (United States) and stock solutions of 5 mg/ml were prepared in DMSO. Aliquots of oxaliplatin were stored at -20 °C until use. Chemosensitivity of 17 CRC cell lines to oxaliplatin was examined using MTT assay. Briefly, cells were harvested in the exponential growth phase, seeded in 96-well plates (3000 cells/well), and incubated overnight. Then, oxaliplatin was added at various concentrations (0, 3, 10, 30 μmol). At the end of treatment period, 10 μL of MTT was added for 4 h and the media gently aspirated. DMSO was used to dissolve the purple crystals and absorbance was determined at 490 nm using a spectrophotometer (Thermo, United States). The inhibition rate (IR) was derived as 1-(A490experimental group-A490blank)/(A490control group-A490blank) × 100%.
DLD1 cells were seeded in 24-well plates and incubated for 24 h before transfection. DLD1 cells at 30% confluence were transfected with 50 nmol/L (final concentration) siRNA per well using Lipofectamine 2000 and Opti-MEM (Invitrogen) media according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were harvested for further analysis.
Total RNA was isolated using the TRIzol reagent (Invitrogen), and 1 μg total RNA was reverse transcribed in 50 μL reaction to yield cDNA, which was stored at -20 °C until use. Girdin mRNA level was assessed by real-time quantitative polymerase chain reaction (PCR) with β-actin used as an internal control. The primer sequences are as follows: β-actin (forward: AGCGAGCATCCCCCAAAGTT; reverse: GGGCACGAGGGCTCATCATT), Girdin (forward: CCAGGCATGAAGCGAACA; reverse: CGAGCATCCGAAAGCAAAT) and TOP2B (forward: AAGAAGAGTCAGAAACTGCCAACA; reverse: ACCCACATGAACTGCGTCAA). Quantitative RT-PCR was carried out using the SYBR Green kit (Qiagen, United States) in a final volume of 25 μL. Each PCR cycle included 15 s of denaturation at 95 °C, 20 s of primer annealing at 58 °C and 20 s of extension at 72 °C.
Based on the interference efficiency of the four siRNAs, the most effective sequence in knocking down Girdin was chosen and converted into shRNA with an online tool (http://www.broadinstitute.org/rnai/public/). Both forward oligonucleotide (CCGGAGGCAAGAGTTGAGGAATTAACTCGAGTTAATTCCTCAACTCTTGCCTTTTTTG) and reverse olignucleotide (AATTCAAAAAAGGCAAGAGTTGAGGAATTAACTCGAGTTAATTCCTCAACTCTTGCCT) were synthesized, annealed and cloned between the BamHI and EcoRI sites of the lentiviral shRNA expression vector LvUCTP derived from FugW (Addgene, United States). The U6 promoter was added for shRNA expression and tdTomato gene for indication of transfection and infection. A scramble shRNA was derived from the scramble siRNA (forward oligonucleotide: CCGGGAGGCGAAACGCTAAATATCTCTCGAGAGATATTTAGCGTTTCGCCTCTTTTTG; reverse oligonucleotide: AATTCAAAAAGAGGCGAAACGCTAA ATATCTCTCGAGAGATATTTAGCGTTTCGCCTC). Lentivirus production was performed using the ViraPower™ Lentiviral Packaging Mix (Invitrogen) following the manufacturer’s instructions. In brief, the lentiviral expression vector was cotransfected with an optimized mixture of three packaging plasmids (pLP1,pLP2 and pLP/VSVG) into 293FT cells. Cell supernatants containing viral particles were harvested 48 h after transfection, filtered through a 0.45 μm filter (Millipore), and stored at -70 °C. Lentivirus preparations containing only the scramble shRNA were used as negative controls.
Total protein was extracted from cells using RIPA buffer (Pierce, United States) and quantified using BCA protein assay kit according to the manufacturer’s instructions (Pierce, United States). Totally 10 μg protein was loaded onto SDS-PAGE gels for electrophoresis. After transfer, membranes were blocked with TBST containing 5% nonfat milk for 0.5 h at room temperature, and incubated with primary polyclonal antibodies raised in rabbits against Girdin, TOP2B and β-tublin (Santa Cruz, United States). Signals were detected after incubation in HRP-conjugated secondary antibodies and addition of ECL substrate kit (Thermo, United States). β-tublin acted as the reference protein for loading control.
