Published online May 14, 2014. doi: 10.3748/wjg.v20.i18.5411
Revised: February 25, 2014
Accepted: March 6, 2014
Published online: May 14, 2014
Processing time: 206 Days and 14 Hours
Non-coding RNAs (ncRNAs) play key roles in development, proliferation, differentiation and apoptosis. Altered ncRNA expression is associated with gastric cancer occurrence, invasion, and metastasis. Moreover, aberrant expression of microRNAs (miRNAs) is significantly related to gastric cancer tumor stage, size, differentiation and metastasis. MiRNAs interrupt cellular signaling pathways, inhibit the activity of tumor suppressor genes, and affect the cell cycle in gastric cancer cells. Some miRNAs, including miR-21, miR-106a and miR-421, could be potential markers for the diagnosis of gastric cancer. Long non-coding RNAs (lncRNAs), a new research hotspot among cancer-associated ncRNAs, play important roles in epigenetic, transcriptional and post-transcriptional regulation. Several gastric cancer-associated lncRNAs, such as CCAT1, GACAT1, H19, and SUMO1P3, have been explored. In addition, Piwi-interacting RNAs, another type of small ncRNA that is recognized by gastroenterologists, are involved in gastric carcinogenesis, and piR-651/823 represents an efficient diagnostic biomarker of gastric cancer that can be detected in the blood and gastric juice. Small interfering RNAs also function in post-transcriptional regulation in gastric cancer and might be useful in gastric cancer treatment.
Core tip: Non-coding RNAs (ncRNAs) are closely associated with gastric cancer occurrence, invasion, and metastasis. MicroRNAs interrupt cellular signaling pathways, inhibit the activity of tumor suppressor genes, and affect the cell cycle in gastric cancer cells. Long non-coding RNAs (lncRNAs) represent a new research hotspot in the field of gastric cancer-associated ncRNAs. The global lncRNA expression profile in gastric cancer has been determined. In addition, Piwi-interacting RNAs are involved in gastric carcinogenesis. Small interfering RNAs function as post-transcriptional regulators in gastric cancer, and they might be used for the treatment of gastric cancer.
- Citation: Li PF, Chen SC, Xia T, Jiang XM, Shao YF, Xiao BX, Guo JM. Non-coding RNAs and gastric cancer. World J Gastroenterol 2014; 20(18): 5411-5419
- URL: https://www.wjgnet.com/1007-9327/full/v20/i18/5411.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i18.5411
Non-coding RNAs (ncRNAs) refer to a class of RNAs with no protein-coding function that are widely expressed in organisms. NcRNAs can be divided into 2 groups: housekeeping ncRNAs and regulatory ncRNAs. According to their size, the latter can further be divided into three types: (1) short ncRNAs, including small interfering RNAs (siRNAs), microRNAs (miRNAs) and Piwi-interacting RNAs (piRNAs); (2) mid-size ncRNAs; and (3) long non-coding RNAs (lncRNAs)[1-3]. Short ncRNAs are shorter than 50 nucleotides (nt), those between 50 nt and 200 nt are referred to as mid-size ncRNAs, and lncRNAs are longer than 200 nt[4-6].
In the past, most ncRNAs were considered to be “junk RNAs”. However, in recent years, advances in molecular biology have demonstrated that ncRNAs play important basic biological roles, and the mutation or aberrant expression of ncRNAs is important in the occurrence and development of diseases, including cancer.
Gastric cancer, one of the most common malignant tumors in the world, accounts for the second most cancer-related death worldwide and is a major cause of cancer-related mortality in China[7]. Gastric cancer originates in the surface of epithelial cells in the stomach. Each site of the stomach can potentially harbor cancer, but the gastric antrum and pylorus display the highest incidence of gastric cancer. The gastric cardia is the second most prevalent site for cancer, and a slightly lower incidence is reported for the gastric body. However, the occurrence of gastric cancer is a complex process of progressive development that involves multiple factors, multiple steps, coding and non-coding genes.
Helicobacter pylori (H. pylori) infection, a major risk factor for gastric cancer, is mainly related to the distal intestinal type of gastric cancer. Although the mechanism of H. pylori carcinogenesis is not clear, gene methylation is an important mechanism that leads to the formation of gastric cancer. H. pylori infection may increase the level of methylation of certain miRNA genes, and aberrant DNA methylation could induce gastric cancer.
Due to the increasing early detection of cancer and the widespread implementation of radical surgery, the overall survival of patients with gastric cancer has improved[8,9]. However, the prognosis of advanced gastric cancer remains poor, and safe and effective adjuvant therapy options are limited. Therefore, identifying valuable early markers of gastric cancer is extremely important for the diagnosis of gastric cancer.
Cancer stem cells (CSCs) are a small fraction of cells that are present in the tumor, and these cells have self-renewal and differentiation potential[10]. Ghaffarzadehgan et al[11] reported that CD44+ gastric cancer stem-like cells (GCSCs) displayed enhanced metastatic capacity and depth of invasion compared to their CD44- counterparts. CSCs display abnormal activation of several signaling pathways, such as Notch, Wnt, and others. Previous reports demonstrate that β-elemene, a naturally occurring compound extracted from Curcumae Radix, suppresses the growth of GCSCs[12] by interfering with Notch-1 expression[12].
