Published online Mar 21, 2014. doi: 10.3748/wjg.v20.i11.3025
Revised: December 5, 2013
Accepted: January 2, 2014
Published online: March 21, 2014
Processing time: 208 Days and 17.5 Hours
AIM: To investigate the potential of serum peptides as a diagnostic tool for hepatocellular carcinoma (HCC) with bone metastasis.
METHODS: Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) was used to characterize the serum peptide profile of HCC patients with bone metastasis. Serum samples from 138 HCC patients (66 cases with and 72 cases without bone metastasis) were randomly assigned into a training set (n = 76) and a test set (n = 62). Differential serum peptides were examined using ClinProt magnetic bead-based purification followed by MALDI-TOF-MS. The sequences of differentially expressed serum peptides were identified using liquid chromatography-mass spectrometry. A diagnostic model was established using a learning algorithm of radial basis function neural network verified by a single blind trial. Receiver operating characteristic (ROC) analysis was performed to evaluate the diagnostic power of the established model.
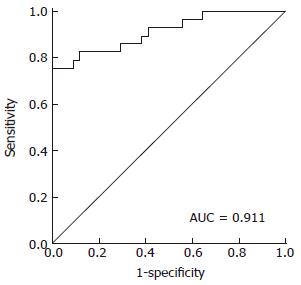
RESULTS: Ten peptide peaks were significantly different between HCC patients with or without bone metastasis (P < 0.001). Sequences of seven peptides with mass to charge ratios (m/z) of 1780.7, 1866.5, 2131.6, 2880.4, 1532.4, 2489.8, and 2234.3 were successfully identified. These seven peptides were derived from alpha-fetoprotein, prothrombin, serglycin, isoform 2 of inter-alpha-trypsin inhibitor heavy chain H4, isoform 1 of autophagy-related protein 16-2, and transthyretin and fibrinogen beta chains, respectively. The recognition rate and predictive power of a diagnostic model established on the basis of six significant peptides (m/z for these six peptides were 1535.4, 1780.7, 1866.5, 2131.6, 2880.4, and 2901.9) were 89.47% and 82.89%, respectively. The sensitivity and specificity of this model based upon a single blind trial were 85.29% and 85.71%, respectively. ROC analysis found that the AUC (area under the ROC curve) value was 0.911.
CONCLUSION: Our study suggested that serum peptides may serve as a diagnosis tool for HCC bone metastasis.
Core tip: Advances in the diagnosis and treatment of hepatocellular carcinoma (HCC) and extension of overall survival rates have led to an increase in the incidence of bone metastasis of HCC, which accounts for approximately 38.5% of all cases of extrahepatic metastasis of HCC. The patient’s quality of life would significantly be affected by the time of diagnosis. However, early diagnostic molecular markers for bone metastasis from HCC have not yet been identified. In this study, we performed differential serum peptide profiling in HCC patients with bone metastasis, which can be used for early prediction of this disorder.
- Citation: He J, Zeng ZC, Xiang ZL, Yang P. Mass spectrometry-based serum peptide profiling in hepatocellular carcinoma with bone metastasis. World J Gastroenterol 2014; 20(11): 3025-3032
- URL: https://www.wjgnet.com/1007-9327/full/v20/i11/3025.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i11.3025
The incidence of primary hepatocellular carcinoma (HCC) in male and female patients with malignant tumors is ranked globally at the fifth and eighth place, respectively[1]. The median survival period for HCC patients with bone metastasis is only 5-7.4 mo[2] due to bone metastasis resulting in severe bone pain, pathologic fracture, neurological deficits, and even paralysis. The long-term outcome of HCC patients with bone metastasis receiving external beam radiotherapy is poor, and most patients die within 1 year[3]. Advances in the diagnosis and treatment of HCC and extension of overall survival rates have led to an increase in the incidence of bone metastasis of HCC, which accounts for approximately 38.5% of all cases of extrahepatic metastasis of HCC[4]. However, molecular markers for bone metastasis from HCC have not yet been identified. Relying primarily on clinical imaging tools, diagnosis of bone metastasis from HCC is based on radiological imaging studies which have both low sensitivity and low specificity. Moreover, by the time bone metastasis images are obtained, patients often already have developed paralysis extensively in the body due to pain, dysfunction, and metastasis. Because the outcomes of palliative treatment for bone metastasis of HCC are dependent on its diagnosis, the patient’s quality of life would significantly be affected by the time of diagnosis. In this study, we investigated differential serum peptide profiling in HCC patients with bone metastasis using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS), to identify potential serum biomarkers for diagnosis of bone metastasis, which can be used for early molecular prediction of this disorder[5].
All serum specimens were obtained from patients with and without bone metastasis from primary HCC between June 2009 and December 2012 at the Department of Radiation Oncology, Zhongshan Hospital, Fudan University. Approval for collecting and processing serum samples was obtained from the research ethics committee of Zhongshan Hospital, and written informed consent was obtained from each patient. Criterion for HCC with bone metastasis was diagnosis with bone metastasis for the first time with no other distant metastasis. Criterion for HCC without bone metastasis was first time diagnosis with HCC with radiological examination showing no bone or other distant metastasis. None of the patients received radiotherapy, chemotherapy, or other intervention treatments before the collection of serum samples. Patient characteristics are summarized in Table 1. Among 66 patients with bone metastasis, 55 bone cases occurred after liver resection, and 11 cases occurred without resection. The cases included 46 men and 20 women, with the age ranging from 23 to 84 years, and with a median age of 58 years. Among the other 72 patients without bone metastasis, 58 underwent resection, and 14 patients did not. These cases included 53 men and 19 women, with an age range of 22-78 years, and a median age of 59 years. One hundred thirty-eight serum samples were randomly assigned into two groups: a training set of 76 cases, including 38 patients with bone metastasis and 38 cases without bone metastasis, and a test set of 62 cases with HCC, including 28 patients with bone metastasis and 34 cases without bone metastasis.
| Variable | With bone metastasis (n = 66) | Without bone metastasis (n = 72) |
| Age, yr | ||
| Range | 23-84 | 22-78 |
| Mean | 58 | 59 |
| Sex | ||
| Male | 46 (70) | 53 (71) |
| Female | 20 (30) | 19 (29) |
| Liver resection | ||
| With | 55 (83) | 58 (81) |
| Without | 11 (17) | 14 (19) |
| Set | ||
| Training set | 38 (58) | 38 (53) |
| Validation set | 28 (42) | 34 (47) |
| TNM stage of HCC | ||
| I | 0 | 0 |
| II | 0 | 8 |
| III | 0 | 64 |
| IV | 66 | 0 |
| Method for | ||
| metastasis detection1 | / | |
| MRI/CT | 54 | |
| ECT/SPET-CT | 66 | |
| PET-CT | 12 |
According to biopsy and postoperative specimen results, the 138 patients with and without bone metastasis were all diagnosed with HCC, of whom 116 were diagnosed by postoperative pathological examination, and 22 by biopsy.
Diagnostic criteria for bone metastasis and bone metastasis-free HCC were as follows: (1) diagnosis of primary tumors based on pathology; and (2) diagnosis of bone metastasis or no bone metastasis based upon clinical symptoms, isotopes and MRI/CT/X-rays, or whether or not SPET-CT/PET-CT detected bone metastasis.
Two milliliters of blood from fasting patients were obtained from each of the 66 HCC cases with bone metastasis, then placed into EDTA anticoagulant tubes at 4 °C, incubated for one hour, and centrifuged at 8000 ×g for 5 min. The supernatant was stored at -80 °C in aliquots of 20 μL. Paired serum samples of 72 patients without bone metastasis as the control group were processed in an identical manner.
Serum peptides were enriched using WCX beads (ClinProt bead Kit, Bruker Daltonics, United States) according to the manufacturer’s instructions, and then subjected to MALDI-TOF-MS analysis.
Seven samples were processed simultaneously with standard serum (Sigma-Aldrich, St. Louis, MO, United States). Standard serum was used as the quality control for experiments using magnetic beads, as well as other experiments throughout the study. Eluent data from standard serum magnetic bead processing were collected using an auto flex mass spectrum instrument (Bruker Daltonics), then compared with data of standard serum, using the same magnetic beads and collection methods. In this manner the instrument parameters were adjusted to obtain a relative standard deviation (CV) < 30%, to ensure that the experimental protocol had good reproducibility and met operational process standards.
Enriched peptide samples were mixed with matrix (alpha-cyano-4-hydroxycinnamic acid (CHCA; Sigma-Aldrich), and after crystallization data were acquired using the auto flex mass spectrometer. Scan range was from 1000 to 20000 Daltons (Da) in positive mode, and laser frequency was 20 Hz. A 50% high energy laser was used to irradiate each crystal point eight times, then a 30% low energy laser was used to capture 50 times. The data was summarized four times and saved.
ClinProTools software (version 2.2, Bruker Daltonics) was used to analyze mass to charge ratio (m/z) and intensity of all peptide spectra, consisting of baseline deductions, normalization of the spectra, and chromatographic peak alignment correction. Wilcoxon-test method in the ClinProTools software was used to determine the m/z of peptides with significant differences [P < 0.001, (Wilcoxon-test)].
Six different peaks showing significant differences were selected between the two groups (38 patients with bone metastasis from HCC and 38 HCC patients without bone metastasis) using the ClinProTools software (ClinProt software version 2.0; Bruker Daltonics), and a diagnostic model was established by RBFNN-based model to filter out patients at a high risk with bone metastasis from HCC. This model was validated by a 10-fold cross validation method, which randomly selected eight case samples (about 10%) as test samples, and utilized other samples as training sets for model validation. The validation process was repeated 10 times. Sera from the validation group of 28 patients with bone metastasis from HCC and 34 HCC patients without bone metastasis underwent single blind experimental verification. Receiver operating characteristic (ROC) analysis was performed to evaluate the diagnostic power of the established model.
Significant differences in peptide characteristics were identified using liquid chromatography-mass spectrometry, which combined Nano Acquity UPLC liquid chromatography (Waters, United States) with an LTQ Orbitrap XL mass spectrometer system (Thermo Fisher Scientific, United States). Peptide trapping was performed using a captrap C18 (2 mm × 0.5 mm) column (Michrom Corporation, United States), and an analytical Magic C18, AQ (100 μm × 150 mm) column (Michrom Corporation). Mobile phase A was a solution of 5% acetonitrile and 0.1% formic acid, and mobile phase B was a solution of 90% acetonitrile and 0.1% formic acid. Peptide mixtures were injected into the trap column at a flow rate of 20 μL/min for 5 min, then eluted with a three step linear gradient, starting from 5% B to 45% B for 40 min, increased to 80% B for 1 min, and then held at 80% B for 4 min. The column was re-equilibrated under the initial conditions for 15 min. Column flow rate was maintained at 500 nL/min and column temperature was maintained at 35 °C.
Electrospray voltage of 1.9 kV versus the inlet of the mass spectrometer was used. LTQ Orbitrap XL mass spectrometer was operated in the data-dependent mode to switch automatically between MS and MS/MS acquisition. Survey full scan MS spectra with two microscans (m/z 400-2000) were acquired in the Obitrap with a mass resolution of 100000 at an m/z of 400, followed by eight sequential LTQ-MS/MS scans. Dynamic exclusion was used with two repeat counts, consisting of a 10 second repeat duration, and a 60 s exclusion duration. For MS/MS, precursor ions were activated using 25% normalized collision energy at the default activation q of 0.25.
All MS/MS spectra were identified using SEQUEST [v.28 (revision 12), Thermo Electron Corp.] that searched the human International Protein Index (IPI) database (IPI human v3.64 fasta with 71983 entries). To minimize false positives, a decoy database containing all of the reverse protein sequences was added to this database. Search parameters were the following: no enzyme digestion, oxidation of methionine as a variable modification, peptide mass tolerance of 20 ppm, and fragment ion tolerance of 1.0 Da.
The resulting filter parameters were the following: ΔCn ≥ 0.10, Xcorr ≥ 2.3 for two charged ions, Xcorr ≥ 2.6 for three charged ions, Xcorr ≥ 3.0 for four or more charged state ions, and peptide probability ≤ 1 × 10-3. The errors were less than 0.1 Da between the m/z of peptide determined by LC-MS and MALDI-TOF-MS.
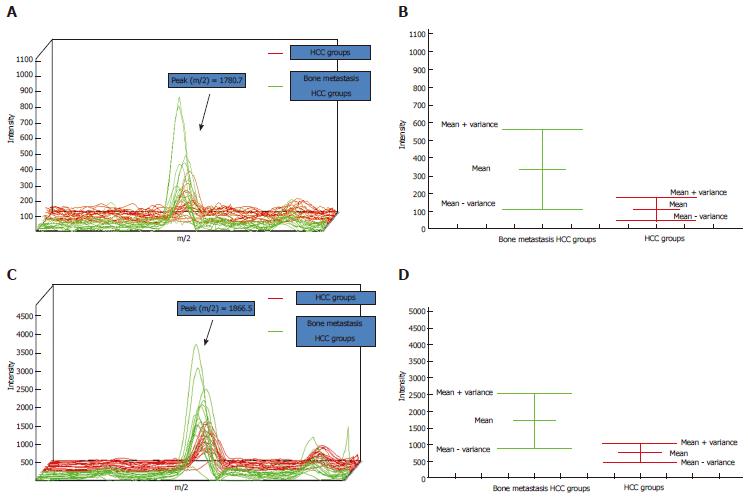
A total of 10 peptides with P < 0.01 (Wilcoxon test P value) were screened. All peptides were upregulated in the bone metastasis group (Table 2). Taking the two most significantly different peptides (m/z 1780.7, 1866.5; P < 3 × 10-4) as an example, the zoom spectrum and the variance comparison chart are shown in Figure 1. The intensities of peaks at m/z of 1780.7 and 1866.5 in the bone metastasis group were significantly higher than those in the bone metastasis-free group. The variance chart showed that the experimental results could fully distinguish between the peptides.
| Serial number | m/z | PTTA (t) | P-W test | PAD_1 | PAD_2 | HCC groups (n = 38) | Bone metastasis HCC groups (n = 38) |
| 1 | 1780.7 | 3.60 × 10-4 | 1.58 × 10-4 | 0.026 | 0.048 | 110.3 | 333.7 |
| 2 | 1866.5 | 8.48 × 10-5 | 2.80 × 10-4 | 0.136 | 0.500 | 750.1 | 1703.5 |
| 3 | 2131.6 | 6.30 × 10-2 | 2.99 × 10-4 | 0.031 | 0.001 | 42.3 | 129.8 |
| 4 | 2880.4 | 2.00 × 10-3 | 3.68 × 10-4 | 0.100 | 0.005 | 142.5 | 294.9 |
| 5 | 1532.4 | 7.30 × 10-2 | 6.32 × 10-4 | 0.001 | 0.001 | 70.8 | 138.1 |
| 6 | 2901.9 | 9.57 × 10-4 | 9.38 × 10-4 | 0.078 | 0.065 | 99.3 | 182.2 |
| 7 | 2489.8 | 7.70 × 10-2 | 2.00 × 10-3 | 0.208 | 0.001 | 31.1 | 70.5 |
| 8 | 3061.1 | 1.02 × 10-1 | 2.00 × 10-3 | 0.010 | 0.001 | 30.1 | 74.9 |
| 9 | 2234.3 | 9.00 × 10-3 | 3.00 × 10-3 | 0.118 | 0.003 | 91.1 | 171.5 |
| 10 | 2864.1 | 7.00 × 10-3 | 3.00 × 10-3 | 0.029 | 0.009 | 135.5 | 337.0 |
Because the MALDI-TOF-MS and the LC-MS have different mass resolution and mass accuracy, to obtain the correct identification result, we used the following rules: the error mass of peptides between the MALDI-TOF-MS data and LC-MS data must be less than 1.0 Da; within this error range, no other precursor of peptide is observed in LC-MS data.
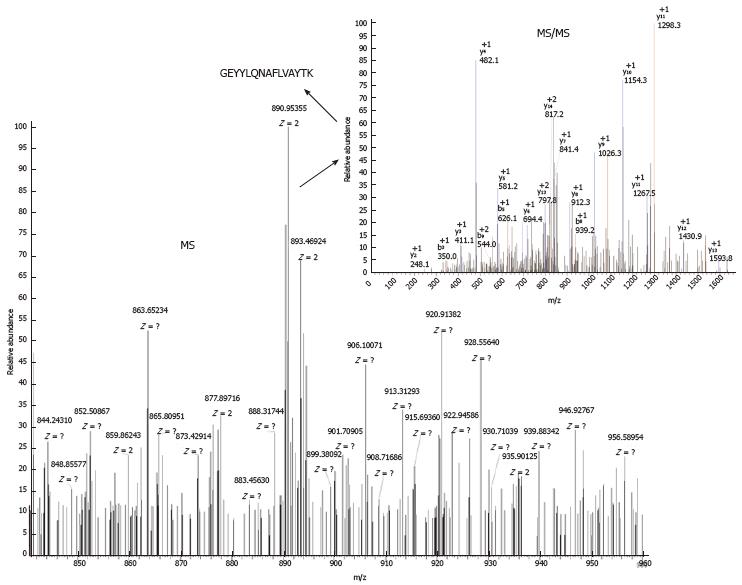
For the 10 peptides that showed significant differences, we successfully identified 7 sequences (m/z 1780.7, 1866.5, 2131.6, 2880.4, 1532.4, 2489.8, and 2234.3) using LC-MS/MS. The sequence of peptide (m/z 1780.7) was GEYYLQNAFLVAYTK, which belonged to alpha-fetoprotein (AFP), as shown in Figure 2. All sequences and their protein identities are shown in Table 3.
| m/za | m/zb | Delta mass | Peptide sequence | Protein name | Accession number |
| 1780.7 | 1779.8952 | 0.8048 | GEYYLQNAFLVAYTK | Alpha-fetoprotein | IPI00022443 |
| 1866.5 | 1866.8657 | -0.3657 | TATSEYQTFFNPRTFG | Prothrombin | IPI00019568 |
| 2131.6 | 2131.9237 | -0.3237 | NLPSDSQDLGQHGLEEDFM | Serglycin | IPI00019372 |
| 2880.4 | 2880.4904 | -0.0904 | RPGVLSSRQLGLPGPPDVPDHAAYHPF | Isoform 2 of inter-alpha-trypsin inhibitor heavy chain H4 | IPI00218192 |
| 1532.4 | 1532.8213 | -0.4213 | CSRDNTLKVIDLR | Isoform 1 of autophagy-related protein 16-2 | IPI00300536 |
| 2489.8 | 2489.2810 | 0.5190 | YTIAALLSPYSYSTTAVVTNPKE | Transthyretin | IPI00022432 |
| 2234.3 | 2235.1993 | -0.8993 | KREEAPSLRPAPPPISGGGYR | Fibrinogen beta chain | IPI00298497 |
As shown in Table 2, a pattern of peaks was selected for RBFNN model building, which was the best model built by this method. The six selected peptides were detected by MALDI-TOF-MS in every patients and each selected peptide showed a peak after the detection by mass spectrum. The area under the peak was subject to RBFNN and finally a model value was generated for each patients. This diagnostic model was able to accurately identify 68 among 76 cases, which were used as the training group, with a recognition rate of 89.47% (68/76). The model could accurately predict an average of 63 cases by using the 10-fold cross validation method, with a predictive ability of 82.89% (63/76). ROC analysis found that the AUC (area under the ROC curve) value was 0.911 (Figure 3).
Of the sera from 28 HCC patients with bone metastasis in the validation group and 34 patients without bone metastasis, 24 patients with bone metastasis and 29 patients without bone metastasis were correctly identified. The model accuracy, specificity and sensitivity were 85.48% (53/62), 85.29% (29/34) and 85.71% (24/28), respectively.
Bone metastasis from HCC seriously affects quality of life, and palliative treatments are the main clinical treatments to maximize the quality of life[6]. Currently, the main diagnostic methods for bone metastasis are imaging such as technetium-99m bone scintigraphy, CT, MRI, and SPECT, which are not the most effective methods for prediction and early diagnosis of this disorder[7]. Colleoni et al[8] and Diel et al[9] made a breakthrough in early intervention of metastatic tumors of the bone resulting from breast cancer, reporting that diphosphate drugs were effective in prevention of bone metastasis. Early prediction of metastasis and early detection of patients with a high risk of bone metastasis are important clinical objectives. Bone metastasis of HCC consists of tumor angiogenesis, matrix degradation, shedding of tumor cells into the circulation, spreading to bone tissue and survival. These cancer cells evade local nonspecific immune functions of the host, and grow in the presence of various growth factors, resulting in bone metastasis. This is a multi-step and multifactorial process, affected by biological characteristics such as genetic background, host environment, and other factors[10]. A timeline model clearly demonstrated that the development of disease is a continuum, with the initial events being molecular perturbations following histomorphological changes and clinical manifestations occurring relatively late in the disease timeline[11]. This theory implied that molecular biomarkers were more likely to be used as early diagnosis/prediction tools than imaging. Furthermore, blood test is convenient and rapid with relative low cost, which also can be used for dynamic monitoring[12]. Abnormal expression of peptides and proteins by tumor cell genes encoding cytokines, reactivity of the organism, and tissue factors released into the blood provide an excellent source of protein markers for prediction and early diagnosis of tumor recurrence[13-17]. Detection of serum markers is technically feasible and of great clinical importance, because multiple characteristics of peptides and proteins in peripheral blood are likely to be important diagnostic predictors of HCC bone metastasis. So it is theoretically feasible to develop procedures to identify different peptides and proteins from serum to establish protein marker models.
Mass spectrometry is the most important analytical method in proteome research, and mass spectrometry based on MALDI-TOF-MS of biomarkers for specific diseases has already played a major role in the development of molecular diagnostics[18,19]. Using mass spectrometry, Liu et al[20] screened HCC-associated molecular markers from sera of 30 patients with hepatitis B-related HCC, from 30 patients with chronic hepatitis B, and from 24 normal control donors, and identified markers for HCC diagnosis. Their protocol accurately distinguished between the three groups, and has been verified in 64 patients with liver cancer and 80 patients with chronic hepatitis B. HCC-associated proteins in that model include prothrombin precursor (fragment), calcium-dependent secretion activator protein-1, and BIRC6. Orvisky et al[21] used MALDI-TOF to screen HCC-associated serum proteins, and found 45 peptide peaks with significant differences (P < 0.001) out of 332 peptide peaks, with an accuracy as high as 90%. Protein sequence analysis revealed that the most significant differences were in peptides with alanine residues. Feng et al[22] used image analysis and MALDI-TOF-MS/MS to identify eight proteins unique to sera of patients with HCC. This was consistent with the discovery that heat shock protein 27 was found in sera in 90% of patients with HCC, but was not detected in normal human serum, showing that this protein can be used as a molecular marker for diagnosis of HCC. Nonetheless, all the above mentioned studies were for diagnosis of HCC, and not for diagnosis of bone metastasis from HCC.
MALDI-TOF-MS uses magnetic beads with different functional surfaces for analysis of HCC serum, to identify potential biomarkers, and to identify single molecular markers with high accuracy, high stability, high sensitivity, high reproducibility, and high flux, whose identification can be automated[23,24]. The present study used a combination of MALDI-TOF-MS and technology-related information to characterize the specific serum peptide spectra of patients with bone metastasis from HCC, obtained from serum samples of 66 patients with bone metastasis from HCC compared to 72 HCC patients without bone metastasis. ClinProt magnetic bead purification and MALDI-TOF-MS ClinProTools bioinformatics methods were used to characterize differences in serum polypeptide spectra. Using analyses and comparison of serum peptide spectra from bone metastasis patients and bone metastasis-free control patients, 10 peptides with P-W test < 0.001 were identified, with a RBFNN recognition rate up to 89.47%. LC-MS was successfully used to identify sequences of seven of the different peptides[25,26], with m/z of 1780.7, 1866.5, 2131.6, 2880.4, 1532.4, 2489.8, and 2234.3. Peptides with m/z of 1780.7 and 1866.5 were from alpha-fetoprotein and prothrombin, respectively. Compared with sera from HCC patients without bone metastasis, these two peptides were significantly upregulated in sera from patients with bone metastasis from HCC. The other five peptides from serglycin, transthyretin, etc. (Table 3) also increased in HCC patients with bone metastasis. We did not find any downregulated peptides in HCC patients with bone metastasis, which might be due to limitations of sample size. Researchers have reported that serglycin is a clinical sign of metastasis from nasopharyngeal cancer (NPC), and high levels of serglycin are closely related to poor prognosis in patients with NPC and metastasis of NPC[27]. Furthermore, studies showed that NPC cells with high metastatic potential had high expression levels of serglycin. The seven peptide sequences identified in our study are protein fragments of molecules, rather than the full protein, therefore the expression levels of peptide may not necessarily reflect the expression of the protein[28].
The 10 peptides that were significantly different in serum of patients with bone metastasis of HCC may possibly be used to establish simple and practical molecular markers for this disorder. They may be used as a potential early clinical predictor of metastasis, or used to monitor palliative treatment and active preventative intervention.
In conclusion, we believe that the serum peptides identified by MALDI-TOF-MS may be adopted as potential tumor makers for bone metastasis of HCC. In addition, their identification facilitates the development of molecular markers to predict bone metastasis of HCC in high risk patients.
We thank our patients for their willingness to take part in this study. We sincerely thank Guoquan Yan for valuable discussions and technical assistance.
Liver cancer is the second leading cause of cancer death in men and the sixth leading cause of cancer death in women worldwide. Advances in diagnosis and treatment of hepatocellular carcinoma (HCC) and extension of overall survival rates have led to an increase in the incidence of bone metastasis of HCC, which accounts for approximately 38.5% of all cases of extrahepatic metastasis of HCC. Bone metastasis often results in severe bone pain, pathologic fracture, neurological deficits, and even paralysis, which reduce quality of life and add health care costs. The diagnosis of bone metastasis from HCC is based on imaging tests which have a low sensitivity. Moreover, by the time bone metastasis images are obtained, patients often already have developed paralysis extensively in the body due to pain, dysfunction, and metastasis. Molecular markers for bone metastasis from HCC have not yet been identified.
Peptidomics is special form of functional proteomics. Since its introduction at the beginning of 2000, peptidomics has defined a new promising “omics” approach for the qualitative-quantitative analysis of endogenous peptides in biological samples, and has experienced a rapid development in the last decade. In this study, the authors tried to screen potential serum biomarkers for diagnosis of bone metastasis from HCC, and this can be used for early molecular prediction of HCC bone metastasis.
A timeline model clearly demonstrated that the development of disease is a continuum, with the initial events being molecular perturbations following histomorphological changes and clinical manifestations occurring relatively late in the disease timeline. This theory implied that molecular biomarkers were more likely to be used as early diagnosis/prediction tools than imaging [such as computed tomography (CT), magnetic resonance imaging (MRI), and single photon emission computed tomography (SPECT)]. Furthermore, blood test is convenient and rapid with relative low cost, which also can be used for dynamic monitoring.
The serum peptides screened by MALDI-TOF-MS may be adopted as potential tumor makers for bone metastasis of HCC. In addition, their identification facilitates the development of molecular markers to predict bone metastasis of HCC in high risk patients.
Peptidomics, as a subset of proteomics, analyzes low molecular weight proteins (< 10 kDa) or endogenous peptides from biological source.
The study design and labor are well organized and should be admired. This work would be more attractive if the authors more clearly presented the advantages of these new biomarkers as compared with the current diagnostic strategy, such as CT, MRI, and PET-CT.
P- Reviewers: Kaplan DE, Tomizawa M S- Editor: Wen LL L- Editor: Wang TQ E- Editor: Liu XM
| 1. | Morgan TR, Mandayam S, Jamal MM. Alcohol and hepatocellular carcinoma. Gastroenterology. 2004;127:S87-S96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 397] [Article Influence: 18.9] [Reference Citation Analysis (0)] |
| 2. | He J, Zeng ZC, Tang ZY, Fan J, Zhou J, Zeng MS, Wang JH, Sun J, Chen B, Yang P. Clinical features and prognostic factors in patients with bone metastases from hepatocellular carcinoma receiving external beam radiotherapy. Cancer. 2009;115:2710-2720. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 3. | He J, Zen ZC, Tang ZY. Radiotherapy for primary liver neoplasms with bone metastases and analysis of related prognostic factors. Tumor. 2002;22:421-423. |
| 4. | Natsuizaka M, Omura T, Akaike T, Kuwata Y, Yamazaki K, Sato T, Karino Y, Toyota J, Suga T, Asaka M. Clinical features of hepatocellular carcinoma with extrahepatic metastases. J Gastroenterol Hepatol. 2005;20:1781-1787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 398] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 5. | Villanueva J, Shaffer DR, Philip J, Chaparro CA, Erdjument-Bromage H, Olshen AB, Fleisher M, Lilja H, Brogi E, Boyd J. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J Clin Invest. 2006;116:271-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 592] [Cited by in RCA: 574] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 6. | Hsieh CH, Wei MC, Lee HY, Hsiao SM, Chen CA, Wang LY, Hsieh YP, Tsai TH, Chen YJ, Shueng PW. Whole pelvic helical tomotherapy for locally advanced cervical cancer: technical implementation of IMRT with helical tomotherapy. Radiat Oncol. 2009;4:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 7. | Nakamoto Y, Cohade C, Tatsumi M, Hammoud D, Wahl RL. CT appearance of bone metastases detected with FDG PET as part of the same PET/CT examination. Radiology. 2005;237:627-634. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 61] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 8. | Colleoni M, O’Neill A, Goldhirsch A, Gelber RD, Bonetti M, Thürlimann B, Price KN, Castiglione-Gertsch M, Coates AS, Lindtner J. Identifying breast cancer patients at high risk for bone metastases. J Clin Oncol. 2000;18:3925-3935. [PubMed] |
| 9. | Diel IJ, Solomayer EF, Costa SD, Gollan C, Goerner R, Wallwiener D, Kaufmann M, Bastert G. Reduction in new metastases in breast cancer with adjuvant clodronate treatment. N Engl J Med. 1998;339:357-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 605] [Cited by in RCA: 559] [Article Influence: 20.7] [Reference Citation Analysis (0)] |
| 10. | Hiratsuka S, Duda DG, Huang Y, Goel S, Sugiyama T, Nagasawa T, Fukumura D, Jain RK. C-X-C receptor type 4 promotes metastasis by activating p38 mitogen-activated protein kinase in myeloid differentiation antigen (Gr-1)-positive cells. Proc Natl Acad Sci USA. 2011;108:302-307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 11. | Anglicheau D, Suthanthiran M. Noninvasive prediction of organ graft rejection and outcome using gene expression patterns. Transplantation. 2008;86:192-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 78] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 12. | Hu J, Wang Z, Tan CJ, Liao BY, Zhang X, Xu M, Dai Z, Qiu SJ, Huang XW, Sun J. Plasma microRNA, a potential biomarker for acute rejection after liver transplantation. Transplantation. 2013;95:991-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 13. | Chen Y, Lin MC, Wang H, Chan CY, Jiang L, Ngai SM, Yu J, He ML, Shaw PC, Yew DT. Proteomic analysis of EZH2 downstream target proteins in hepatocellular carcinoma. Proteomics. 2007;7:3097-3104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Ren P, Yu ZT, Xiu L, Wang M, Liu HM. Elevated serum levels of human relaxin-2 in patients with esophageal squamous cell carcinoma. World J Gastroenterol. 2013;19:2412-2418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 16] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 15. | Su BB, Shi H, Wan J. Role of serum carcinoembryonic antigen in the detection of colorectal cancer before and after surgical resection. World J Gastroenterol. 2012;18:2121-2126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 67] [Cited by in RCA: 80] [Article Influence: 6.2] [Reference Citation Analysis (1)] |
| 16. | Cao XY, Jia ZF, Jin MS, Cao DH, Kong F, Suo J, Jiang J. Serum pepsinogen II is a better diagnostic marker in gastric cancer. World J Gastroenterol. 2012;18:7357-7361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 29] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Dong QM, Zhang JQ, Li Q, Bracher JC, Hendricks DT, Zhao XH. Clinical significance of serum expression of GROβ in esophageal squamous cell carcinoma. World J Gastroenterol. 2011;17:2658-2662. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 9] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 18. | Melle C, Ernst G, Scheibner O, Kaufmann R, Schimmel B, Bleul A, Settmacher U, Hommann M, Claussen U, von Eggeling F. Identification of specific protein markers in microdissected hepatocellular carcinoma. J Proteome Res. 2007;6:306-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 19. | Yang MH, Tyan YC, Jong SB, Huang YF, Liao PC, Wang MC. Identification of human hepatocellular carcinoma-related proteins by proteomic approaches. Anal Bioanal Chem. 2007;388:637-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 20. | Liu T, Xue R, Huang X, Zhang D, Dong L, Wu H, Shen X. Proteomic profiling of hepatitis B virus-related hepatocellular carcinoma with magnetic bead-based matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Acta Biochim Biophys Sin (Shanghai). 2011;43:542-550. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Orvisky E, Drake SK, Martin BM, Abdel-Hamid M, Ressom HW, Varghese RS, An Y, Saha D, Hortin GL, Loffredo CA. Enrichment of low molecular weight fraction of serum for MS analysis of peptides associated with hepatocellular carcinoma. Proteomics. 2006;6:2895-2902. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 109] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 22. | Feng JT, Liu YK, Song HY, Dai Z, Qin LX, Almofti MR, Fang CY, Lu HJ, Yang PY, Tang ZY. Heat-shock protein 27: a potential biomarker for hepatocellular carcinoma identified by serum proteome analysis. Proteomics. 2005;5:4581-4588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 156] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 23. | Takashima M, Kuramitsu Y, Yokoyama Y, Iizuka N, Harada T, Fujimoto M, Sakaida I, Okita K, Oka M, Nakamura K. Proteomic analysis of autoantibodies in patients with hepatocellular carcinoma. Proteomics. 2006;6:3894-3900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Zinkin NT, Grall F, Bhaskar K, Otu HH, Spentzos D, Kalmowitz B, Wells M, Guerrero M, Asara JM, Libermann TA. Serum proteomics and biomarkers in hepatocellular carcinoma and chronic liver disease. Clin Cancer Res. 2008;14:470-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 456] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 25. | Kapp EA, Schütz F, Reid GE, Eddes JS, Moritz RL, O’Hair RA, Speed TP, Simpson RJ. Mining a tandem mass spectrometry database to determine the trends and global factors influencing peptide fragmentation. Anal Chem. 2003;75:6251-6264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 194] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 26. | Chen J, Anderson M, Misek DE, Simeone DM, Lubman DM. Characterization of apolipoprotein and apolipoprotein precursors in pancreatic cancer serum samples via two-dimensional liquid chromatography and mass spectrometry. J Chromatogr A. 2007;1162:117-125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 27. | Li XJ, Ong CK, Cao Y, Xiang YQ, Shao JY, Ooi A, Peng LX, Lu WH, Zhang Z, Petillo D. Serglycin is a theranostic target in nasopharyngeal carcinoma that promotes metastasis. Cancer Res. 2011;71:3162-3172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 759] [Reference Citation Analysis (0)] |
| 28. | Koomen JM, Li D, Xiao LC, Liu TC, Coombes KR, Abbruzzese J, Kobayashi R. Direct tandem mass spectrometry reveals limitations in protein profiling experiments for plasma biomarker discovery. J Proteome Res. 2005;4:972-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 172] [Article Influence: 8.6] [Reference Citation Analysis (0)] |