DLD1 cells were infected with the recombinant lentivirus for 90 min, incubated for an additional 24 h, and seeded in 96-well plates at 4000 cells/well. Seventy-two hours after infection, cells were treated with oxaliplatin at the indicated concentrations for 72 h and analyzed by MTT assay. Analysis of combinational inhibition of oxaliplatin and adriamycin was conducted at indicated concentrations and cell viability was assessed 72 h after treatment.
The mRNA extracted from lentivirus mediated Girdin shRNA (Lv-Girdin shRNA) infected DLD1 cells was reverse transcribed into cDNA using Cy5-dUTP labeling, while the mRNA from the Lv-scramble shRNA infected DLD1 cells was processed with Cy3-dUTP labeling, following the manufacturer’s protocols. The labeled probes were then hybridized to the cDNA microarray chips from Affimetrix (United States). Ratios of Cy5: Cy3 greater than 1.2 or less than 0.8 were chosen for further analysis.
Values obtained from real-time PCR and MTT assay and IC50 are expressed as mean ± SD. Statistical analysis was carried out by one-way ANOVA using the statistical package SPSS 19.0 (Chicago, IL, United States). Differences were considered statistically significant at P < 0.05. For microarray analysis, P < 0.01 was considered statistically significant.
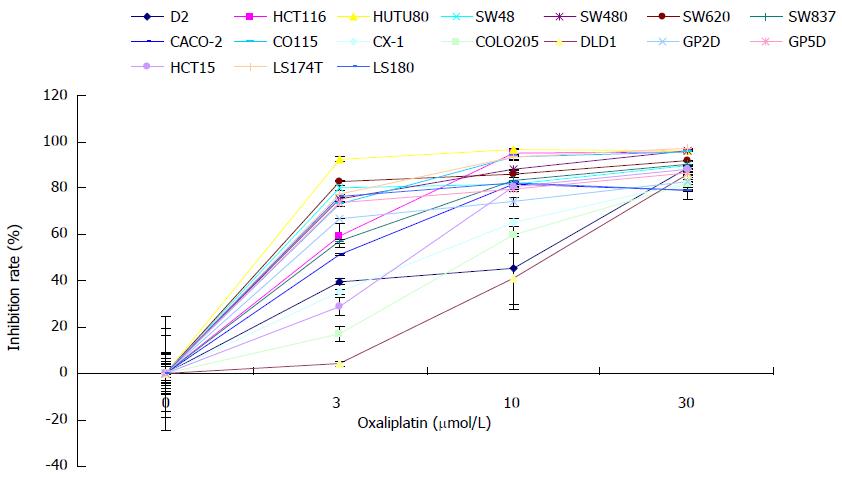
To explore the mechanisms by which CRC cells resist cytotoxicity of oxaliplatin, it is of prime importance to identify at least one cell line resistant to the drug. We examined the chemosensitivity of 17 CRC cell lines to oxaliplatin at various concentrations as previously described[18]. All CRC cells were inhibited by oxaliplatin in a dose-dependent manner (Figure 1). Of the 17 cell lines, DLD1 cells displayed the highest resistance to oxaliplatin at various concentrations. Based on these findings, DLD1 was chosen to further assess oxaliplatin resistance.
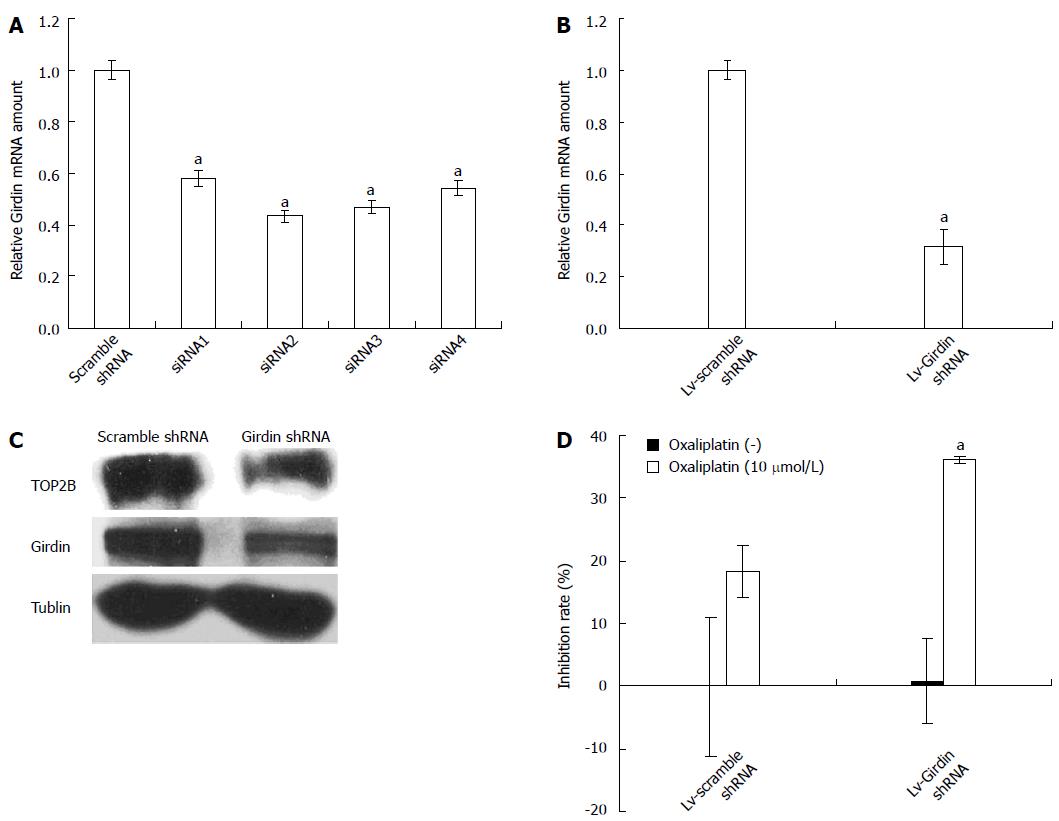
We then examined cell viability of Girdin knockdown DLD1 cells after treatment with oxaliplatin to determine the effect of Girdin on resistance. Four pairs of siRNAs targeting different sites of Girdin mRNA were tested for knockdown efficacy. Real-time PCR was performed to evaluate Girdin mRNA levels after siRNA transfections. The relative Girdin mRNA levels in DLD1 cells were 58% ± 3%, 43% ± 2%, 47% ± 3% and 54% ± 3% for siRNA1, siRNA2, siRNA3 and siRNA4, respectively, compared with the scramble siRNA (P < 0.05) (Figure 2A), indicating that siRNA2 was the most effective sequence.
To enhance the transfection efficiency and maintain the knockdown efficacy for a relatively long time, siRNA2 was converted to shRNA sequence for the production of lentivirus mediated shRNA. Real-time PCR and Western blotting assays were performed to evaluate Girdin mRNA and protein expression after transfection. As shown in Figure 2B, Girdin relative mRNA levels in the Lv-Girdin shRNA group was 32% ± 7% compared to the scramble control (P < 0.05). In agreement, Western blotting data demonstrated that the Girdin protein was less expressed in the Lv-Girdin shRNA group compared with the scramble control (Figure 2C). These findings suggested that lentivirus-mediated RNAi effectively silenced Girdin expression in transfected cells.
Next, DLD1 cells were infected with the Lv-Girdin shRNA and scramble control for 72 h and exposed to 10 μmol/L oxaliplatin. We chose the concentration of 10 μM for the reason that single treatment with Girdin knockdown or with oxaliplatin less than 10 μmol/L did not show detectable inhibition effect on DLD1. As shown in Figure 2D, Lv-Girdin shRNA transfected DLD1 cells showed increased chemosensitivity to oxaliplatin (P < 0.05), compared with those transfected with the scramble control, indicating that Girdin silencing enhanced DLD1 cells sensitivity.
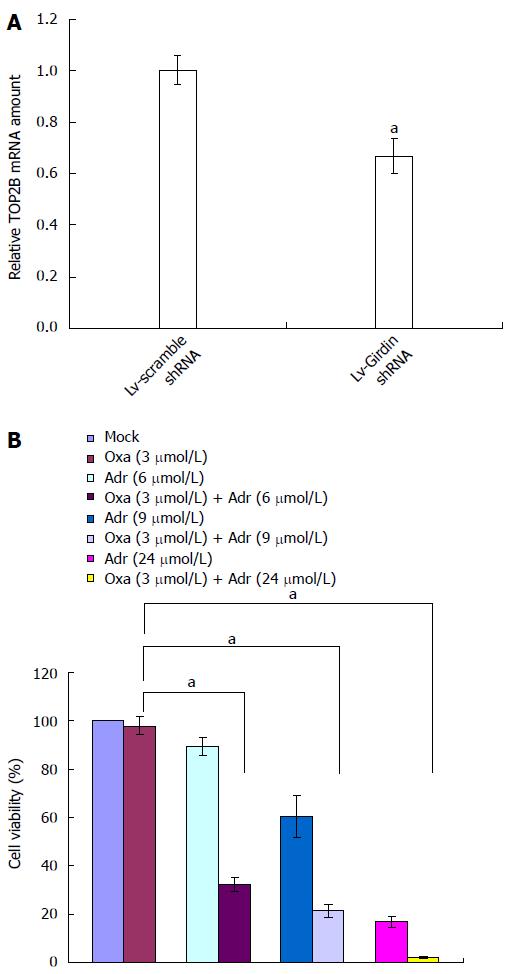
To identify the downstream genes of Girdin responsible for oxaliplatin resistance, we performed a global analysis of gene expression after Girdin silencing. In comparison to the scramble control, 162 and 381 genes were downregulated and upregulated in response to Girdin reduction, respectively, with ratios > 1.2 or < 0.8 (P < 0.01). The top 10 differentially expressed genes are listed in Table 2. Of note, TOP2B appeared in the list as downregulated with the lowest P value, indicating that Girdin reduction resulted in TOP2B loss and enhanced sensitivity to oxaliplatin. TOP2B downregulation after Girdin silencing was confirmed by real-time PCR and protein levels were also reduced as shown by Western blotting (Figure 3A and 2C).
| Gene name | P value | Ratio |
| Downregulated | ||
| TOP2B | 0.000148 | 0.780296 |
| EIF3E | 0.000154 | 0.794333 |
| SCARNA9 | 0.000166 | 0.768316 |
| SEMA3A | 0.000227 | 0.781830 |
| IKZF2 | 0.000296 | 0.712167 |
| KIT | 0.000311 | 0.792173 |
| MIR34A | 0.000358 | 0.783264 |
| LRRC37A4 | 0.000366 | 0.661987 |
| LRRC37A4 | 0.000391 | 0.66294 |
| ARHGAP18 | 0.000400 | 0.756169 |
| Upregulated | ||
| PPP1R15A | 0.000114 | 1.504745 |
| TAP2 | 0.000117 | 1.217997 |
| FOSL2 | 0.000151 | 1.228042 |
| KRTAP2-1 | 0.000170 | 1.588156 |
| TNFRSF9 | 0.000170 | 5.843542 |
| BCL3 | 0.000185 | 1.369511 |
| LCN2 | 0.000195 | 5.641935 |
| CYB561D2 | 0.000196 | 1.203703 |
| SERPINE2 | 0.000204 | 1.247110 |
| RAB15 | 0.000205 | 1.353464 |
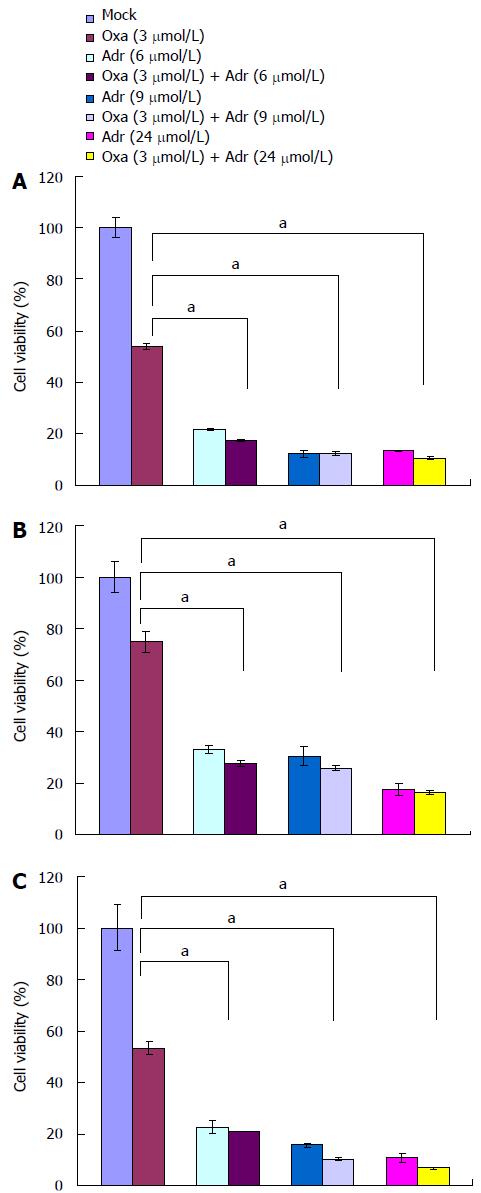
Since the action of TOP2B leads to a reversible double strand breakage (DSB) in DNA, it is used as a drug target to generate DNA damage for cancer therapy[19]. To confirm TOP2B as a sensitizing target for oxaliplatin, we examined the effect of adriamycin, a TOP2B inhibitor, on the sensitivity of DLD1 cells to oxaliplatin. As addition of adriamycin only showed an inhibitory effect on DLD1, we chose 3 μmol oxaliplatin to determine the effect of their combinational treatment. Treatment of DLD1 cells with a combination of 3 umol oxaliplatin and varying concentrations of adriamycin resulted in a shift of the dose response curve and enhanced cytotoxicity compared with single agents (Figure 3B). Similar results were also observed in other three CRC cell lines (Figure 4)
Oxaliplatin-based chemotherapy is widely used in CRC adjuvant and metastatic settings[2-4]. However, typical tumor resistance processes are the main reasons for treatment failure[20]. Thus, it is of paramount importance to search for novel strategies to enhance the chemosensitivity of CRC cells to oxaliplatin.
Induction of apoptosis is the principal mechanism by which the majority of chemotherapeutic agents exert their effects. Consequently, failure to undergo apoptosis is the likely mechanism mediating drug resistance in tumors[21]. Girdin, a nonreceptor GEF, can interact with Gαi resulting in Gβγ release and subsequently activates PI3K/AKT signaling events, which have been recognized as an important pathway for chemoresistance through its antiapoptotic effects in cancers[22]. Moreover, Girdin family proteins were shown to suppress apoptosis[15]. These findings raised the possibility that Girdin might be involved in chemoresistance of CRC cells to oxaliplatin.
In this study, lentivirus-mediated shRNA was constructed to investigate the role of Girdin in chemoresistance of CRC cells to oxaliplatin. After successful transfection with a lentivirus-mediated Girdin shRNA, Girdin mRNA and protein levels were virtually reduced and the downregulation of Girdin significantly increased chemosensitivity of CRC cells to oxaliplatin. These findings suggested that Girdin is a modulator of CRC cell chemoresistance, and therefore a potential therapeutic target. To our knowledge, this is the first study that demonstrates the ability of Girdin to enhance the chemoresistance of CRC cells to oxaliplatin.
To identify the underlying mechanisms by which Girdin contributes to the chemoresistance of CRC cells, we performed a global analysis of gene expression after Girdin silencing by microarrays. We found that TOP2B appeared in the list of downregulated genes, with the lowest P value. TOP2B belongs to the TOP2 family which possesses both double strand cleavage and DNA ligation activities involved in DNA replication and DNA repair[19,23,24]. TOP2B is capable of uncoiling DNA and thus introduces a reversible DSB in DNA molecules. The DSB is stabilized by intercalative agents such as adriamycin which is a TOP2B inhibitor leading to apoptosis in a Fas-dependent or independent manner[25,26]. The microarray data described herein showed that TOP2B expression decreased upon Girdin downregulation. In addition, the combination of adriamycin and oxaliplatin greatly enhanced the cancer cell killing effect compared with treatment with oxaliplatin alone. These results demonstrate that TOP2B reduction is involved in the enhancement of chemosensitivity to oxaliplatin by Girdin suppression, and oxaliplatin/adriamycin combination resulted in increased cell death compared with treatments with individual agents.
In conclusion, Girdin plays a significant role in chemoresistance of CRC cells. Indeed, lentivirus-mediated shRNA targeting Girdin efficiently inhibits Girdin expression and enhances chemosensitivity to oxaliplatin in CRC. These findings provide a strong evidence for the use of Girdin knockdown in combination with oxaliplatin as a novel therapeutic strategy for CRC.
Oxaliplatin-based chemotherapy is widely used in the adjuvant and metastatic setting of colorectal cancer (CRC). However, treatment efficacy has reached a plateau with a response rate of 40%, which highlights the need for novel strategies to enhance the chemosensitivity of CRC cells to oxaliplatin.
Girdin, a nonreceptor guanine nucleotide exchange factor (GEF), can interact with Gαi, resulting in Gβγ release and subsequent PI3K/AKT activation which has been recognized as an important pathway for chemoresistance through its antiapoptotic effects in cancers. Moreover, Girdin family proteins were found to suppress apoptosis. All of these raised the possibility that Girdin was involved in chemoresistance of CRC cells to oxaliplatin.
Previous studies have focused on the role of Girdin in the progression and metastasis of CRC cells. However, studies assessing the contribution of Girdin to chemoresistance in CRC are inexistent. Herein, the authors report for the first time that Girdin knockdown enhances chemosensitivity of CRC cells to oxaliplatin via reduction of TOP2B.
Lentivirus-mediated shRNA targeting Girdin efficiently inhibits Girdin expression and enhances chemosensitivity of CRC cells to oxaliplatin. These findings provide a strong evidence for the use of Girdin suppression in combination with oxaliplatin as a novel therapeutic strategy for CRC.
Girdin, also called coiled-coil domain-containing protein 88A, is a nonreceptor guanine nucleotide exchange factor that can interact with Gαi resulting in Gβγ release and subsequently activates PI3K/AKT signaling pathway, which has been recognized as an important pathway for chemoresistance through its antiapoptotic effects in cancers. Topoisomerase 2-beta belongs to the TOP2 family which possesses both double strand cleavage and DNA ligation activities involved in DNA replication and DNA repair.
This article is of interest as resistance to oxaliplatin is a major problem in the treatment of colorectal cancer. The study is well designed and properly developed.
P- Reviewers: Chan EC, Feliu J, Mo H S- Editor: Qi Y L- Editor: Wang TQ E- Editor: Zhang DN
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. [PubMed] |
| 2. | André T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, Topham C, Zaninelli M, Clingan P, Bridgewater J. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350:2343-2351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2653] [Cited by in RCA: 2732] [Article Influence: 130.1] [Reference Citation Analysis (0)] |
| 3. | Rothenberg ML, Oza AM, Bigelow RH, Berlin JD, Marshall JL, Ramanathan RK, Hart LL, Gupta S, Garay CA, Burger BG. Superiority of oxaliplatin and fluorouracil-leucovorin compared with either therapy alone in patients with progressive colorectal cancer after irinotecan and fluorouracil-leucovorin: interim results of a phase III trial. J Clin Oncol. 2003;21:2059-2069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 504] [Cited by in RCA: 494] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 4. | de Gramont A, Buyse M, Abrahantes JC, Burzykowski T, Quinaux E, Cervantes A, Figer A, Lledo G, Flesch M, Mineur L. Reintroduction of oxaliplatin is associated with improved survival in advanced colorectal cancer. J Clin Oncol. 2007;25:3224-3229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 105] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 5. | Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3435] [Cited by in RCA: 3686] [Article Influence: 204.8] [Reference Citation Analysis (0)] |
| 6. | Huang WC, Hung MC. Induction of Akt activity by chemotherapy confers acquired resistance. J Formos Med Assoc. 2009;108:180-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 98] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 7. | Wang G, Reed E, Li QQ. Molecular basis of cellular response to cisplatin chemotherapy in non-small cell lung cancer (Review). Oncol Rep. 2004;12:955-965. [PubMed] |
| 8. | Ali AY, Farrand L, Kim JY, Byun S, Suh JY, Lee HJ, Tsang BK. Molecular determinants of ovarian cancer chemoresistance: new insights into an old conundrum. Ann N Y Acad Sci. 2012;1271:58-67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 9. | Johnson-Holiday C, Singh R, Johnson EL, Grizzle WE, Lillard JW, Singh S. CCR9-CCL25 interactions promote cisplatin resistance in breast cancer cell through Akt activation in a PI3K-dependent and FAK-independent fashion. World J Surg Oncol. 2011;9:46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 10. | Yoon H, Min JK, Lee JW, Kim DG, Hong HJ. Acquisition of chemoresistance in intrahepatic cholangiocarcinoma cells by activation of AKT and extracellular signal-regulated kinase (ERK)1/2. Biochem Biophys Res Commun. 2011;405:333-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 71] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 11. | Ripka S, Neesse A, Riedel J, Bug E, Aigner A, Poulsom R, Fulda S, Neoptolemos J, Greenhalf W, Barth P. CUX1: target of Akt signalling and mediator of resistance to apoptosis in pancreatic cancer. Gut. 2010;59:1101-1110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 12. | Hsu CP, Kao TY, Chang WL, Nieh S, Wang HL, Chung YC. Clinical significance of tumor suppressor PTEN in colorectal carcinoma. Eur J Surg Oncol. 2011;37:140-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 56] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 13. | Dunkel Y, Ong A, Notani D, Mittal Y, Lam M, Mi X, Ghosh P. STAT3 protein up-regulates Gα-interacting vesicle-associated protein (GIV)/Girdin expression, and GIV enhances STAT3 activation in a positive feedback loop during wound healing and tumor invasion/metastasis. J Biol Chem. 2012;287:41667-41683. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Garcia-Marcos M, Kietrsunthorn PS, Pavlova Y, Adia MA, Ghosh P, Farquhar MG. Functional characterization of the guanine nucleotide exchange factor (GEF) motif of GIV protein reveals a threshold effect in signaling. Proc Natl Acad Sci USA. 2012;109:1961-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 15. | Matsushita E, Asai N, Enomoto A, Kawamoto Y, Kato T, Mii S, Maeda K, Shibata R, Hattori S, Hagikura M. Protective role of Gipie, a Girdin family protein, in endoplasmic reticulum stress responses in endothelial cells. Mol Biol Cell. 2011;22:736-747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Jun BY, Kim SW, Jung CK, Cho YK, Lee IS, Choi MG, Choi KY, Oh ST. Expression of girdin in human colorectal cancer and its association with tumor progression. Dis Colon Rectum. 2013;56:51-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Mao JZ, Jiang P, Cui SP, Ren YL, Zhao J, Yin XH, Enomoto A, Liu HJ, Hou L, Takahashi M. Girdin locates in centrosome and midbody and plays an important role in cell division. Cancer Sci. 2012;103:1780-1787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 18. | Ehrsson H, Wallin I, Yachnin J. Pharmacokinetics of oxaliplatin in humans. Med Oncol. 2002;19:261-265. [PubMed] |
| 19. | Zhang A, Lyu YL, Lin CP, Zhou N, Azarova AM, Wood LM, Liu LF. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J Biol Chem. 2006;281:35997-36003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 114] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 20. | Rabik CA, Dolan ME. Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev. 2007;33:9-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1096] [Cited by in RCA: 1205] [Article Influence: 63.4] [Reference Citation Analysis (0)] |
| 21. | Lowe SW, Lin AW. Apoptosis in cancer. Carcinogenesis. 2000;21:485-495. [PubMed] |
| 22. | Garcia-Marcos M, Ghosh P, Farquhar MG. GIV is a nonreceptor GEF for G alpha i with a unique motif that regulates Akt signaling. Proc Natl Acad Sci USA. 2009;106:3178-3183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 158] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 23. | Ali-Osman F, Berger MS, Rajagopal S, Spence A, Livingston RB. Topoisomerase II inhibition and altered kinetics of formation and repair of nitrosourea and cisplatin-induced DNA interstrand cross-links and cytotoxicity in human glioblastoma cells. Cancer Res. 1993;53:5663-5668. [PubMed] |
| 24. | Gupta KP, Swain U, Rao KS, Kondapi AK. Topoisomerase IIβ regulates base excision repair capacity of neurons. Mech Ageing Dev. 2012;133:203-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 25. | Friesen C, Fulda S, Debatin KM. Induction of CD95 ligand and apoptosis by doxorubicin is modulated by the redox state in chemosensitive- and drug-resistant tumor cells. Cell Death Differ. 1999;6:471-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 58] [Article Influence: 2.2] [Reference Citation Analysis (0)] |