Similar to CSC studies, research on ncRNAs is becoming a new field for gastric cancer.
miRNAs are a class of small ncRNAs containing approximately 19-24 nt. Most genes encoding miRNAs are single copy, multiple copies or gene clusters; other forms exist in the spacer region of protein coding genes or introns. They are highly conserved, temporal and tissue-specific[13]. Although miRNAs do not code for proteins, they regulate gene expression at the post-transcriptional level. Through complete or incomplete complementary binding to the 3’-untranslated regions (3’-UTRs) of target mRNAs, miRNAs promote targeted-mRNA degradation or translational suppression and negatively regulate the expression of target genes[14-16].
Growing evidence indicates that miRNAs are involved in crucial biological processes, including development, differentiation, proliferation, apoptosis, invasion and metastasis[17-19]. Various aberrantly expressed miRNAs have been identified in gastric cancer (Table 1). Several studies have demonstrated that miRNAs can be used not only as biomarkers but also as potential therapeutic targets for cancer[20,21].
| Expression | miRNAs |
| Up | miR-9, miR-21, miR-25, miR-106a, miR-106b, miR-130b, miR-191, miR-214, miR-421, miR-650 |
| Down | miR-31, miR-29a, miR-148a, miR-155, miR-195, miR-218, miR-375, miR-378, miR-429 |
In 1993, using a classical positional cloning approach, Lee et al[22] identified lin-4, the first miRNA molecule. In 2000, Reinhart et al[23] identified let-7, another miRNA, and discovered its role in the post-transcriptional regulation of gene expression.
For miRNA synthesis, the primary transcript (pri-miRNA), which is hundreds to thousands of nt in length, is first transcribed from genomic DNA by RNA polymerase II in the nucleus. Then, the pri-miRNA is cut by the Drosha enzyme of RNase 3 endonuclease enzyme family into hairpin precursors of miRNA (pre-miRNA), which are approximately 70 nt and present in the nucleus[24]. Finally, the synergistic effect of Ran-GTP and transporter protein Exportin 5 transports pre-miRNA out of the nucleus, and the enzyme Dicer cuts it to produce the approximately 22 nt mature miRNA[25].
With the use of microarray technology, bioinformatics and other genetics methods, the ectopic expression of miRNAs in gastric cancer has been found to be closely related to its occurrence, development, invasion, and metastasis. By decreasing the expression of tumor suppressor genes or enhancing the expression of oncogenes, miRNAs play an important role in the regulation of cancer-related genes[26].
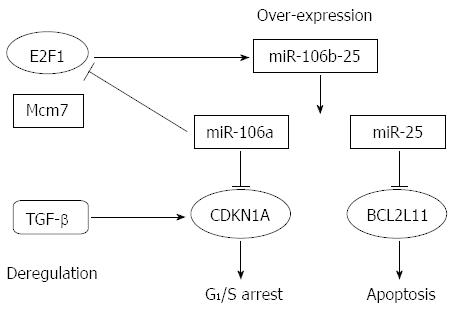
Abnormal regulation of the transcription factor E2F1 and transforming growth factor-β (TGF-β) plays a critical role in gastric carcinogenesis. E2F1 activates its own promoter. Petrocca et al[27] reported that miR-106b-25 cluster expression is activated by E2F1 in parallel with its host gene, Mcm7; it is also involved in E2F1 post-transcriptional regulation. Furthermore, the TGF-β tumor suppressor pathway was impaired by over-expression of the miR-106b-25 cluster, and the expression of CDKN1A (p21Waf1/Cip1) and BCL2L11 (Bim) was effected as well. Finally, CDKN1A and BCL2L11 disrupted the G1/S checkpoint and conferred resistance to TGF-β-dependent apoptosis, respectively (Figure 1).
In another study, miR-375 suppressed tumor growth by targeting Janus kinase 2 (JAK2), influencing the proliferation of gastric cancer cells[28]. miR-375 markedly inhibited JAK2 expression via its mRNA 3’-UTR region[28]. However, over-expression of miR-375 in gastric cancer significantly reduced the protein level of JAK2, while the level of JAK2 mRNA was not greatly affected[28]. These results suggest that miR-375 affects JAK2 expression at the post-transcriptional level.
miR-9 is down-regulated in human gastric adenocarcinoma. Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 (NF-κB1) is a direct target of miR-9. To demonstrate the role of miR-9 in tumor cell proliferation, Wan et al[29] transfected gastric cancer cells with a vector containing pri-miR-9. They reported that cell growth and proliferation were significantly inhibited. Meanwhile, over-expression of miR-9 not only inversely regulated endogenous NF-κB1 protein expression but also decreased endogenous NF-κB1 mRNA levels[29].
miR-214 is up-regulated in several human gastric cancer cell lines, including BGC-823, MKN45 and SGC-7901, compared with the normal gastric mucosal cell line, GES-1[30]. Phosphatase and tensin homolog deleted on chromosome ten (PTEN), a famous tumor suppressor gene, is a target of miR-214, and its expression is inversely correlated with miR-214 expression. miR-214 promoted the proliferation of cancer cells, and the down-regulation of miR-214 induced G1 phase arrest in cancer cells[30].
Zhang et al[31] used a luciferase reporter assay to demonstrate that inhibitor of growth 4 (ING4), a tumor suppressor, is a direct target of miR-650. Over-expression of miR-650 in gastric cancer cells may inhibit the expression of ING4 through post-transcriptional repression. Moreover, ectopic expression of miR-650 significantly promoted tumorigenesis and proliferation in gastric cancer cells, at least partially through ING4[31].
Runt-related transcription factor 3 (RUNX3) is a newly discovered tumor suppressor gene that is inactivated in a variety of tumors. Research showed that miR-130b was significantly higher in gastric cancer tissues than in normal tissues. RUNX3 is a target of miR-130[32].
Altered expression of miR-106a was related to the expression of the tumor suppressor gene Rb1 and the transcription factor E2F1[33]. More importantly, miR-106a expression is positively associated with TNM stage, indicating that miR-106a could be used as a novel diagnosis biomarker of gastric carcinoma[33]. Other miRNAs, such as miR-31 and miR-421, might also be potential tumor biomarkers. miR-31 is located on 9p21.3. Our group found that in gastric cancer, the expression of miR-31 was significantly down-regulated and could be used as an efficient biomarker for the diagnosis of gastric carcinoma[34]. In contrast to miR-31, miR-421 is a miRNA with oncogenic properties. Our group reported that miR-421 was up-regulated in gastric cancer via three targets: RBMXL1 (RNA binding motif protein, X-linked-like 1), CNTN-1 (contactin 1) and CBX7 (chromobox homolog)[35]. Furthermore, the expression of miR-421 in the gastric juice might be used as a potential marker for the diagnosis of early gastric cancer[36].
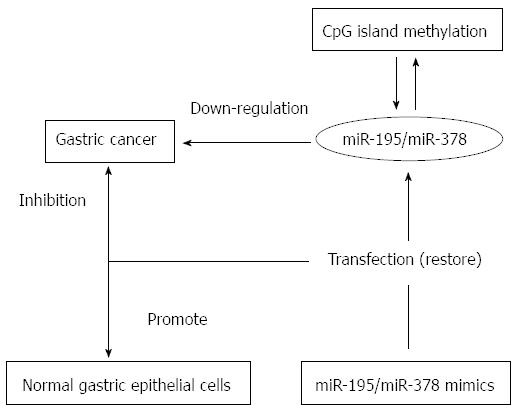
DNA methylation, as an important type of epigenetic modification, plays a critical role in the process of gastric cancer by regulating gene transcription. Our group demonstrated that miR-195 and miR-378 expression levels were down-regulated in gastric cancer compared to those in non-tumor tissue[37] due to CpG island methylation in their promoters. Furthermore, treatment with their mimics restored the expression of these miRNAs[37]. The mimics inhibited tumor cell growth and promoted the growth of normal gastric epithelial cells (Figure 2).
As a novel gene, p42.3 encodes a 42.3 kDa protein of 389 amino acid residues that is associated with cell proliferation and tumorigenicity by regulating the M-phase of the cell cycle. p42.3 expression is up-regulated in human gastric cancer and is a direct target of miR-29a. Experiments in gastric cancer cells and tissue samples showed that miR-29a reduced the expression of p42.3, indicating that miR-29a may play a role in human gastric cancer[38].
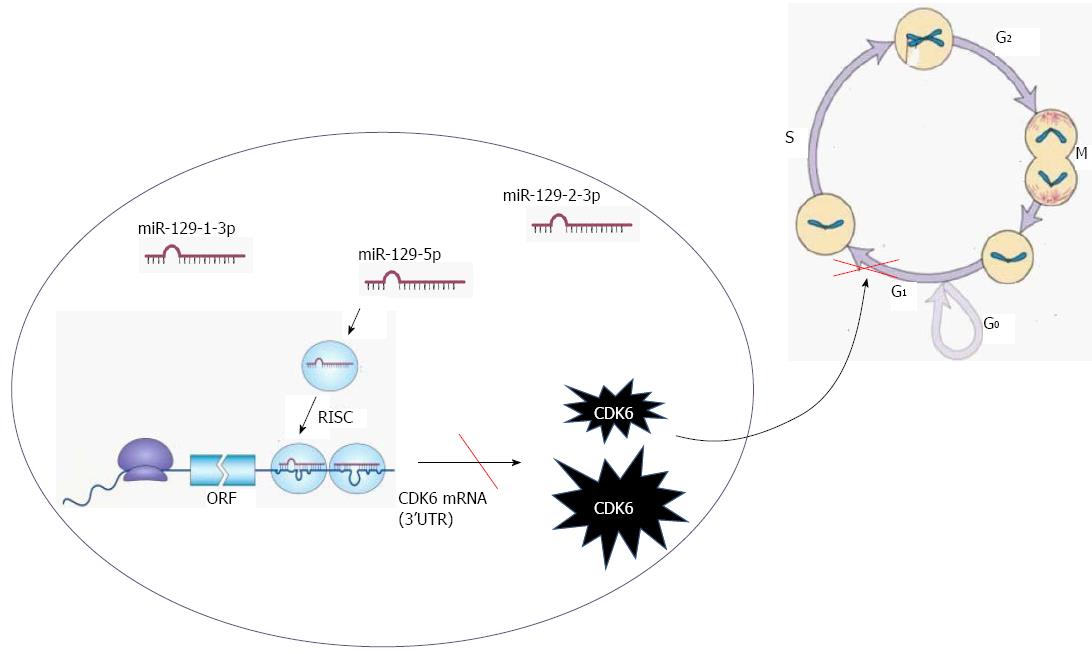
We recently demonstrated that all miR-129 family members, miR-129-1-3p, miR-129-2-3p, and miR-129-5p, were down-regulated in gastric cancer cell lines compared with normal gastric epithelial cells[39]. Furthermore, all mature products of miR-129 displayed tumor suppressor activity. To elucidate the molecular mechanisms underlying the down-regulation of miR-129 in gastric cancer, we analyzed the effects of miR-129 mimics on the cell cycle. We found that increased miR-129 levels in gastric cancer cells resulted in significant G0/G1 phase arrest, which is associated with cyclin dependent kinase 6 (CDK6), a cell cycle-associated protein involved in G1-S transition (Figure 3). Additionally, we demonstrated that CDK6 and sex determining region Y-box 4 (SOX4) are the targets of miR-129[39]. In addition, miR-429 and miR-148a inhibit cell growth and proliferation in gastric cancer by targeting c-Myc and p27, respectively[40,41]. These proteins are also cell cycle regulators.
Some miRNAs are involved in tumor invasion and metastasis. Reversion-inducing-cysteine-rich protein with kazal motifs (RECK), which is a target of miR-21, inhibits tumor invasion, metastasis and angiogenesis. Zhang et al[42] demonstrated that over-expression of miR-21 in the gastric cancer cell line AGS enhanced the proliferation and invasive ability of cells, whereas knockdown of miR-21 suppressed cell proliferation, invasion and metastasis, while increasing apoptosis. These processes are associated with RECK.
Tie et al[43] confirmed that miR-218 inhibited the invasion and metastasis of gastric cancer in vivo and in vitro by targeting the Robo1 receptor. miR-218 expression in gastric cancer was closely related to clinical stage, lymph node metastasis and prognosis[43].
miR-155, a typical multifunctional miRNA, plays an important role in the process of gastric cancer. It was reported that miR-155 was remarkably down-regulated in gastric cancer[44]. Li et al[44] reported that transfection of gastric cancer cells with synthetic miR-155 mimics decreased the migratory and invasive capacity of cells compared to the control sample, as determined by scratch wound healing assay, transwell migration assay and transwell invasion assay. Thus, miR-155 may act as a tumor suppressor, and it plays a pivotal function in gastric cancer metastasis. miR-155 expression levels were restored after treatment with 5-aza-dC, a DNA-demethylating agent[44]. These data suggest that dysregulation of miR-155 expression in gastric cancer may be partially due to DNA methylation.
piRNAs are a class of newly discovered small non-coding RNAs that are approximately 24-33 nt in length. piRNAs only bind specifically with Piwi protein family members. Similar to miRNAs, the 5’ ends of piRNAs also contain a strong uracil bias. To date, piRNAs are thought to mainly exist in the intergenic region and rarely in the gene region and repeat region. In July, 2006, piRNAs were first identified as being closely related with germ cell development[45].
Studies have shown that piRNAs mainly exist in mammalian germ cells and stem cells by binding to the Piwi subfamily protein to form the piRNA compound (piRC) to regulate gene silencing pathways[46]. Genetic analysis and time characteristics of piRNA accumulation revealed that piRC plays an important role in gametogenesis. Only 17% to 20% of mammalian piRNA repeat sequences correspond to transposons and retrotransposons[47]. Thus, piRNAs may have different functions in epigenetic programming, repressing transcription and post-transcriptional regulation.
According to the known function of Piwi proteins, scientists speculate that piRNAs may have three aspects of function[48,49]: transcriptional gene silencing, maintaining germline stem cell function and regulating translation and mRNA stability.
There are several reports investigating the association between piRNAs and gastric cancer. Our study demonstrated that piR-823 was significantly down-regulated in gastric cancer tissues[50]. In vitro studies also revealed that piR-823 analogues effectively inhibited gastric cancer cell proliferation[50]. The expression of another piRNA, piR-651, was significantly up-regulated in gastric cancer, and transfection with piR-651 inhibitors blocked gastric cancer cell growth and induced G2/M phase arrest[51]. Our study also illustrated that piR-651 was up-regulated in other types of cancers. To explore the potential clinical application of piRNAs, we used real-time reverse transcription-polymerase chain reaction (RT-PCR) to detect circulating tumor cells (CTCs) in patients with gastric cancer and found that piRNAs, such as piR-651 and piR-823, were more sensitive than serum carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9)[52]. These results indicate that piRNAs may be promising molecular markers for the diagnosis of gastric cancer.
lncRNAs are transcripts longer than 200 nt that are rarely involved in protein coding[53]. lncRNAs play roles in epigenetic, transcriptional and post-transcription regulation[54,55]. They have important physiological and biochemical functions, such as X-inactivation, development and differentiation[56-58]. In structural characteristics and sequence composition, lncRNAs are similar to protein-coding RNAs, with highly tissue specific and time specific properties[59]. lncRNAs participate in gene imprinting (genetic imprinting), chromatin modification, transcriptional activation, interference, cell cycle regulation, splicing and translation regulation[60-62]. Researchers are paying more attention to lncRNAs in various biological processes and their roles in the disease process. Some studies found that ectopic expression of lncRNAs was associated with disease occurrence, development, proliferation, metastasis, and invasion[63,64]. Moreover, researchers observed the abnormal expression of various lncRNAs in gastric cancer (Table 2).
| Expression | lncRNAs |
| Up | H19, CCAT1, H19, HMlincRNA717, BM709340, BQ213083, AK054978, DB077273, SUMO1P3 |
| Down | GACAT1, FER1L4, uc001lsz, BG491697, AF131784, uc009ycs, BG981369, AF147447, HMlincRNA1600, AK054588 |
Our group investigated the global lncRNA expression profile in gastric cancer[65]. In total, we identified 135 lncRNAs whose expression levels between tumor and non-tumorous tissues were altered more than two-fold (GEO accession numbers is 47850; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE47850). The most down-regulated lncRNAs in gastric cancer tissues were FER1L4, uc001lsz, BG491697, AF131784, uc009ycs, BG981369, AF147447, HMlincRNA1600, and AK054588; while the most up-regulated lncRNAs were H19, HMlincRNA717, BM709340, BQ213083, AK054978, and DB077273[65]. Furthermore, the expression level of gastric cancer-associated transcript 1 (GACAT1), or AC096655.1-002, was associated with lymph node metastasis, distant metastasis, tumor-node-metastasis stage, and differentiation, suggesting that GACAT1 might serve as a potential biological marker for the clinical diagnosis of gastric cancer[66,67].
H19 is an lncRNA widely expressed from the maternal allele and is only separated from the neighboring insulin like growth factor 2 (IGF2) gene by 90 kb. H19 expression is up-regulated in various human cancers, including hepatocellular carcinoma, bladder cancer, gastric cancer and breast cancer, suggesting that it has oncogenic properties[65,68]. Using real-time RT-PCR, flow cytometry, RNA immunoprecipitation and other methods, Yang et al[69] confirmed that H19 expression in gastric cancer accelerated cell proliferation. Furthermore, siRNA-mediated knockdown of H19 induced apoptosis and inactivated P53[69]. These results suggest that H19 may have a potential application for the treatment of gastric cancer.
Another study reported that lncRNA CCAT1 was up-regulated in gastric carcinoma tissues, and its expression was closely related to the transcription factor c-Myc[70]. Abnormal expression of c-Myc increased the activity of the CCAT1 promoter[70]. In the CCAT1 promoter region, c-Myc interacts with the E-box and up-regulates CCAT1 expression. CCAT1 up-regulation was correlated with primary tumor growth, lymph node metastasis, and metastatic disease[70]. Therefore, these results indicate that CCAT1 functions as an oncogene, and it may be used as a biomarker and a therapeutic target in gastric cancer.
Pseudogene-expressed lncRNAs are a major member of the lncRNA family. Recently, our group reported that small ubiquitin-like modifier (SUMO) 1 pseudogene 3, SUMO1P3, was markedly up-regulated in gastric cancer tissues compared with paired adjacent non-tumor tissues[71]. Its expression level was significantly correlated with tumor size, differentiation, lymphatic metastasis, and invasion. These results indicate that SUMO1P3 may be a potential biomarker for the diagnosis of gastric cancer.
siRNAs are exogenous RNA molecules that are double stranded with a length of approximately 21-25 nt, including the sub replication of the RNA virus, transposon or transgenic target, etc. siRNAs are formed by Dicer, a specific enzyme for double-stranded RNA in the RNase III family[72], and they play a role by loading to AGO proteins[73,74]. siRNAs silence RNA by causing target gene degradation via perfect complementarity with the target mRNA. Recent studies have indicated that siRNA-mediated DNA methylation and histone modification lead to transcriptional gene silencing in mammalian cells[75-79].
Studies demonstrated that the proto-oncogene astrocyte elevated gene 1 (AEG-1) is involved in many biological processes, including cell proliferation, survival, apoptosis, invasion and metastasis[80]. AEG-1 regulates tumor cell proliferation through pre-proliferative and anti-apoptotic effects[80]. In gastric cancer tissues, AEG-1 expression was significantly higher than that in normal tissues[81], suggesting that it may play an important role in the occurrence and development of gastric cancer. Therefore, compared with non-transfected cells and control siRNA transfected cells, AEG-1 siRNA markedly down-regulated endogenous AEG-1 expression at the mRNA and protein levels in gastric cancer cells. Expression of AEG-1 siRNA inhibited cell proliferation and changed the cell cycle distribution of gastric cancer cells.
The B lymphocyte/leukemia-2 (Bcl-2) gene is an oncogene that effectively inhibits cell apoptosis and prolongs cell vitality. Liu et al[82] reported that a specific siRNA against Bcl-2 increased the apoptosis of gastric cancer BGC-823 cells. Gao et al[83] recently demonstrated that siRNA-mediated knockdown of the phosphatase of regenerating liver-3 gene effectively inhibited gastric carcinoma invasion and metastasis. These results indicate that siRNA mediated-gene knockdown may be promising for clinical applications.
In the mammalian genome, ncRNAs account for approximately 98% of all transcripts. ncRNAs are a critical component of highly complex gene regulatory networks in cells. Their functions include adjusting the timing of development, cell proliferation, early embryonic development, tumor development, and stem cell differentiation. Various aberrantly expressed ncRNAs have been identified in gastric cancer (Tables 1 and 2). These ncRNAs may not only be new biological markers for the diagnosis of gastric cancer but also new targets for gastric cancer therapy.
P- Reviewers: Ding SZ, Jung YD, Wei PK S- Editor: Wen LL L- Editor: Wang TQ E- Editor: Zhang DN
| 1. | Place RF, Noonan EJ. Non-coding RNAs turn up the heat: an emerging layer of novel regulators in the mammalian heat shock response. Cell Stress Chaperones. 2014;19:159-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 66] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 2. | Gomes AQ, Nolasco S, Soares H. Non-coding RNAs: multi-tasking molecules in the cell. Int J Mol Sci. 2013;14:16010-16039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 171] [Cited by in RCA: 196] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 3. | Zhang XM, Ma ZW, Wang Q, Wang JN, Yang JW, Li XD, Li H, Men TY. A new RNA-seq method to detect the transcription and non-coding RNA in prostate cancer. Pathol Oncol Res. 2014;20:43-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Hong J, Zhang H, Kawase-Koga Y, Sun T. MicroRNA function is required for neurite outgrowth of mature neurons in the mouse postnatal cerebral cortex. Front Cell Neurosci. 2013;7:151. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 5. | Srivastava SP, Koya D, Kanasaki K. MicroRNAs in kidney fibrosis and diabetic nephropathy: roles on EMT and EndMT. Biomed Res Int. 2013;2013:125469. [PubMed] |
| 6. | Lv J, Liu H, Huang Z, Su J, He H, Xiu Y, Zhang Y, Wu Q. Long non-coding RNA identification over mouse brain development by integrative modeling of chromatin and genomic features. Nucleic Acids Res. 2013;41:10044-10061. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 52] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 7. | Smith MG, Hold GL, Tahara E, El-Omar EM. Cellular and molecular aspects of gastric cancer. World J Gastroenterol. 2006;12:2979-2990. [PubMed] |
| 8. | Ishihara R. Infrared endoscopy in the diagnosis and treatment of early gastric cancer. Endoscopy. 2010;42:672-676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 9. | Tanizawa Y, Terashima M. Lymph node dissection in the resection of gastric cancer: review of existing evidence. Gastric Cancer. 2010;13:137-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 50] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 10. | Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells--perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339-9344. [PubMed] |
| 11. | Ghaffarzadehgan K, Jafarzadeh M, Raziee HR, Sima HR, Esmaili-Shandiz E, Hosseinnezhad H, Taghizadeh-Kermani A, Moaven O, Bahrani M. Expression of cell adhesion molecule CD44 in gastric adenocarcinoma and its prognostic importance. World J Gastroenterol. 2008;14:6376-6381. [PubMed] |
| 12. | Yan B, Zhou Y, Feng S, Lv C, Xiu L, Zhang Y, Shi J, Li Y, Wei P, Qin Z. β -Elemene-Attenuated Tumor Angiogenesis by Targeting Notch-1 in Gastric Cancer Stem-Like Cells. Evid Based Complement Alternat Med. 2013;2013:268468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Kim VN, Nam JW. Genomics of microRNA. Trends Genet. 2006;22:165-173. [PubMed] |
| 14. | Singh TR, Gupta A, Suravajhala P. Challenges in the miRNA research. Int J Bioinform Res Appl. 2013;9:576-583. [PubMed] |
| 15. | Wang C, Leng X, Zhang Y, Kayesh E, Zhang Y, Sun X, Fang J. Transcriptome-wide analysis of dynamic variations in regulation modes of grapevine microRNAs on their target genes during grapevine development. Plant Mol Biol. 2014;84:269-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 32] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 16. | Zhang Y, Wang Z, Gemeinhart RA. Progress in microRNA delivery. J Control Release. 2013;172:962-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 460] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 17. | Zhuo Y, Gao G, Shi JA, Zhou X, Wang X. miRNAs: biogenesis, origin and evolution, functions on virus-host interaction. Cell Physiol Biochem. 2013;32:499-510. [PubMed] |
| 18. | Zhao X, Li X, Yuan H. microRNAs in gastric cancer invasion and metastasis. Front Biosci (Landmark Ed). 2013;18:803-810. [PubMed] |
| 19. | Gaál Z, Oláh E. [MicroRNA-s and their role in malignant hematologic diseases]. Orv Hetil. 2012;153:2051-2059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 20. | Petri A, Lindow M, Kauppinen S. MicroRNA silencing in primates: towards development of novel therapeutics. Cancer Res. 2009;69:393-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 60] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 21. | Wu Y, Crawford M, Yu B, Mao Y, Nana-Sinkam SP, Lee LJ. MicroRNA delivery by cationic lipoplexes for lung cancer therapy. Mol Pharm. 2011;8:1381-1389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 143] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 22. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [PubMed] |
| 23. | Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901-906. [PubMed] |
| 24. | Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415-419. [PubMed] |
| 25. | Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95-98. [PubMed] |
| 26. | Ayyanar M, Subash-Babu P, Ignacimuthu S. Syzygium cumini (L.) Skeels., a novel therapeutic agent for diabetes: folk medicinal and pharmacological evidences. Complement Ther Med. 2013;21:232-243. [PubMed] |
| 27. | Petrocca F, Visone R, Onelli MR, Shah MH, Nicoloso MS, de Martino I, Iliopoulos D, Pilozzi E, Liu CG, Negrini M. E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell. 2008;13:272-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 678] [Cited by in RCA: 693] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 28. | Ding L, Xu Y, Zhang W, Deng Y, Si M, Du Y, Yao H, Liu X, Ke Y, Si J. MiR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010;20:784-793. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 281] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 29. | Wan HY, Guo LM, Liu T, Liu M, Li X, Tang H. Regulation of the transcription factor NF-kappaB1 by microRNA-9 in human gastric adenocarcinoma. Mol Cancer. 2010;9:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 123] [Cited by in RCA: 142] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 30. | Xiong X, Ren HZ, Li MH, Mei JH, Wen JF, Zheng CL. Down-regulated miRNA-214 induces a cell cycle G1 arrest in gastric cancer cells by up-regulating the PTEN protein. Pathol Oncol Res. 2011;17:931-937. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 31. | Zhang X, Zhu W, Zhang J, Huo S, Zhou L, Gu Z, Zhang M. MicroRNA-650 targets ING4 to promote gastric cancer tumorigenicity. Biochem Biophys Res Commun. 2010;395:275-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 32. | Lai KW, Koh KX, Loh M, Tada K, Subramaniam MM, Lim XY, Vaithilingam A, Salto-Tellez M, Iacopetta B, Ito Y. MicroRNA-130b regulates the tumour suppressor RUNX3 in gastric cancer. Eur J Cancer. 2010;46:1456-1463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 132] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 33. | Xiao B, Guo J, Miao Y, Jiang Z, Huan R, Zhang Y, Li D, Zhong J. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin Chim Acta. 2009;400:97-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 127] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 34. | Zhang Y, Guo J, Li D, Xiao B, Miao Y, Jiang Z, Zhuo H. Down-regulation of miR-31 expression in gastric cancer tissues and its clinical significance. Med Oncol. 2010;27:685-689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 35. | Jiang Z, Guo J, Xiao B, Miao Y, Huang R, Li D, Zhang Y. Increased expression of miR-421 in human gastric carcinoma and its clinical association. J Gastroenterol. 2010;45:17-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 115] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 36. | Zhang X, Cui L, Ye G, Zheng T, Song H, Xia T, Yu X, Xiao B, Le Y, Guo J. Gastric juice microRNA-421 is a new biomarker for screening gastric cancer. Tumour Biol. 2012;33:2349-2355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 37. | Deng H, Guo Y, Song H, Xiao B, Sun W, Liu Z, Yu X, Xia T, Cui L, Guo J. MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene. 2013;518:351-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 124] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 38. | Cui Y, Su WY, Xing J, Wang YC, Wang P, Chen XY, Shen ZY, Cao H, Lu YY, Fang JY. MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation of p42.3 in human gastric cancer. PLoS One. 2011;6:e25872. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 39. | Yu X, Song H, Xia T, Han S, Xiao B, Luo L, Xi Y, Guo J. Growth inhibitory effects of three miR-129 family members on gastric cancer. Gene. 2013;532:87-93. [PubMed] |
| 40. | Sun T, Wang C, Xing J, Wu D. miR-429 modulates the expression of c-myc in human gastric carcinoma cells. Eur J Cancer. 2011;47:2552-2559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 41. | Guo SL, Peng Z, Yang X, Fan KJ, Ye H, Li ZH, Wang Y, Xu XL, Li J, Wang YL. miR-148a promoted cell proliferation by targeting p27 in gastric cancer cells. Int J Biol Sci. 2011;7:567-574. [PubMed] |
| 42. | Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 375] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 43. | Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S, Guo X, Wang B, Gang Y, Zhang Y. MiR-218 inhibits invasion and metastasis of gastric cancer by targeting the Robo1 receptor. PLoS Genet. 2010;6:e1000879. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 350] [Cited by in RCA: 370] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 44. | Li CL, Nie H, Wang M, Su LP, Li JF, Yu YY, Yan M, Qu QL, Zhu ZG, Liu BY. microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol Rep. 2012;27:1960-1966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 45. | Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203-207. [PubMed] |
| 46. | Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE. Characterization of the piRNA complex from rat testes. Science. 2006;313:363-367. [PubMed] |
| 47. | Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 2007;129:69-82. [PubMed] |
| 48. | Saito K. The epigenetic regulation of transposable elements by PIWI-interacting RNAs in Drosophila. Genes Genet Syst. 2013;88:9-17. [PubMed] |
| 49. | Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14:523-534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 247] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 50. | Cheng J, Deng H, Xiao B, Zhou H, Zhou F, Shen Z, Guo J. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315:12-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 221] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 51. | Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, Li QN. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621-1625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 239] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 52. | Cui L, Lou Y, Zhang X, Zhou H, Deng H, Song H, Yu X, Xiao B, Wang W, Guo J. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem. 2011;44:1050-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 125] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 53. | Gao L, Mai A, Li X, Lai Y, Zheng J, Yang Q, Wu J, Nan A, Ye S, Jiang Y. LncRNA-DQ786227-mediated cell malignant transformation induced by benzo(a)pyrene. Toxicol Lett. 2013;223:205-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 49] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 54. | Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 823] [Cited by in RCA: 955] [Article Influence: 79.6] [Reference Citation Analysis (0)] |
| 55. | Zhu J, Fu H, Wu Y, Zheng X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci China Life Sci. 2013;56:876-885. [PubMed] |
| 56. | Froberg JE, Yang L, Lee JT. Guided by RNAs: X-inactivation as a model for lncRNA function. J Mol Biol. 2013;425:3698-3706. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 57. | Scheuermann JC, Boyer LA. Getting to the heart of the matter: long non-coding RNAs in cardiac development and disease. EMBO J. 2013;32:1805-1816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 58. | Zhang YC, Chen YQ. Long noncoding RNAs: new regulators in plant development. Biochem Biophys Res Commun. 2013;436:111-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 59. | Niazi F, Valadkhan S. Computational analysis of functional long noncoding RNAs reveals lack of peptide-coding capacity and parallels with 3’ UTRs. RNA. 2012;18:825-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 134] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 60. | Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470:284-288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1072] [Cited by in RCA: 1005] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 61. | Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689-693. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2754] [Cited by in RCA: 2649] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 62. | Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621-629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 983] [Cited by in RCA: 958] [Article Influence: 68.4] [Reference Citation Analysis (0)] |
| 63. | Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A. Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull. 2013;97:69-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 291] [Article Influence: 24.3] [Reference Citation Analysis (1)] |
| 64. | Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013;34:613-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 320] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 65. | Song H, Sun W, Ye G, Ding X, Liu Z, Zhang S, Xia T, Xiao B, Xi Y, Guo J. Long non-coding RNA expression profile in human gastric cancer and its clinical significances. J Transl Med. 2013;11:225. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 166] [Cited by in RCA: 197] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 66. | Sun W, Wu Y, Yu X, Liu Y, Song H, Xia T, Xiao B, Guo J. Decreased expression of long noncoding RNA AC096655.1-002 in gastric cancer and its clinical significance. Tumour Biol. 2013;34:2697-2701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 76] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 67. | Xiao B, Guo J. Long noncoding RNA AC096655.1-002 has been officially named as gastric cancer-associated transcript 1, GACAT1. Tumour Biol. 2013;34:3271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 68. | Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu-lail R, Hochberg A, Galun E. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007;2:e845. [PubMed] |
| 69. | Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, Fang G. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159-3165. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 376] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 70. | Yang F, Xue X, Bi J, Zheng L, Zhi K, Gu Y, Fang G. Long noncoding RNA CCAT1, which could be activated by c-Myc, promotes the progression of gastric carcinoma. J Cancer Res Clin Oncol. 2013;139:437-445. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 228] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 71. | Mei D, Song H, Wang K, Lou Y, Sun W, Liu Z, Ding X, Guo J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med Oncol. 2013;30:709. [PubMed] |
| 72. | Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642-655. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4017] [Cited by in RCA: 3701] [Article Influence: 231.3] [Reference Citation Analysis (0)] |
| 73. | Jinek M, Doudna JA. A three-dimensional view of the molecular machinery of RNA interference. Nature. 2009;457:405-412. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 526] [Cited by in RCA: 528] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 74. | Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413-420. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 665] [Cited by in RCA: 557] [Article Influence: 34.8] [Reference Citation Analysis (0)] |
| 75. | Bayne EH, Allshire RC. RNA-directed transcriptional gene silencing in mammals. Trends Genet. 2005;21:370-373. [PubMed] |
| 76. | Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA. 2007;104:12422-12427. [PubMed] |
| 77. | Kawasaki H, Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211-217. [PubMed] |
| 78. | Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289-1292. [PubMed] |
| 79. | Kawasaki H, Taira K, Morris KV. siRNA induced transcriptional gene silencing in mammalian cells. Cell Cycle. 2005;4:442-448. [PubMed] |
| 80. | Ying Z, Li J, Li M. Astrocyte elevated gene 1: biological functions and molecular mechanism in cancer and beyond. Cell Biosci. 2011;1:36. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 81. | Jian-bo X, Hui W, Yu-long H, Chang-hua Z, Long-juan Z, Shi-rong C, Wen-hua Z. Astrocyte-elevated gene-1 overexpression is associated with poor prognosis in gastric cancer. Med Oncol. 2011;28:455-462. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 82. | Liu HT, Lu CL. Effect of silencing Bcl-2 expression by small interfering RNA on radiosensitivity of gastric cancer BGC823 cells. Asian Pac J Trop Med. 2013;6:49-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 83. | Cao Y, Tu Y, Mei J, Li Z, Jie Z, Xu S, Xu L, Wang S, Xiong Y. RNAi-mediated knockdown of PRL-3 inhibits cell invasion and downregulates ERK 1/2 expression in the human gastric cancer cell line, SGC-7901. Mol Med Rep. 2013;7:1805-1811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |