Published online Apr 28, 2014. doi: 10.3748/wjg.v20.i16.4761
Revised: February 25, 2014
Accepted: March 7, 2014
Published online: April 28, 2014
Processing time: 152 Days and 21.7 Hours
AIM: To investigate the rate of Helicobacter pylori (H. pylori) resistance to clarithromycin among ethnic minority patients in Guangxi, explore the underlying mechanisms, and analyze factors influencing genotype distribution of H. pylori isolates.
METHODS: H. pylori strains were isolated, cultured and subjected to drug sensitivity testing. The 23S rRNA gene of H. pylori isolates was amplified by PCR and analyzed by PCR-RFLP and direct sequencing to detect point mutations. REP-PCR was used for genotyping of H. pylori isolates, and NTsys_2 software was used for clustering analysis based on REP-PCR DNA fingerprints. Factors potentially influencing genotype distribution of H. pylori isolates were analyzed.
RESULTS: The rate of clarithromycin resistance was 31.3%. A2143G and A2144G mutations were detected in the 23S rRNA gene of all clarithromycin-resistant H. pylori isolates. At a genetic distance of 78%, clarithromycin-resistant H. pylori isolates could be divided into six groups. Significant clustering was noted among H. pylori isolates from patients with peptic ulcer or gastritis.
CONCLUSION: The rate of clarithromycin resistance is relatively high in ethnic minority patients in Guangxi. Main mechanisms of clarithromycin resistance are A2143G and A2144G mutations in the 23S rRNA gene. Clarithromycin-resistant H. pylori isolates can be divided into six groups based on REP-PCR DNA fingerprints. Several factors such as disease type may influence the genotype distribution of H. pylori isolates.
Core tip: The present study showed that the rate of Helicobacter pylori (H. pylori) resistance to clarithromycin was 31.3% in ethnic minority patients in Guangxi, slightly higher than that in Dongguan in 2009 but significantly higher than the reported resistance rate in 2008 in the same region (Guangxi). The significant increase in the rate of H. pylori resistance to clarithromycin in this region may be caused by the long-term and/or wide use of clarithromycin, which can decrease the populations of sensitive bacteria and promote the propagation of drug-resistant bacteria. Of note, this study also found that there existed multidrug resistant H. pylori strains (resistant to amoxicillin, metronidazole, tetracycline, and levofloxacin).
-
Citation: Zhao LJ, Huang YQ, Chen BP, Mo XQ, Huang ZS, Huang XF, Wei LD, Wei HY, Chen YH, Tang HY, Huang GR, Qin YC, Li XH, Wang LY.
Helicobacter pylori isolates from ethnic minority patients in Guangxi: Resistance rates, mechanisms, and genotype. World J Gastroenterol 2014; 20(16): 4761-4770 - URL: https://www.wjgnet.com/1007-9327/full/v20/i16/4761.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i16.4761
Helicobacter pylori (H. pylori) is a frequent cause of human chronic gastritis and peptic ulcer, and is also involved in the pathogenesis of gastric cancer and mucosa-associated lymphoid tissue lymphoma[1-4]. The wide application of antibiotics in H. pylori eradication therapy has led to the increasing prevalence of H. pylori resistance to antibiotics. Antibiotic resistance is a major cause of treatment failure. Due to the differences in doctors’ prescribing habits, patients’ medical history, and diets, the rates of H. pylori infection and antibiotic resistance vary among different countries or regions[5-10]. Therefore, monitoring and research of antibiotic-resistant H. pylori strains from different regions can help not only understand the status of antibiotic resistance and guide clinical medication, but also overcome antibiotic resistance, increase the rate of eradication and avoid the emergence of drug-resistant strains. Guangxi is home to many ethnic minorities besides the Han Chinese, including Zhuang, Yao and Miao. Particularly, Zhuang has the largest population. These ethnic minorities have distinct living and eating habits. Additionally, the level of economic development and the standard of living are relatively low in this region, and there are fewer types of antibiotics available. Therefore, it is possible that the rates of H. pylori infection and antibiotic resistance in this region are significantly different from those in other regions of China. In the present study, we investigated the rate of H. pylori resistance to clarithromycin among ethnic minority patients in this region, explored the mechanism of clarithromycin resistance, and analyzed factors potentially influencing clarithromycin resistance, with an aim to reduce the rate of H. pylori resistance to antibiotics and improve the effect of treatment.
Between May 2011 and May 2012, 164 gastric mucosal biopsies were collected from patients with gastritis or peptic ulcer at the Department of Gastroscopy, the Affiliated Hospital of Youjiang Medical University for Nationalities. The samples were inoculated into Columbia medium and cultured for 5-7 d in a microaerobic bag at 37 °C. The suspected strains were confirmed as H. pylori by Gram-staining and urease, oxidase and catalase tests.
The bacterial suspension was adjusted to a density of 1.0 × 108 CFU/mL and plated on the Columbia sheep blood agar. After the clarithromycin discs were plated, the plates were incubated at 37 °C for 5 d. The diameter of inhibition zone was measured. According to the criteria recommended by the 2012 Clinical and Laboratory Standards Institute (CLSI), strains were considered sensitive to clarithromycin when the diameter of inhibition zone was ≥ 17 mm and resistant to clarithromycin when the diameter was ≤ 13 mm.
Genomic DNA was isolated from H. pylori cells using a commercial kit (Generay Biotech, Shanghai, China) according to the manufacturer’s instructions. The A2144G and A2143G loci were amplified by PCR in a 50-μL reaction system consisting of 29.5 μL ddH2O, 6.3 μL 10 × PCR buffer, 5.0 μL dNTPs (25 mmol/L), 0.5 μL Taq polymerase (5 kU/L), 3.7 μL of each forward and reverse primer (10 μmol/L), and 1.3 μL DNA template. The primers were designed as previously described[5] and their sequences were 5′-CCA CAG CGA TGT GGT CTCAG-3′ (forward) and 5′-CTC CAT AAG AGC CAA AGCCC-3′ (reverse), which yields a fragment of 425 bp. PCR cycling parameters were pre-denaturation at 94.0 °C for 4 min, 32 cycles of denaturation at 94.0 °C for 40 s, annealing at 61.5 °C for 1 min, and extension at 72.0 °C for 1 min, and a final extension at 72.0 °C for 7 min. PCR products were resolved by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining under ultraviolet light.
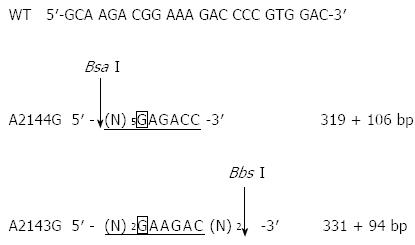
The two most common mutations associated with clarithromycin resistance (A2143G and A2144G) result in the generation of two new restriction sites for Bbs I and Bsa I. To examine whether these two mutations were present, the above PCR products (8 μL) were incubated with Bbs I at 37 °C for 24 h or with Bsa I at 50 °C for 24 h. After enzyme digestion, the reaction products (10 μL) were resolved by 1% agarose gel electrophoresis and visualized by ethidium bromide staining under ultraviolet light.
PCR products for 1 sensitive strain and 5 clarithromycin-resistant strains were randomly selected for DNA sequencing. DNA sequencing was performed by Generay Biotech (Shanghai, China). DNATool 6.0 program was used to analyze the 23S rRNA gene sequences of clarithromycin-sensitive and -resistant strains, and the sequences were compared with that of HPJ99 strain (NC-000921) deposited in the genome database (National Center for Biotechnology Information, NCBI).
REP-PCR was performed in a 25-μL reaction system consisting of 14.25 μL ddH2O, 2.5 μL 10 × PCR buffer, 0.5 μL dNTPs, 0.5 μL Taq polymerase (2 U), 0.5 μL of each forward and reverse primer, 0.25 μL MgC12 and 5 μL DNA template. The primers were designed as previously described[6] and their sequences were 5′-CGGICTAcIGCIGcIIII-3′ (forward) and 5′-ICGICITFATCIGGCCTAC-3′ (reverse), where I represents inosine. PCR cycling parameters were pre-denaturation at 95.0 °C for 30 s, 80 °C for 2 min, 65 cycles of denaturation at 95.0 °C for 30 s, annealing at 40 °C for 1 min, and extension at 65.0 °C for 8 min, and a final extension at 65.0 °C for 8 min. PCR products were resolved by 1.0% agarose gel electrophoresis and visualized by ethidium bromide staining under ultraviolet light.
Clustering analysis
Based on REP-PCR DNA fingerprints, the band was scored 1 if there was a mobility shift and 0 if there was not. NTsys_2 software was used for clustering analysis.
The rate of clarithromycin resistance is expressed as a percentage (%).
A total of 115 clinical isolates were identified as H. pylori, of which 82 were isolated from patients with peptic ulcer and 33 from patients with gastritis.
Drug sensitivity testing revealed that there were 36 H. pylori strains that were resistant to clarithromycin, and the rate of clarithromycin resistance was 31.3% (36/115).
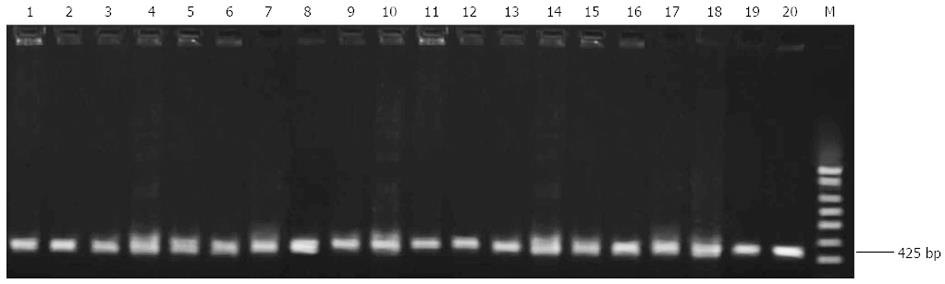
Ten each of clarithromycin-sensitive and -resistant strains were randomly selected for PCR-RFLP analysis of the 23S rRNA gene. A 425-bp fragment of interest was amplified in all clarithromycin-sensitive and -resistant strains (Figure 1).
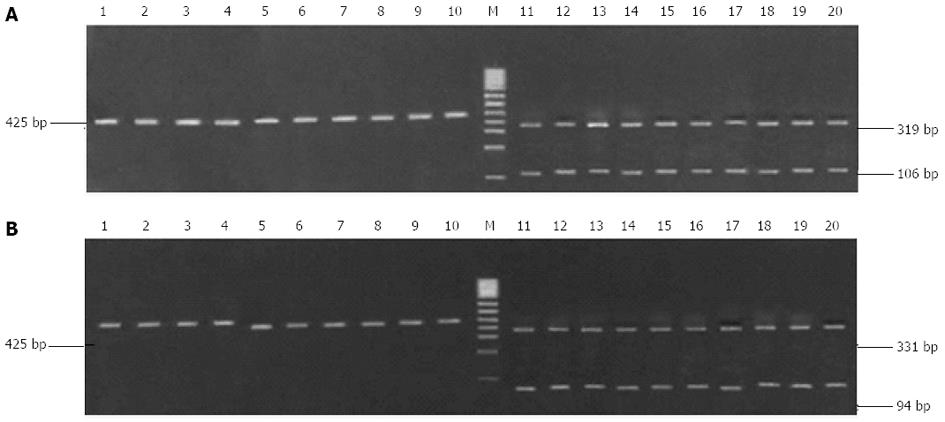
Clarithromycin resistance-associated mutations A2143G and A2144G result in the generation of two new restriction sites for Bbs I and Bsa I (Figure 2). Although the 425-bp fragment could be amplified from all H. pylori strains, only the fragment from the 10 clarithromycin-resistant strains could be digested by Bbs I and Bsa I, and the digestion resulted in the generation of two bands (319 bp and 106 bp for Bsa I digestion, and 331 bp and 94 bp for Bbs I digestion, Figure 3). This finding suggests the presence of A2143G and A2144G mutations in clarithromycin-resistant strains. In contrast, the fragment from the 10 clarithromycin-sensitive strains could not be digested by Bbs I and Bsa I, indicating the absence of A2143G and A2144G mutations in clarithromycin-sensitive strains (Figure 3). Of note, the 425-bp fragment could not be completely digested by both enzymes in all clarithromycin-resistant strains.
PCR products for 1 sensitive strain and 5 clarithromycin-resistant strains were randomly selected for DNA sequencing. The obtained DNA sequences were compared with that of HPJ99 strain deposited in the NCBI genome database. The results confirmed the presence of A2143G and A2144G mutations in clarithromycin-resistant strains, but not in clarithromycin-sensitive strains (Figure 4).
Twenty-six each of randomly selected clarithromycin-resistant and -sensitive H. pylori isolates and the standard strain ATCC43504 were subjected to REP-PCR analysis, and the results are shown in Tables 1 and 2 and Figure 5.
| No. | Strain | Gender | Age | Ethnicity | Region | Disease type | History of medication | Family history of gastric disease | Antibiotic resistance |
| 1 | GXHP039 | F | 46 | H | LY | PU | + | - | CA |
| 2 | GXHP051 | F | 46 | H | BS | PU | + | + | CA |
| 3 | GXHP071 | F | 39 | Ch | TY | CG | - | - | C |
| 4 | GXHP080 | F | 54 | Ch | BS | CG | + | - | CAS |
| 5 | GXHP043 | F | 47 | H | BS | PU | + | - | CA |
| 6 | GXHP093 | M | 46 | Ch | TL | CG | - | - | CASG |
| 7 | GXHP004 | F | 49 | Ch | TY | PU | - | - | C |
| 8 | GXHP014 | F | 46 | Ch | BS | PU | + | - | CAS |
| 9 | GXHP016 | F | 57 | Ch | BS | PU | - | + | CASG |
| 10 | GXHP019 | M | 52 | Ch | LY | PU | - | + | C |
| 11 | GXHP021 | M | 51 | Ch | TL | PU | + | - | CASG |
| 12 | GXHP023 | M | 48 | Ch | LY | PU | + | - | CAG |
| 13 | GXHP026 | M | 41 | Ch | TY | PU | - | - | C |
| 14 | GXHP121 | M | 46 | H | TY | CG | + | - | C |
| 15 | GXHP085 | M | 53 | Ch | TY | CG | - | + | C |
| 16 | GXHP034 | M | 51 | Ch | BS | PU | - | - | CAG |
| 17 | GXHP090 | M | 41 | Ch | BS | CG | - | - | C |
| 18 | GXHP067 | M | 41 | H | BS | PU | + | - | C |
| 19 | GXHP056 | M | 39 | H | TY | PU | - | + | CA |
| 20 | GXHP100 | F | 47 | H | LY | CG | - | - | CA |
| 21 | GXHP104 | F | 46 | H | TY | CG | + | - | CG |
| 22 | GXHP107 | F | 56 | H | BS | CG | - | + | CAS |
| 23 | GXHP110 | F | 59 | H | BS | CG | + | + | CAG |
| 24 | GXHP113 | M | 46 | H | TL | CG | - | + | C |
| 25 | GXHP115 | M | 53 | H | LY | CG | - | + | CAG |
| 26 | GXHP120 | M | 46 | H | BS | CG | + | + | C |
| No. | Strain | Gender | Age | Ethnicity | Region | Disease type | History of medication | Family history of gastric disease |
| 1 | GXHP069 | M | 46 | Ch | TL | CG | + | + |
| 2 | GXHP070 | M | 43 | Ch | LY | CG | + | + |
| 3 | GXHP096 | F | 36 | Ch | BS | CG | - | + |
| 4 | GXHP119 | F | 48 | H | TY | CG | - | - |
| 5 | GXHP094 | F | 46 | Ch | BS | CG | + | + |
| 6 | GXHP074 | M | 47 | Ch | TL | CG | + | - |
| 7 | GXHP116 | F | 46 | H | BS | CG | - | + |
| 8 | GXHP075 | M | 45 | Ch | LY | CG | - | - |
| 9 | GXHP076 | M | 48 | Ch | TY | CG | - | + |
| 10 | GXHP079 | M | 53 | Ch | TL | CG | + | + |
| 11 | GXHP078 | M | 52 | Ch | BS | CG | + | + |
| 12 | GXHP101 | M | 42 | H | BS | CG | - | + |
| 13 | GXHP102 | M | 56 | H | BS | CG | + | - |
| 14 | GXHP005 | M | 35 | Ch | BS | PU | - | + |
| 15 | GXHP006 | M | 41 | Ch | TL | PU | - | + |
| 16 | GXHP007 | M | 60 | Ch | TL | PU | + | + |
| 17 | GXHP008 | M | 38 | Ch | BS | PU | - | - |
| 18 | GXHP024 | F | 50 | Ch | TL | PU | + | + |
| 19 | GXHP025 | F | 39 | Ch | BS | PU | - | - |
| 20 | GXHP027 | F | 59 | Ch | BS | PU | - | - |
| 21 | GXHP040 | M | 51 | H | BS | PU | + | + |
| 22 | GXHP041 | M | 52 | H | BS | PU | + | + |
| 23 | GXHP052 | F | 53 | H | BS | PU | - | - |
| 24 | GXHP053 | F | 47 | H | BS | PU | + | + |
| 25 | GXHP054 | F | 49 | H | BS | PU | - | - |
| 26 | GXHP050 | M | 72 | H | LY | PU | - | + |
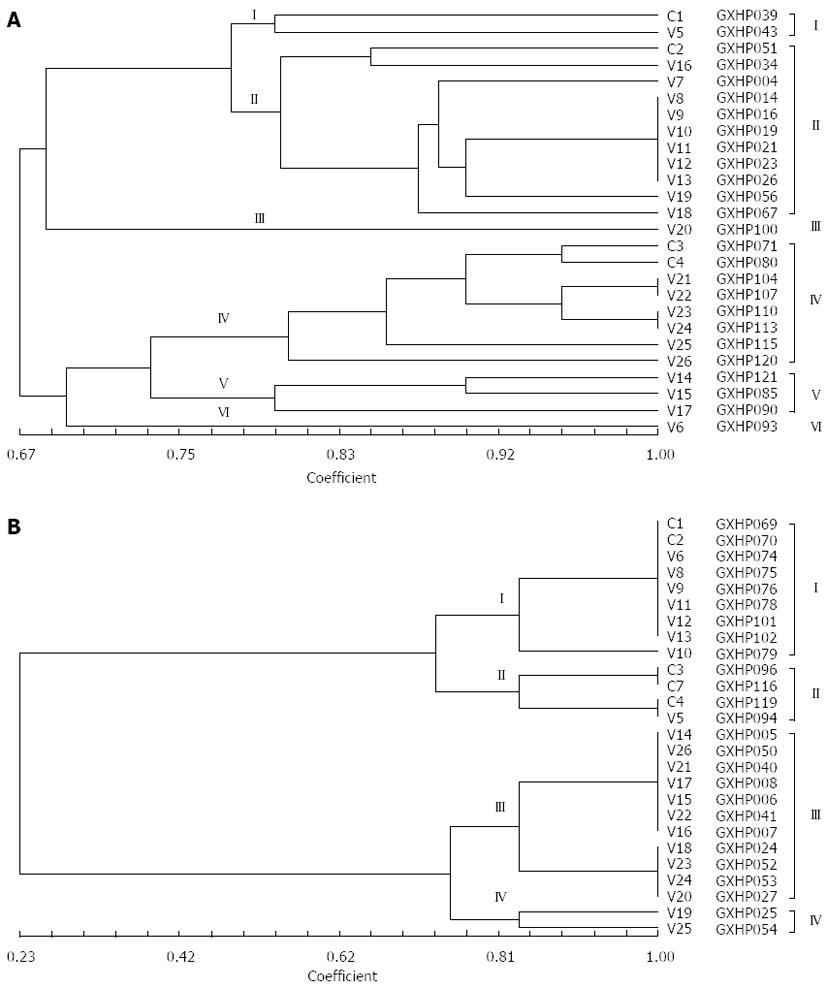
The NTsys-2 software was used to analyze the similarity among the randomly selected clarithromycin-resistant and -sensitive H. pylori isolates. At a genetic distance of 78%, 26 clarithromycin-resistant H. pylori isolates could be classified into 6 genotypes (Figure 6), and 26 clarithromycin-sensitive H. pylori isolates could be classified into 4 genotypes (Figure 6).
Factors influencing genotype distribution of H. pylori isolates may include disease type, ethnicity, gender, age, region, history of antibiotic medication, history of gastric diseases, and multidrug resistance. The frequencies of the presence of these factors in the patients are presented in Tables 3 and 4.
| CG | PU | Ch | H | M | F | TY | TL | LY | BS | HMR | HM | FHGD | 30-40 yr | 41-50 yr | 51-60 yr | |
| Group I | 0 | 2 | 0 | 2 | 2 | 0 | 0 | 0 | 1 | 1 | 2 | 2 | 0 | 0 | 2 | 0 |
| Group II | 0 | 11 | 8 | 3 | 4 | 7 | 3 | 1 | 2 | 5 | 4 | 5 | 4 | 1 | 6 | 4 |
| Group III | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 1 | 0 | 1 | 0 |
| Group IV | 8 | 0 | 2 | 6 | 5 | 3 | 2 | 1 | 1 | 4 | 4 | 4 | 5 | 1 | 3 | 4 |
| Group V | 3 | 0 | 2 | 1 | 0 | 3 | 2 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 2 | 1 |
| Group VI | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 |
| CG | PU | Ch | H | M | F | TY | TL | LY | BS | HM | FHGD | 30-40 yr | 41-50 yr | 51-60 yr | |
| Group I | 9 | 0 | 7 | 2 | 0 | 9 | 1 | 2 | 2 | 4 | 2 | 6 | 0 | 6 | 3 |
| Group II | 4 | 0 | 2 | 2 | 4 | 0 | 1 | 0 | 0 | 3 | 1 | 3 | 1 | 3 | 0 |
| Group III | 0 | 11 | 5 | 6 | 4 | 7 | 1 | 2 | 1 | 7 | 3 | 7 | 3 | 3 | 5 |
| Group IV | 0 | 2 | 1 | 1 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 |
Antibiotic resistance is a main factor affecting therapeutic effects in patients with H. pylori infection. Particularly, the rates of H. pylori resistance to clarithromycin and metronidazole have been increasing year by year, and accordingly, the rate of H. pylori eradication achieved with regimens containing either of the two antibiotics becomes lower and lower. The rate of H. pylori resistance to antibiotics varies among different countries or regions, although it shows an upward trend[11-15]. From 1996 to 2004 in Japan, the prevalence of H. pylori resistance to clarithromycin has increased to 30%[16]. In Vietnam in 2008[17], the rate of clarithromycin resistance was 33%, and the rate in Ho Chi Minh City (49%) was obviously higher than that in Hanoi (18.5%). In China, the rate of H. pylori resistance to antibiotics varies among different regions. For example, a study on H. pylori resistance to antibiotics in Beijing, Shanghai and Wenzhou[18] showed that the rate of resistance to metronidazole was highest, followed by clarithromycin. In 2009 in Dongguan, the rate of H. pylori resistance to clarithromycin was 27.6%[19].
The present study showed that the rate of H. pylori resistance to clarithromycin was 31.3% in ethnic minority patients in Guangxi, slightly higher than that in Dongguan in 2009 but significantly higher than the reported resistance rate in 2008 in the same region (Guangxi). The significant increase in the rate of H. pylori resistance to clarithromycin in this region may be caused by the long-term and/or wide use of clarithromycin, which can decrease the populations of sensitive bacteria and promote the propagation of drug-resistant bacteria. Of note, this study also found that there existed multidrug resistant H. pylori strains (resistant to amoxicillin, metronidazole, tetracycline, and levofloxacin)[20]. Therefore, we recommend that, in order to improve the rate of H. pylori eradication, tests for H. pylori antibiotic resistance should be established in this region, sensitive antibiotics should be selected based on antibiotic sensitivity of H. pylori strains, and the research on the mechanisms of antibiotic resistance should be enhanced.
Clarithromycin, as a new generation of macrolide antibiotic, is acid-stable and can be dissolved in gastric juice that has a low pH level. Therefore, it has good oral bioavailability and its concentration is high in the gastric mucosa. Additionally, clarithromycin has few adverse reactions. Since clarithromycin monotherapy can achieve an eradication rate between 42%-54%, it is one of the currently known antibiotics that have the strongest effect on H. pylori. Compared with triple therapy without clarithromycin, clarithromycin-containing triple therapy can increase the rate of H. pylori eradication by 10%-20%. Therefore, clarithromycin is the main antibiotic used in regimens for H. pylori. However, significant resistance of H. pylori to clarithromycin has been observed, and the rate of clarithromycin resistance has increased, especially after an initial failure[21-23]. Clarithromycin exerts antibacterial effects by penetrating the bacteria cell wall, binding to the domain V of the 23S ribosomal RNA of the 50S subunit of the bacterial ribosome, inhibiting peptidyl transferase activity, interfering with amino acid translocation and thereby suppressing bacterial protein synthesis. With regard to the mechanisms of H. pylori resistance to clarithromycin, the consensus view is point mutations in the domain V of the 23S ribosomal RNA, which reduce the binding force between clarithromycin and the ribosome. Most of the discovered mutations are A to G transition mutations, and currently known mutations include A2144G, A2143G, A2142G, A2142C, G2115A, G214lA, A2142T and A2143C. Some mutations can occur even in other chromosomal segments. Since the minimum inhibitory concentrations of clarithromycin are higher in H. pylori strains bearing A2144G and A2143G mutations, these strains have more stable resistance and higher growth rate[24-26]. Therefore, A2143G and A2144G mutations are most common in clinically isolated clarithromycin-resistant H. pylori strains. However, point mutations in clarithromycin-resistant H. pylori strains can vary among different regions and different ethnicity groups.
This study detected the A2143G and A2144G mutations in the 23S rRNA gene in H. pylori isolates using PCR-RFLP, which is a simple and high-sensitivity method. Both mutations were detected in all 10 randomly selected clarithromycin-resistant isolates, but not in clarithromycin-sensitive isolates. Direct DNA sequencing of the 23S rRNA gene in 5 randomly selected clarithromycin-resistant isolates confirmed the above finding. Therefore, A2143G and A2144G mutations are closely related to H. pylori resistance to clarithromycin in ethnic minority patients in Guangxi. Future studies should develop methods to repair or avoid these mutations to reduce the rate of clarithromycin resistance and improve the effectiveness of prevention and control.
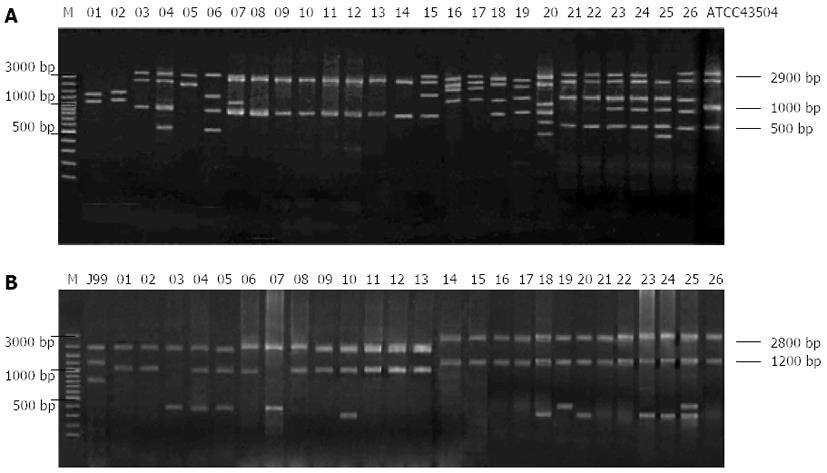
To investigate the association of genotypes of H. pylori isolates from ethnic minority patients in Guangxi with disease type, ethnicity and multidrug resistance, REP-PCR was used to genotype clarithromycin-resistant H. pylori isolates from patients of different ethnicity or with different disease types, and the results showed that strains isolated from different patients had different DNA fingerprints (Figure 5A).
The NTsys_2 software was used to perform clustering analysis. The dendrogram of REP-PCR DNA fingerprints for clarithromycin-resistant H. pylori strains (Figure 6A) showed a similarity of 100% among GXHP014, GXHP016, GXHP019, GXHP021, GXHP023 and GXHP026; between GXHP104 and GXHP107; and between GXHP110 and GXHP113. These three groups shared a similarity of 90%. In addition, GXHP104 and GXHP107 had a similarity of 95.2% to GXHP110 and GXHP113. At a genetic distance of 78%, the 26 strains of clarithromycin-resistant H. pylori were divided into 6 groups, which are as follows: (1) group I: This group includes GXHP039 and GXHP043, which were isolated from a woman with peptic ulcer in Baise and a woman with the same disease in Lingyun, respectively. The two women, ranging in age between 41 and 50 years old, had a history of clarithromycin use, but their family history of gastric disease was unknown. The two strains were also resistant to amoxicillin, streptomycin, and gentamicin; (2) group II: This group includes GXHP051, GXHP034, GXHP004, GXHP014, GXHP016, GXHP019, GXHP021, GXHP023, GXHP026, GXHP056 and GXHP067. All the 11 strains were isolated from patients with peptic ulcer. GXHP034, GXHP004, GXHP014, GXHP016, GXHP019, GXHP021, GXHP023, GXHP026 and GXHP056 came from 9 Zhuang patients (6 females and 3 males) in Baise, Tianlin, Lingyun, or Tianyang. They had an unknown history of medication or a family history of gastric diseases. GXHP014, GXHP019, GXHP021 and GXHP026 were also resistant to amoxicillin and streptomycin. GXHP051, GXHP056 and GXHP067 came from 3 Han patients in Baise or Tianlin, all of whom had a history of clarithromycin use and a family history of gastric disease. These three strains were also resistant to amoxicillin, streptomycin and gentamicin. In this group, one patient was in the 30-40 age group, 6 in the 41-50 age group, and 4 in the 51-60 age group; (3) group III: This group includes only GXHP100. This strain came from a Han male with chronic gastritis in Lingyun. He had a history of clarithromycin use and family history of gastric disease. The strain was also resistant to amoxicillin. The patient was in the 41-50 age group; (4) group IV: This group includes GXHP071, GXHP080, GXHP104, GXHP107, GXHP110, GXHP113, GXHP113, GXHP115 and GXHP120. The 8 strains came from 8 patients with chronic gastritis (5 males and 3 females) in Baise, Tianlin, Lingyun, or Tianyang. There were 6 Han patients and 2 Zhuang patients. Four patients had a history of clarithromycin use and five patients had a family history of gastric diseases. Four strains were also resistant to amoxicillin and gentamicin. In this group, one patient was in the 30-40 age group, 3 in the 41-50 age group, and 4 in the 51-60 age group; (5) group V: This group includes GXHP085, GXHP090 and GXHP121. All 3 strains came from 3 females with chronic gastritis in Baise or Tianlin. There were 1 Han patient and 2 Zhuang patients. One patient had both a history of clarithromycin use and a family history of gastric diseases. One strain was also resistant to amoxicillin and gentamicin. In this group, 2 patients were in the 41-50 age group, and 1 in the 51-60 age group; and (6) group VI: This group includes only GXHP093. This strain came from a Zhuang female with chronic gastritis in Tianlin. She had an unknown history of clarithromycin use or family history of gastric diseases. The strain was also resistant to amoxicillin and streptomycin. The patient was in the 41-50 age group.
Based on the above data, it can be found that factors influencing genotype distribution include disease type, ethnicity, gender, age, region, history of medication, family history of gastric disease, and multidrug resistance. The frequencies of the presence of these factors influencing genotype distribution of clarithromycin-resistant strains are shown in Table 3. Main factors influencing genotype distribution of clarithromycin-resistant strains may be: (1) disease type: Gastritis and peptic ulcer were main influencing factors, because group II and group IV strains were isolated from patients with gastritis and those with peptic ulcer, respectively; (2) ethnicity: Zhuang and Han were main influencing factors, because GXHP014, GXHP016, GXHP019, GXHP021, GXHP023 and GXHP026, which shared a similarity of 100%, were all isolated from Zhuang patients; GXHP104 and GXHP107, which shared a similarity of 100%, as well as GXHP110 and GXHP113, which also shared a similarity of 100%, were all isolated from Han patients; (3) family history of gastric disease: Since GXHP104 and GXHP107, which shared a similarity of 100%, were isolated from Han patients with a family history of gastric diseases, while GXHP110 and GXHP113, which also shared a similarity of 100%, were isolated from Han patients without a family history of gastric diseases, we speculate that family history of gastric disease may be associated with the genotype distribution; (4) multidrug resistance: Since the frequency of the presence of multidrug resistance is high (50%-100%) in H. pylori isolates of various genotypes, there may exist an association between multidrug resistance and the genotype distribution.
We also analyzed the genotypes of clarithromycin-sensitive H. pylori strains using the same method and found that the main factors influencing the genotype distribution of clarithromycin-sensitive strains were disease type and patient gender (Figures 5B and 6B, Table 2). Compared with clarithromycin-resistant strains, the genotype distribution of clarithromycin-sensitive strains was influenced by fewer factors. This discrepancy may be partly explained by the genetic mutations in clarithromycin-resistant strains. In addition, we found that region, age and history of medication had relatively small impact on genotype distribution. However, since the number of strains analyzed in the present study is relatively small, we could not use statistical methods to analyze the influence of various factors on genotype distribution. Future studies should carefully address this issue.
Helicobacter pylori (H. pylori) is a frequent cause of human chronic gastritis and peptic ulcer, and is also involved in the pathogenesis of gastric cancer and mucosa-associated lymphoid tissue lymphoma. The wide application of antibiotics in H. pylori eradication therapy has led to the increasing prevalence of H. pylori resistance to antibiotics. Antibiotic resistance is a major cause of treatment failure.
Antibiotic resistance is a main factor affecting therapeutic effects in patients with H. pylori infection. Particularly, the rates of H. pylori resistance to clarithromycin and metronidazole have been increasing year by year, and accordingly, the rate of H. pylori eradication achieved with regimens containing either of the two antibiotics becomes lower and lower.
The rate of clarithromycin resistance is relatively high in ethnic minority patients in Guangxi. Main mechanisms of clarithromycin resistance are A2143G and A2144G mutations in the 23S rRNA gene. Clarithromycin-resistant H. pylori isolates can be divided into six groups based on REP-PCR DNA fingerprints. Several factors such as disease type may influence the genotype distribution of H. pylori isolates.
The manuscript is very interesting. The authors try to investigate the rate of H. pylori resistance to clarithromycin among ethnic minority patients in Guangxi, the home to many ethnic minorities besides the Han Chinese. The research is well designed. The data are very interesting.
P- Reviewers: Chorny M, Wenzel SE S- Editor: Qi Y L- Editor: Logan S E- Editor: Wang CH
| 1. | Gisbert JP. Helicobacter pylori-related diseases. Gastroenterol Hepatol. 2012;35 Suppl 1:12-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 2. | Cid TP, Fernández MC, Benito Martínez S, Jones NL. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2013;18 Suppl 1:12-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 3. | Ueda J, Gosho M, Inui Y, Matsuda T, Sakakibara M, Mabe K, Nakajima S, Shimoyama T, Yasuda M, Kawai T. Prevalence of Helicobacter pylori Infection by Birth Year and Geographic Area in Japan. Helicobacter. 2014;19:105-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 73] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 4. | Adlekha S, Chadha T, Krishnan P, Sumangala B. Prevalence of helicobacter pylori infection among patients undergoing upper gastrointestinal endoscopy in a medical college hospital in kerala, India. Ann Med Health Sci Res. 2013;3:559-563. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 5. | O’Connor A, O’Moráin C. Helicobacter pylori infection in Europe: current perspectives. Expert Rev Gastroenterol Hepatol. 2013;7:541-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 12] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 6. | Thirumurthi S, Graham DY. Helicobacter pylori infection in India from a western perspective. Indian J Med Res. 2012;136:549-562. [PubMed] |
| 7. | Vilaichone RK, Yamaoka Y, Shiota S, Ratanachu-ek T, Tshering L, Uchida T, Fujioka T, Mahachai V. Antibiotics resistance rate of Helicobacter pylori in Bhutan. World J Gastroenterol. 2013;19:5508-5512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 11] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | Altman E, Harrison BA, Chandan V, Slinger R. Lipopolysaccharide glycotyping of clarithromycin-resistant and clarithromycin-susceptible Canadian isolates of Helicobacter pylori. Can J Microbiol. 2014;60:35-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 9. | Ierardi E, Giorgio F, Losurdo G, Di Leo A, Principi M. How antibiotic resistances could change Helicobacter pylori treatment: A matter of geography? World J Gastroenterol. 2013;19:8168-8180. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 80] [Cited by in RCA: 88] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 10. | Kim BJ, Kim JG. Substitutions in penicillin-binding protein 1 in amoxicillin-resistant Helicobacter pylori strains isolated from Korean patients. Gut Liver. 2013;7:655-660. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 11. | An B, Moon BS, Kim H, Lim HC, Lee YC, Lee G, Kim SH, Park M, Kim JB. Antibiotic resistance in Helicobacter pylori strains and its effect on H. pylori eradication rates in a single center in Korea. Ann Lab Med. 2013;33:415-419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 12. | Abdollahi H, Savari M, Zahedi MJ, Moghadam SD, Hayatbakhsh Abasi M. Detection of A2142C, A2142G, and A2143G Mutations in 23s rRNA Gene Conferring Resistance to Clarithromycin among Helicobacter pylori Isolates in Kerman, Iran. Iran J Med Sci. 2011;36:104-110. [PubMed] |
| 13. | Vega AE, Cortiñas TI, Puig ON, Silva HJ. Molecular characterization and susceptibility testing of Helicobacter pylori strains isolated in western Argentina. Int J Infect Dis. 2010;14 Suppl 3:e85-e92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Kim MS, Kim N, Kim SE, Jo HJ, Shin CM, Park YS, Lee DH. Long-term follow up Helicobacter Pylori reinfection rate after second-line treatment: bismuth-containing quadruple therapy versus moxifloxacin-based triple therapy. BMC Gastroenterol. 2013;13:138. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 15. | Molina-Infante J, Gisbert JP. Update on the efficacy of triple therapy for Helicobacter pylori infection and clarithromycin resistance rates in Spain (2007-2012). Gastroenterol Hepatol. 2013;36:375-381. [PubMed] |
| 16. | Horiki N, Omata F, Uemura M, Suzuki S, Ishii N, Iizuka Y, Fukuda K, Fujita Y, Katsurahara M, Ito T. Annual change of primary resistance to clarithromycin among Helicobacter pylori isolates from 1996 through 2008 in Japan. Helicobacter. 2009;14:86-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 89] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 17. | Binh TT, Shiota S, Nguyen LT, Ho DD, Hoang HH, Ta L, Trinh DT, Fujioka T, Yamaoka Y. The incidence of primary antibiotic resistance of Helicobacter pylori in Vietnam. J Clin Gastroenterol. 2013;47:233-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 47] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 18. | Wang YJ, Wang L, Du JM, Wang XH. Analysis of antibiotic resistance of Helicobacter pylori isolates in Wenzhou. Zhejiang Linchuang Yixue Zazhi. 2008;10:410-411. |
| 19. | Cai YF, Chen LQ, Yin L, Shen YJ, Liu Y. Analysis of antibiotic resistance of Helicobacter pylori isolates in Dongguan. Dangdai Yixue. 2010;16:90-91. |
| 20. | Li XH, Huang ZS, Huang YQ, Zhou XH, Wu MH, Liu L, Qin YC, Huang GR. Status of multidrug resistance of Helicobacter pylori in Guixi and analysis of treatment regimens. Dangdai Yixue. 2013;42:1578-157. |
| 21. | Wu JY, Liou JM, Graham DY. Evidence-based recommendations for successful Helicobacter pylori treatment. Expert Rev Gastroenterol Hepatol. 2014;8:21-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 22. | Vilaichone RK, Gumnarai P, Ratanachu-Ek T, Mahachai V. Nationwide survey of Helicobacter pylori antibiotic resistance in Thailand. Diagn Microbiol Infect Dis. 2013;77:346-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 23. | Giorgio F, Principi M, De Francesco V, Zullo A, Losurdo G, Di Leo A, Ierardi E. Primary clarithromycin resistance to Helicobacter pylori: Is this the main reason for triple therapy failure? World J Gastrointest Pathophysiol. 2013;4:43-46. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 24. | Ho SL, Tan EL, Sam CK, Goh KL. Clarithromycin resistance and point mutations in the 23S rRNA gene in Helicobacter pylori isolates from Malaysia. J Dig Dis. 2010;11:101-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Lee HJ, Kim JI, Cheung DY, Kim TH, Jun EJ, Oh JH, Chung WC, Kim BW, Kim SS, Park SH. Eradication of Helicobacter pylori according to 23S ribosomal RNA point mutations associated with clarithromycin resistance. J Infect Dis. 2013;208:1123-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 26. | Zhen-Hua Z, De-Qiang H, Yong X, Lin-Lin L, Nong-Hua L. Characterization of 23S rRNA gene mutation in primary and secondary clarithromycin-resistant Helicobacter pylori strains from East China. Turk J Gastroenterol. 2013;24:5-9. [PubMed] |