Published online May 21, 2014. doi: 10.3748/wjg.v20.i19.5700
Revised: November 4, 2013
Accepted: January 3, 2014
Published online: May 21, 2014
Processing time: 261 Days and 21.2 Hours
miRNAs are endogenous 19- to 25-nt noncoding RNAs that can negatively regulate gene expression by directly cleaving target mRNA or by inhibiting its translation. Recent studies have revealed that miRNA plays a significant role in gastric cancer development either as a tumor suppressor gene or oncogene. miRNA-single-nucleotide polymorphisms (SNPs), as a novel class of functional SNPs/polymorphisms, have been identified as candidate biomarkers for gastric cancer susceptibility. On the basis of recent data, the present review summarizes current knowledge of the functional effects of miRNA-SNPs and their importance as candidate gastric cancer biomarkers. Additionally, this review also includes a meta-analysis of the most frequently studied miRNA-SNPs in gastric cancer.
Core tip: Host genetic susceptibility plays an important role in gastric carcinogenesis. In this review, we summarize current knowledge about the functional effects of miRNA-single-nucleotide polymorphisms (SNPs) and their importance in gastric cancer susceptibility. Furthermore, we also conduct a simple meta-analysis about most frequently studied miRNA-SNPs (rs11614913 in miR-196a-2, rs895819 in miR-27a and rs2910164 in miR-146a) in gastric cancer.
- Citation: Hua HB, Yan TT, Sun QM. miRNA polymorphisms and risk of gastric cancer in Asian population. World J Gastroenterol 2014; 20(19): 5700-5707
- URL: https://www.wjgnet.com/1007-9327/full/v20/i19/5700.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i19.5700
miRNAs are small endogenous noncoding RNAs of about 22 nucleotides that negatively regulate post-transcriptional gene expression by pairing with complementary binding sites located in the 3’-untranslated region (UTR) of mRNA[1]. In 1993, Wightman et al[2] and Lee et al[3] found the first miRNA, lin-4, which controls developmental timing and cell fate specification in Caenorhabditis elegans. To date, thousands of miRNA have been characterized in animal and plant genomes[4]. In human malignancies, miRNA have shown important roles in a variety of cellular processes including apoptosis, differentiation, angiogenesis and proliferation[5]. It has been suggested that miRNA regulates almost one-third of the human genes although it represents only a small part of the genome[6]. Moreover, recent evidence shows that more than half of the known human miRNAs are located within cancer-associated genomic regions or fragile sites[7]. Therefore, miRNA can be considered as a potential and ideal biomarker for cancer.
Gastric cancer remains one of the commonest malignant tumors worldwide. In 2013, it was estimated that about 21600 patients were diagnosed with gastric cancer and 10990 died of the disease in the United States[8]. In East Asia, especially in China, gastric cancer has the highest rates of incidence and mortality[9]. Epidemiological studies have identified environmental and lifestyle risk factors that contribute to the development of gastric cancer, such as Helicobacter pylori (H. pylori) infection or high salt intake[10]. However, only a minority of patients exposed to risk factors such as H. pylori infection ultimately develop gastric cancer, which indicates that host genetic susceptibility also plays an important role in gastric carcinogenesis[11-13]. In recent decades, many correlations between single nucleotide polymorphisms (SNPs) in the genome and the risk of various diseases, including gastric cancer, were reported[14,15]. Recently, a class of novel functional polymorphisms in miRNA or its binding sites is the most interesting. Our previous epidemiological studies also provided evidence that the risk of gastric cancer is associated with miRNA-SNPs[16,17]. Therefore, miRNA polymorphisms can be used as specific markers of predisposition for gastric cancer prevention.
In this review, we provide a comprehensive list of potentially functional miRNA-SNPs and have a brief description of miRNA biogenesis, biology and the underlying mechanism of miRNA-SNPs in gastric cancer susceptibility.
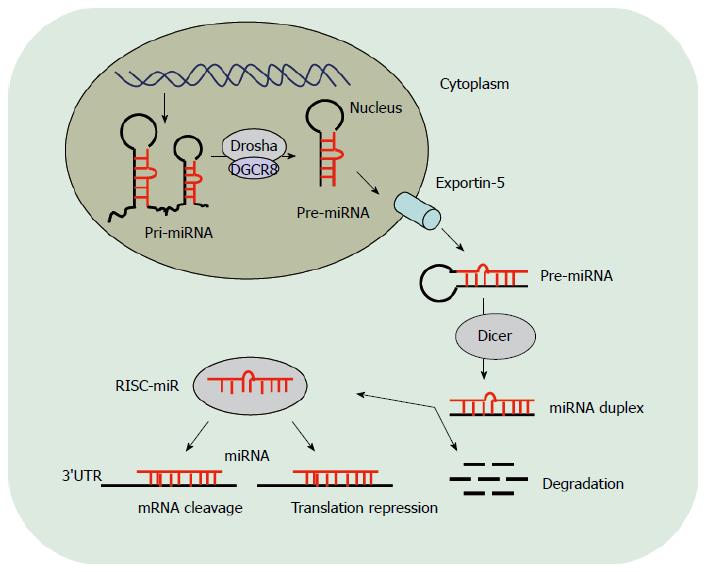
miRNA biogenesis is a multistage process[18,19]. Most human miRNA are first transcribed by RNA polymerase II and are encoded by introns[20]. The primary miRNA (pri-miRNA) is excised from the primary transcript of the intron[21]. Then, the pri-miRNA is processed by the RNase Drosha-DGCR8 complex into 70-nucleotide-long precursor hairpin structures (pre-miRNA)[22]. The pre-miRNA is subsequently exported to the cytoplasm by the nuclear membrane protein exportin-5 and cleaved by the RNase III enzyme Dicer to produce a transient miRNA duplex composed of a mature miRNA sequence (about 22 nucleotides in length) and its complementary sequence miRNA*[23]. The miRNA duplex is unwound by an RNA helicase and the mature miRNA is incorporated into the targeting miRNA containing RNA induced silencing complex (RISC) together with the argonate (Ago) protein[24,25]. The other strand (or miRNA*) is often degraded. Ago is considered as the heart of the miRNA-induced silencing complex (RISC-miR), which induce the mature miRNA to bind the complementary elements of target mRNA 3’ UTR. Lastly, the RISC-miR exerting negatively regulates gene expression function by either mRNA cleavage or inhibiting translation[26] (Figure 1).
As described above, miRNA exert the functionality by sequence-dependent regulation of post-transcriptional gene expression by targeting mRNA for cleavage or translational repression. Target selection is dependent on the extent of Watson-Crick base pairing between the miRNA and mRNA. Nucleotides 2-8 (from the 5’ end of miRNA), also referred to as the “seed sequence”, are a major determinant of mRNA target selection[27]. Mutation in either the seed or seed-complementary site could inhibit miRNA activity, which highlights the importance of seed sequence complementarity[28]. Additionally, Grimson et al[29] also reported that more than four contiguous Watson-Crick base pairs between nucleotides 12-17 at the 3’ end of the miRNA could enhance target recognition. Therefore, miRNA biogenesis and post-transcriptional regulation is highly sequence dependent, and sequence variants (such as SNPs) in either the miRNA sequence or miRNA-target site can have a significant effect on miRNA function. So far, > 1000 miRNAs have been found in humans, which are indicated to regulate up to 30% of all protein-coding genes in the human genome[30]. Moreover, Calin et al[7] have suggested that more than half of the human miRNAs are located in cancer-associated fragile regions. This shows that miRNA regulation plays a key regulatory role during development and in various cellular processes such as differentiation, growth and death. These processes are frequently dysregulated in carcinogenesis, implicating miRNA function as oncogenes or tumor suppressors.
Accumulating evidence has strongly indicated that aberrant miRNA expression is an important feature of gastric cancer[31]. In 2012, Wang et al[32] summarized 13 miRNA expression profiling studies and suggested that there were 139 miRNAs differentially expressed in gastric cancer tissues compared with neighboring noncancerous or normal gastric tissues. They also found that the expression levels of miR-21, miR-18a, miR-17 and miR-20a in gastric cancer were significantly upregulated in most studies. Conversely, miR-375 and miR-378 expression levels were downregulated in gastric cancer tissues. Also, expression levels of 29 miRNAs were reported to be inconsistent. In addition, recent evidence using quantitative real-time reverse transcriptase polymerase chain reaction (qRT-PCR) also confirmed that miR-17-5p/20a, miR-181a-5p, miR-214 and miR-23a expression levels were upregulated in human gastric cancer tissues, and overexpression of miR-17-5p/20a could promote gastric cancer cell progression and inhibit apoptosis[31,33-35]. Recent studies have indicated that gastric cancer tissue has significantly lower expression of miR-148a and miR-22 compared to non-tumor tissue, and miR-148a and miR-22 inhibit gastric cancer cell migration and invasion. Moreover, low miR-148a levels are associated with lymph node metastasis, N stage, and blood vessel invasion in gastric cancer patients[36,37]. These miRNA expression profiles analyzed by microarray or qRT-PCR could help us understand better the molecular mechanisms of tumorigenesis and contribute to the development of diagnosis and therapy for gastric cancer.
SNPs in the miRNA regulatory pathway, as a novel class of functional polymorphisms in the human genome, have been widely implicated in cancer development[38]. In 2005, Calin et al[39] reported the first evidence that mutations in miRNA genes are common and might have functional importance in chronic lymphocytic leukemia (CLL). They identified a germ-line mutation in the miR-16-1-miR-15a primary precursor, which causes low levels of miRNA expression and is associated with prognostic factors and disease progression in CLL[39]. Since then, a series of studies has used systematic sequencing or in silico approaches to identify more SNPs in miRNA. In our present study, we focused on the miRNA SNPs in gastric cancer susceptibility. Therefore, we searched PubMed to August 2013 to identify all relevant papers, using the key words miRNA, miRNA polymorphism or variant in combination with gastric cancer or gastric tumor. There were 35 epidemiological studies with a focus on the importance of miRNA-related SNPs in gastric cancer susceptibility. We scanned the titles and abstracts and excluded the studies that were clearly irrelevant to the current topic. The remaining articles were read to determine whether they contained information of interest. In addition, to be eligible, studies had to fulfill the following criteria: (1) a case-control study design or prospective study design; (2) number of subjects with each allele or genotype in cases and controls reported; (3) risk estimates OR and 95%CI, or raw data that allowed us to calculate them; and (4) risk estimate was only with miRNA polymorphism, and not with miRNA binding site SNPs. All searches were performed independently by two investigators. Thirteen epidemiological studies were included in our study. As summarized in Table 1, Peng et al[40] suggested a significantly increased risk of gastric cancer in subjects homozygous for the variant C of miR-196a-2 (rs11614913) compared with wild-type homozygote TT and heterozygote CT carriers in a Chinese population (adjusted OR = 1.57, 95%CI: 1.03-2.39). Stratified analyses showed that the subjects homozygous for the variant C genotype of miR-196a-2 had a strong association with lymph node metastasis of gastric cancer (adjusted OR = 2.25, 95%CI: 1.21-4.18). Subsequently, Okubo et al[41] evaluated the associations of three SNPs (rs11614913, rs2910164 and rs3746444) in pre-miRNA (miR-196a2, miR-146a and miR-499) with the risk of gastric cancer in the Japanese population. The strongest association was observed only in the miR-146a rs2910164 (G > C) SNP. Our research also indicated that miR-27a A/G (rs895819) and miR-146a C/G (rs2910164) polymorphisms had important significance in gastric cancer susceptibility[16,17]. In addition, an increased risk of gastric cancer was obtained with AA genotype in pre-miR-30c compared with GG genotype in the Chinese population (adjusted OR = 1.83, 95%CI: 1.07-3.15). Moreover, the gastric cancer risk was especially elevated in older individuals (aged > 60 years), men, non-smokers, and H. pylori-infected individuals[42]. More interestingly, Zhang et al[43] found that lifestyle-related factors, including tea consumption and smoking, might have the potential to modify the associations between miR-149 and miR-605 polymorphisms and gastric cancer risk. One study also suggested that miR-938 polymorphism (rs2505901) C locus carried a decreased risk overall for gastric cancer (OR = 0.73, 95%CI: 0.56-0.99)[44]. Additionally, many epidemiological studies also reported that miRNA binding site polymorphisms were associated with the risk of gastric cancer[45-47]. As shown in Table 1, the most frequently studied SNPs are rs11614913 in miR-196a-2, rs895819 in miR-27a, and rs2910164 in miR-146a. Therefore, we discuss and perform a meta-analysis to evaluate the association between the three polymorphisms and gastric cancer susceptibility based on all eligible case-control published data. We calculated the pooled OR and 95%CI based on the most frequently adopted genetic model in original literature. The Z test was used to determine the statistical significance of the pooled OR, and P < 0.05 was considered statistically significant. Additionally, Q and I2 statistics were used to examine possible heterogeneity of study results[48]. If significant heterogeneity existed (P < 0.10 was considered representative of statistically significant heterogeneity), the pooled OR estimate of each study was calculated by the random effects model, otherwise the fixed-effects model was used.
| First author year | Population | miRNA | SNP ID | Allele | Case/control | Risk1 (OR and 95%CI) |
| Peng et al[40] 2010 | Chinese | miR-196a-2 | rs11614913 | T/C | 213/213 | 1.57 (1.03-2.39) |
| Okubo et al[41] 2010 | Japanese | miR-146a | rs2910164 | G/C | 552/697 | 1.30 (1.02-1.66) |
| miR-196a-2 | rs11614913 | T/C | 552/697 | NS | ||
| miR-499 | rs3746444 | A/G | 552/697 | NS | ||
| Sun et al[16] 2010 | Chinese | miR-27a | rs895819 | A/G | 304/304 | 1.48 (1.06-2.05) |
| Zeng et al[17] 2010 | Chinese | miR-146a | rs2910164 | C/G | 304/304 | 1.58 (1.11-2.20) |
| Hishida et al[56] 2011 | Japanese | miR-146a | rs2910164 | C/G | 583/540 | NS |
| Mu et al[42] 2012 | Chinese | miR-30c | rs928508 | A/G | 240/240 | 1.83 (1.07-3.15) |
| Zhang et al[43] 2012 | Chinese | miR-149 | rs2292832 | T/C | 762/757 | NS |
| miR-605 | rs2043556 | A/G | 762/757 | NS | ||
| Arisawa et al[44] 2012 | Japanese | miR-938 | rs2505901 | T/C | 333/574 | 0.73 (0.55-0.99) |
| Ahn et al[50] 2012 | South Korean | miR-146a | rs2910164 | C/G | 461/447 | NS |
| miR-149 | rs2292832 | T/C | 461/447 | NS | ||
| miR-196a-2 | rs11614913 | T/C | 461/447 | NS | ||
| miR-499 | rs3746444 | A/G | 461/447 | NS | ||
| Zhou et al[53] 2012 | Chinese | miR-27a | rs895819 | A/G | 295/413 | 0.28 (0.12-0.66) |
| Yang et al[54] 2012 | Chinese | miR-27a | rs11671784 | G/A | 892/978 | 0.76 (0.63–0.92) |
| miR-27a | rs895819 | A/G | 892/978 | NS | ||
| Zhou et al[57] 2012 | Chinese | miR-146a | rs2910164 | C/G | 1686/1895 | 1.26 (1.01-1.56) |
| Wang et al[51] 2013 | Chinese | miR-196a-2 | rs11614913 | T/C | 1689/1946 | 0.71 (0.60-0.83) |
The SNP (rs11614913) in the pre-miRNA region of miR-196a-2 was first identified in a case-control study of 1009 breast cancer cases and 1093 cancer-free controls in a population of Chinese women[49]. In gastric cancer, Peng et al[40] suggested a significantly increased risk of gastric cancer in subjects homozygous for the variant C of miR-196a-2 (rs11614913) compared with TT + CT carriers. However, one study conducted in a Japanese population only found the rs11614913 SNP in the miR-196a2 was associated with the degree of H. pylori-induced mononuclear cell infiltration, and not gastric cancer risk[41]. To understand this effect better, the SNP was genotyped again in a study conducted on 461 Korean patients with gastric cancer and 447 matched controls. They revealed that miR-196a-2 CC genotype was associated with elevated gastric cancer risk among women (adjusted OR = 1.86, 95%CI: 1.09-3.19)[50]. On the contrary, Wang et al[51] recently found that the CC genotype was significantly associated with a reduced risk of gastric cancer compared with the CT + TT genotypes in larger samples of the Chinese population (adjusted OR = 0.71, 95%CI: 0.60-0.83). Therefore, to determine further whether there is an association between rs11614913 and risk of gastric cancer, we summarized the published data from these studies using a recessive model, and showed no significant association between SNP rs11614913 in miR-196a-2 and gastric cancer risk (OR = 1.06, 95%CI: 0.74-1.50, Z = 0.30, P = 0.77; Q = 18.85, P < 0.01, Table 2).
| First author year | Study design | Case/control | Case | Control | Risk1 (OR and 95%CI) | ||||
| TT | CT | CC | TT | CT | CC | ||||
| Peng et al[40] 2010 | HB | 213/213 | 43 | 94 | 76 | 50 | 107 | 56 | 1.56 (1.03-2.35) |
| Okubo et al[41] 2010 | HB | 552/697 | 166 | 281 | 105 | 223 | 350 | 124 | 1.09 (0.81-1.45) |
| Ahn et al[50] 2012 | HB | 461/447 | 119 | 242 | 100 | 128 | 232 | 87 | 1.15 (0.83-1.58) |
| Wang et al[51] 2013 | HB | 1689/1946 | 519 | 851 | 319 | 524 | 940 | 482 | 0.71 (0.60-0.83) |
| Overall | 2915/3303 | 847 | 1468 | 600 | 925 | 1629 | 749 | 1.06 (0.74-1.50) | |
A recent study has suggested that miR-27a expression is significantly upregulated in gastric cancer tissues, and high expression levels of miR-27a are associated with poor tumor histological grade[52]. These findings indicate the important role of miR-27a in the development and progression of gastric cancer. Our recent research including 304 gastric cancer cases and 304 cancer-free controls indicated that miR-27a A/G polymorphism (rs895819) polymorphism was associated with an elevated risk of gastric cancer. We observed that the patients with variant genotypes (AG + GG) of miR-27a showed a significantly increased risk of gastric cancer relative to AA carriers (adjusted OR = 1.48, 95%CI: 1.06-2.05). The elevated risk was especially evident in older subjects (age > 58 years), men, non-smokers and rural residents. Moreover, we also found that polymorphism of miR-27a might be responsible for elevated miR-27a levels and reduced target gene zinc finger and BTB domain containing 10 (ZBTB10) mRNA[16]. However, another study showed that the SNP rs895819 in miR-27a with the minor allele C (C corresponds to G in the opposite strand) presented a significantly reduced risk of gastric cancer (adjusted OR = 0.77, 95%CI: 0.60-0.99)[53]. Similarly, in the Chinese population, Yang et al[54] found that the G/A polymorphism in miR-27a (rs11671784) reduced gastric cancer risk, but not the association with rs895819. Therefore, we extracted the genotypes from the above studies, and calculated the overall OR based on the dominant model (Table 3). There was no significant risk association in the overall pooled analysis (OR = 1.04, 95%CI: 0.76-1.42, Z = 0.25, P = 0.81; Q = 7.82, P = 0.02).
| First author year | Study design | Case/control | Case | Control | Risk1 (OR and 95%CI) | ||||
| AA | AG | GG | AA | AG | GG | ||||
| Sun et al[16] 2010 | HB | 304/304 | 115 | 135 | 54 | 145 | 119 | 40 | 1.50 (1.08-2.07) |
| Zhou et al[53] 2012 | HB | 295/413 | 166 | 122 | 7 | 214 | 167 | 32 | 0.84 (0.62-1.12) |
| Yang et al[54] 2012 | HB | 892/978 | 349 | 437 | 106 | 367 | 472 | 139 | 0.93 (0.76-1.13) |
| Overall | 1491/1695 | 630 | 694 | 167 | 726 | 758 | 211 | 1.04 (0.76-1.42) | |
A G to C polymorphism (rs2910164) located within the sequence of an miR-146a precursor was identified by Shen et al[55]. They revealed that the variant predisposed to an earlier age of onset of familial breast and ovarian cancers. In a Japanese population study, Okubo et al[41] revealed that rs2910164 CC genotype held a significantly higher risk of gastric cancer when compared to non-cancer subjects (adjusted OR = 1.30, 95%CI: 1.02-1.66). However, we showed that subjects with the variant genotypes (GC + GG) in miR-146a had a significantly higher risk of gastric cancer when compared with CC genotype carriers in the Chinese population (adjusted OR = 1.58, 95%CI: 1.11-2.20)[17]. Additionally, in other stratified analyses for gastric cancer risk, Ahn et al[50] found that subjects with miR-146a GG and (CG + GG) genotypes were associated with increased risk of gastric cancer among the non-smokers in a Korean population. In contrast, the study conducted by Hishida et al[56] on 583 gastric cancer cases and 540 gastric atrophy cases suggested a lack of association between SNP rs2910164 and gastric cancer risk. These observations were validated by Zhou et al[57] in a large sample of the Chinese population (1686 gastric cancer patients and 1895 cancer-free subjects). Their logistic regression suggested a significant association between rs2910164 polymorphism and increased gastric cancer risk (OR = 1.26, 95%CI = 1.01-1.56; GG vs CC + CG]. Apparently, these conclusions of the relevant studies are inconsistent, in part because of the small sample size, and the different genetic model and ethnicity of the patients. So, we performed a comprehensive analysis based on published data using a recessive model (Table 4). The combined results indicated that subjects homozygous for the variant C genotype in miR-146a had no statistically significant risk of gastric cancer when compared with (CC + CG) genotypes carrier (OR = 1.08, 95%CI: 0.87-1.33, Z = 0.68, P = 0.50; Q = 10.13, P = 0.04).
| First author year | Study design | Case/control | Case | Control | Risk1 (OR and 95%CI) | ||||
| CC | CG | GG | CC | CG | GG | ||||
| Okubo et al[41] 2010 | HB | 552/697 | 236 | 243 | 73 | 254 | 322 | 121 | 0.73 (0.53-0.99) |
| Zeng et al[17] 2010 | HB | 304/304 | 89 | 153 | 62 | 119 | 132 | 53 | 1.21 (0.81-1.82) |
| Hishida et al[56] 2011 | HB | 583/540 | 230 | 271 | 82 | 215 | 254 | 71 | 1.08 (0.77-1.52) |
| Ahn et al[50] 2012 | HB | 461/447 | 159 | 231 | 71 | 164 | 221 | 62 | 1.13 (0.78-1.63) |
| Zhou et al[57] 2012 | HB | 1686/1895 | 286 | 822 | 578 | 393 | 951 | 551 | 1.27 (1.10-1.47) |
| Overall | 3586/3913 | 1000 | 1720 | 866 | 1145 | 1880 | 858 | 1.08 (0.87-1.33) | |
This review includes a summary and discussion of the current findings evaluating the role of SNPs of miRNA in gastric cancer occurrence and development. Furthermore, we performed a meta-analysis of the most frequently studied miRNA-SNPs (rs11614913 in miR-196a-2, rs895819 in miR-27a and rs2910164 in miR-146a) in gastric cancer. Based on current data, there was no significant association between these miRNA-SNPs and gastric cancer risk in our meta-analysis. Future studies based on larger samples and independent cohorts are still needed to confirm the predictive signature of miRNA-SNPs in gastric cancer, for a better understanding of gastric cancer carcinogenesis.
P- Reviewers: Bashashati M, Dai Q, Jamil K, Pirola CJ, Reddy P S- Editor: Cui XM L- Editor: Kerr C E- Editor: Liu XM
| 1. | Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25833] [Cited by in RCA: 27829] [Article Influence: 1325.2] [Reference Citation Analysis (0)] |
| 2. | Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2890] [Cited by in RCA: 2949] [Article Influence: 92.2] [Reference Citation Analysis (0)] |
| 3. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8672] [Cited by in RCA: 8864] [Article Influence: 277.0] [Reference Citation Analysis (0)] |
| 4. | Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 510] [Cited by in RCA: 568] [Article Influence: 47.3] [Reference Citation Analysis (0)] |
| 5. | Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2463] [Cited by in RCA: 2547] [Article Influence: 115.8] [Reference Citation Analysis (0)] |
| 6. | Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 349] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 7. | Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999-3004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2908] [Cited by in RCA: 3110] [Article Influence: 148.1] [Reference Citation Analysis (0)] |
| 8. | Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9215] [Cited by in RCA: 9856] [Article Influence: 821.3] [Reference Citation Analysis (4)] |
| 9. | Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17-20. [PubMed] |
| 10. | Fock KM, Talley NJ, Fass R, Goh KL, Katelaris P, Hunt R, Hongo M, Ang TL, Holtmann G, Nandurkar S. Asia-Pacific consensus on the management of gastroesophageal reflux disease: update. J Gastroenterol Hepatol. 2008;23:8-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 121] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 11. | Kennemann L, Didelot X, Aebischer T, Kuhn S, Drescher B, Droege M, Reinhardt R, Correa P, Meyer TF, Josenhans C. Helicobacter pylori genome evolution during human infection. Proc Natl Acad Sci USA. 2011;108:5033-5038. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 204] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 12. | Xue H, Lin B, An J, Zhu Y, Huang G. Interleukin-10-819 promoter polymorphism in association with gastric cancer risk. BMC Cancer. 2012;12:102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 49] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Xue H, Ni P, Lin B, Xu H, Huang G. X-ray repair cross-complementing group 1 (XRCC1) genetic polymorphisms and gastric cancer risk: A HuGE review and meta-analysis. Am J Epidemiol. 2011;173:363-375. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 14. | Tsigris C, Chatzitheofylaktou A, Xiromeritis C, Nikiteas N, Yannopoulos A. Genetic association studies in digestive system malignancies. Anticancer Res. 2007;27:3577-3587. [PubMed] |
| 15. | Gu H, Yang L, Sun Q, Zhou B, Tang N, Cong R, Zeng Y, Wang B. Gly82Ser polymorphism of the receptor for advanced glycation end products is associated with an increased risk of gastric cancer in a Chinese population. Clin Cancer Res. 2008;14:3627-3632. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 16. | Sun Q, Gu H, Zeng Y, Xia Y, Wang Y, Jing Y, Yang L, Wang B. Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. Cancer Sci. 2010;101:2241-2247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 17. | Zeng Y, Sun QM, Liu NN, Dong GH, Chen J, Yang L, Wang B. Correlation between pre-miR-146a C/G polymorphism and gastric cancer risk in Chinese population. World J Gastroenterol. 2010;16:3578-3583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 71] [Cited by in RCA: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 18. | Shafi G, Jamil K, Kapley A, Purohit HJ, Vamsy MC. RNAi as a novel therapeutic platform technology for oncological solutions. Biotec Mol Bio Rev. 2008;4:55-70. |
| 19. | Kaiser J. MiRNAs rewrite the rules of molecular biology. Research J Bio Sec. 2007;2:3-5. |
| 20. | Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902-1910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1501] [Cited by in RCA: 1454] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 21. | Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775-783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 577] [Cited by in RCA: 611] [Article Influence: 33.9] [Reference Citation Analysis (0)] |
| 22. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3513] [Cited by in RCA: 3629] [Article Influence: 165.0] [Reference Citation Analysis (0)] |
| 23. | Lowery AJ, Miller N, McNeill RE, Kerin MJ. MicroRNAs as prognostic indicators and therapeutic targets: potential effect on breast cancer management. Clin Cancer Res. 2008;14:360-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 117] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 24. | Salzman DW, Shubert-Coleman J, Furneaux H. P68 RNA helicase unwinds the human let-7 microRNA precursor duplex and is required for let-7-directed silencing of gene expression. J Biol Chem. 2007;282:32773-32779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 25. | Salzman DW, Weidhaas JB. SNPing cancer in the bud: microRNA and microRNA-target site polymorphisms as diagnostic and prognostic biomarkers in cancer. Pharmacol Ther. 2013;137:55-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 26. | Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1545] [Cited by in RCA: 1594] [Article Influence: 83.9] [Reference Citation Analysis (0)] |
| 27. | Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787-798. [PubMed] |
| 28. | Vaucheret H, Vazquez F, Crété P, Bartel DP. The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev. 2004;18:1187-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 707] [Cited by in RCA: 683] [Article Influence: 32.5] [Reference Citation Analysis (0)] |
| 29. | Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2851] [Cited by in RCA: 2956] [Article Influence: 164.2] [Reference Citation Analysis (0)] |
| 30. | Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 3742] [Article Influence: 187.1] [Reference Citation Analysis (0)] |
| 31. | Chen G, Shen ZL, Wang L, Lv CY, Huang XE, Zhou RP. Hsa-miR-181a-5p expression and effects on cell proliferation in gastric cancer. Asian Pac J Cancer Prev. 2013;14:3871-3875. [PubMed] |
| 32. | Wang F, Sun GP, Zou YF, Hao JQ, Zhong F, Ren WJ. MicroRNAs as promising biomarkers for gastric cancer. Cancer Biomark. 2012;11:259-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 33. | Wang M, Gu H, Qian H, Zhu W, Zhao C, Zhang X, Tao Y, Zhang L, Xu W. miR-17-5p/20a are important markers for gastric cancer and murine double minute 2 participates in their functional regulation. Eur J Cancer. 2013;49:2010-2021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 34. | Yang TS, Yang XH, Wang XD, Wang YL, Zhou B, Song ZS. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013;13:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 35. | Liu X, Ru J, Zhang J, Zhu LH, Liu M, Li X, Tang H. miR-23a targets interferon regulatory factor 1 and modulates cellular proliferation and paclitaxel-induced apoptosis in gastric adenocarcinoma cells. PLoS One. 2013;8:e64707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 53] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 36. | Wang SH, Li X, Zhou LS, Cao ZW, Shi C, Zhou CZ, Wen YG, Shen Y, Li JK. microRNA-148a suppresses human gastric cancer cell metastasis by reversing epithelial-to-mesenchymal transition. Tumour Biol. 2013;34:3705-3712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 37. | Guo MM, Hu LH, Wang YQ, Chen P, Huang JG, Lu N, He JH, Liao CG. miR-22 is down-regulated in gastric cancer, and its overexpression inhibits cell migration and invasion via targeting transcription factor Sp1. Med Oncol. 2013;30:542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 38. | Slaby O, Bienertova-Vasku J, Svoboda M, Vyzula R. Genetic polymorphisms and microRNAs: new direction in molecular epidemiology of solid cancer. J Cell Mol Med. 2012;16:8-21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 100] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 39. | Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793-1801. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1837] [Cited by in RCA: 1796] [Article Influence: 89.8] [Reference Citation Analysis (0)] |
| 40. | Peng S, Kuang Z, Sheng C, Zhang Y, Xu H, Cheng Q. Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a Chinese population. Dig Dis Sci. 2010;55:2288-2293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 135] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 41. | Okubo M, Tahara T, Shibata T, Yamashita H, Nakamura M, Yoshioka D, Yonemura J, Ishizuka T, Arisawa T, Hirata I. Association between common genetic variants in pre-microRNAs and gastric cancer risk in Japanese population. Helicobacter. 2010;15:524-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 130] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 42. | Mu YP, Su XL. Polymorphism in pre-miR-30c contributes to gastric cancer risk in a Chinese population. Med Oncol. 2012;29:1723-1732. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 43. | Zhang MW, Jin MJ, Yu YX, Zhang SC, Liu B, Jiang X, Pan YF, Li QI, Ma SY, Chen K. Associations of lifestyle-related factors, hsa-miR-149 and hsa-miR-605 gene polymorphisms with gastrointestinal cancer risk. Mol Carcinog. 2012;51 Suppl 1:E21-E31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 44. | Arisawa T, Tahara T, Shiroeda H, Matsue Y, Minato T, Nomura T, Yamada H, Hayashi R, Saito T, Matsunaga K. Genetic polymorphisms of IL17A and pri-microRNA-938, targeting IL17A 3’-UTR, influence susceptibility to gastric cancer. Hum Immunol. 2012;73:747-752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 45. | Li ZH, Pan XM, Han BW, Guo XM, Zhang Z, Jia J, Gao LB. A let-7 binding site polymorphism rs712 in the KRAS 3’ UTR is associated with an increased risk of gastric cancer. Tumour Biol. 2013;34:3159-3163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 46. | Xu Y, Ma H, Yu H, Liu Z, Wang LE, Tan D, Muddasani R, Lu V, Ajani JA, Wang Y. The miR-184 binding-site rs8126 T& gt; C polymorphism in TNFAIP2 is associated with risk of gastric cancer. PLoS One. 2013;8:e64973. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 47. | Wang W, Li F, Mao Y, Zhou H, Sun J, Li R, Liu C, Chen W, Hua D, Zhang X. A miR-570 binding site polymorphism in the B7-H1 gene is associated with the risk of gastric adenocarcinoma. Hum Genet. 2013;132:641-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 48. | Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539-1558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21630] [Cited by in RCA: 25760] [Article Influence: 1120.0] [Reference Citation Analysis (0)] |
| 49. | Hu Z, Liang J, Wang Z, Tian T, Zhou X, Chen J, Miao R, Wang Y, Wang X, Shen H. Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat. 2009;30:79-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 305] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 50. | Ahn DH, Rah H, Choi YK, Jeon YJ, Min KT, Kwack K, Hong SP, Hwang SG, Kim NK. Association of the miR-146aC& gt; G, miR-149T& gt; C, miR-196a2T& gt; C, and miR-499A& gt; G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog. 2013;52 Suppl 1:E39-E51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 72] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 51. | Wang S, Tao G, Wu D, Zhu H, Gao Y, Tan Y, Wang M, Gong W, Zhou Y, Zhou J. A functional polymorphism in MIR196A2 is associated with risk and prognosis of gastric cancer. Mol Carcinog. 2013;52 Suppl 1:E87-E95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 52. | Liu D, Sun Q, Liang S, Xu L, Luo X, Zhao X, Wang X, Yang L. MicroRNA-27a inhibitors alone or in combination with perifosine suppress the growth of gastric cancer cells. Mol Med Rep. 2013;7:642-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 53. | Zhou Y, Du WD, Chen G, Ruan J, Xu S, Zhou FS, Zuo XB, Lv ZJ, Zhang XJ. Association analysis of genetic variants in microRNA networks and gastric cancer risk in a Chinese Han population. J Cancer Res Clin Oncol. 2012;138:939-945. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 54. | Yang Q, Jie Z, Ye S, Li Z, Han Z, Wu J, Yang C, Jiang Y. Genetic variations in miR-27a gene decrease mature miR-27a level and reduce gastric cancer susceptibility. Oncogene. 2014;33:193-202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 77] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 55. | Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 271] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 56. | Hishida A, Matsuo K, Goto Y, Naito M, Wakai K, Tajima K, Hamajima N. Combined effect of miR-146a rs2910164 G/C polymorphism and Toll-like receptor 4 +3725 G/C polymorphism on the risk of severe gastric atrophy in Japanese. Dig Dis Sci. 2011;56:1131-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 57. | Zhou F, Zhu H, Luo D, Wang M, Dong X, Hong Y, Lu B, Zhou Y, Zhou J, Zhang Z. A functional polymorphism in Pre-miR-146a is associated with susceptibility to gastric cancer in a Chinese population. DNA Cell Biol. 2012;31:1290-1295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |