Published online Mar 21, 2014. doi: 10.3748/wjg.v20.i11.2839
Revised: November 19, 2013
Accepted: January 6, 2014
Published online: March 21, 2014
Processing time: 180 Days and 12.4 Hours
Hepatitis C virus (HCV) infection is a global health problem that affects more than 170 million people worldwide. It is a major cause of cirrhosis and hepatocellular carcinoma, making the virus the most common cause of liver failure and transplantation. The standard-of-care treatment for chronic hepatitis C (CHC) has been changed during the last decade and direct acting antiviral drugs have already been used. Besides, understanding of the pathogenesis of CHC has evolved rapidly during the last years and now several host factors are known to affect the natural history and response to treatment. Recent genome-wide association studies have shown the important role of interleukin-28B and inosine triphosphatase in HCV infection. The present review article attempts to summarize the current knowledge on the role of host factors towards individualization of HCV treatment.
Core tip: Hepatitis C virus (HCV) is a major health problem with personal, social and economic implications. In the past few years, advances in HCV molecular virology and host genetics revealed a complex interplay between the virus and the host that influence the natural history of chronic hepatitis C (CHC) and response to treatment. Besides, the management of CHC has evolved with the development of direct acting antiviral agents (DAAs). In this review, we have summarized the knowledge regarding host determinants of HCV treatment outcome and consider how this knowledge might help to individualize clinical management in the era of DAAs.
- Citation: Gatselis NK, Zachou K, Saitis A, Samara M, Dalekos GN. Individualization of chronic hepatitis C treatment according to the host characteristics. World J Gastroenterol 2014; 20(11): 2839-2853
- URL: https://www.wjgnet.com/1007-9327/full/v20/i11/2839.htm
- DOI: https://dx.doi.org/10.3748/wjg.v20.i11.2839
Chronic hepatitis C (CHC) is one of the most important health issues problem worldwide with more than 170 million people infected and significant personal, social, and economic impact[1]. The long-term sequelae of CHC include cirrhosis, decompensation, hepatocellular carcinoma and liver related death[2,3].
The standard-of-care (SOC) treatment for CHC during the last decade has been the combination therapy of pegylated-interferon-α (PEG-IFN) with ribavirin (RBV), a guanosine analogue that interrupts the viral RNA metabolism[4,5]. However, the rate of sustained virological response (SVR), defined as having undetectable serum hepatitis C virus (HCV) RNA at week 24 after treatment discontinuation, is suboptimal for genotype 1 hepatitis C virus (HCV-1), with less than 50% of patients achieving viral eradication[4,6,7]. Moreover, PEG-IFN/RBV therapy is associated with significant adverse effects, which increase morbidity, while treatment is prolonged ranging from 24 to 48 wk or more, according to EASL and AASLD guidelines[8,9]. As a result clinicians often have to weigh the various viral and host characteristics for each patient before initiating treatment[10].
Antiviral therapy is currently changing and various direct acting antiviral drugs (DAAs) have been developed, which are directed against essential components of viral replication[11,12]. In addition, understanding of the pathogenesis of CHC has evolved rapidly, leading to development of novel compounds, targeting host factors that are potentially modifiable[13]. Given the high cost of treatment and the increased possibility of adverse events, identification of factors predicting SVR and the need to individualize HCV therapy by incorporating new predictive data in clinical decision-making is urgent.
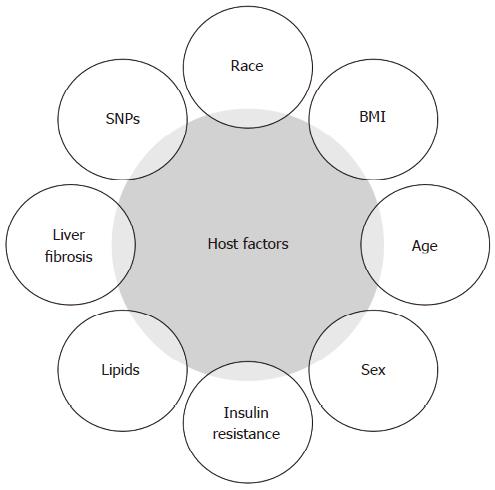
Several host factors such as age, gender, race-ethnicity, fibrosis stage, obesity, hepatic steatosis, low-density lipoprotein cholesterol (LDLc), insulin resistance, and genetic variances of the host[14,15], are known to affect the spontaneous clearance and treatment outcomes (Figure 1).
Recently, genome-wide association studies (GWAS) have identified several clinically important genetic determinants of PEG-IFN/RBV treatment outcomes. The most important are single nucleotide polymorphisms (SNPs) in interleukin 28B (IL28B) gene, which is associated with spontaneous clearance and response to anti-HCV treatment[16-18] and in inosine triphosphatase (ITPA) gene, which protects against ribavirin-related hemolytic anaemia and subsequent dose reductions[19,20].
In this review, we present in brief the major host factors that can modify the response to treatment of CHC, focusing on the clinical utility of IL28B and ITPA genotyping for pretreatment counseling and individualization of therapy modalities.
It is well documented that race is associated with treatment response either with SOC or triple (SOC plus DAAs) therapy[21-31]. In several controlled trials, it was demonstrated that SOC in African-American patients has a reduced likelihood of SVR, ranging between 19% and 28%, compared to non-African-Americans in whom SVR rate was 39%-52%, especially when HCV-1 is taken into account[22,23,25]. The same was true for triple therapy in naïve patients infected with HCV-1; SPRINT-2 trial (boceprevir-based therapy)[30] as well as ADVANCED study (telaprevir-based therapy)[29] showed that Black ethnic origin negatively affected SVR. In addition, HCV infected individuals of Asian origin seem to achieve better SVR rates in comparison to Caucasians[26,31]. Differences in population frequency of the favorable IL28B genotype may explain the recognized ethnic disparity in treatment response rates[16,32]. However, the response to treatment in Black populations was poorer across all IL28B genotypes, suggesting that there may be other viral and/or host factors influencing SVR[27,28]. Hispanics also tended to have poorer SVR rates compared to Caucasian patients[21,24,27].
Although female gender was considered a positive predictor of SVR in IFN plus RBV era[33], in PEG-IFN/RBV trials no statistically significant correlation was found on multivariate analysis between gender and SVR[4,6,34-36]. Age has also been considered a significant factor for predicting response to treatment. In particular, large prospective studies of PEG-IFN/RBV combination therapy showed that patients younger than 40-45 years achieved significantly higher SVR rates compared to older patients[4,6,33,34]. In triple therapy trials with boceprevir, age was also significant[30,37] regarding the achievement of SVR in univariate investigation but not after multivariate analysis, especially when IL28B genotype was taken into account[38].
Obesity is traditionally considered a significant predictor of disease progression in CHC, probably due to increased inflammatory milieu, which promotes liver injury in overweight individuals[39]. In a prospective trial[40], fibrosis progression was associated with body mass index (BMI). A high BMI was also inversely correlated with SVR in both IFN and SOC treated HCV patients[41,42]. Furthermore, a lower baseline body weight was significantly associated with SVR across all genotypes in the era of PEG-IFN/RBV combination therapy[4,6,34,42]. However, when RBV weight-based dosing was implicated, BMI and body weight were no longer significant parameters of SVR[43,44]. On the contrary, in a retrospective analysis of SPRINT-2[30] and RESPOND-2[37] studies by Poordad et al[38], low BMI proved to be one of the significant factors for achieving SVR in boceprevir-based therapy of treatment-naïve patients.
In two recent meta-analyses, insulin resistance (IR) was strongly associated with the probability of achieving SVR to dual treatment[45,46]. In Spanish[47] as well as Japanese[48] patients, IR seemed to be an independent predictor of response irrespective of the IL28B genotype. Concerning triple therapy, IR did not have any effect on SVR when telaprevir was used in the combination[49].
Recent studies suggest that pretreatment serum lipid levels may be important predictors of treatment response. Several studies indicate that high pretreatment LDLc and total cholesterol levels are associated with higher rates of SVR to dual PEG-IFN/RBV therapy in multivariate analysis[50-56]. In addition, pretreatment triglyceride levels may also play a role in SVR[56]. LDLc remained a significant independent predictor of SVR in post hoc analysis of the telaprevir-based REALIZE study[57] in treatment experienced patients[58]. The abovementioned associations between serum lipids and treatment response are supported by potential biological mechanisms. In vitro studies suggest relationships between lipoproteins and HCV that are important for viral entry into hepatocytes, viral replication and secretion. Indeed, several studies have shown that HCV may combine with lipoproteins in the serum, obscuring the virus from the host immune response, which may in turn help viral entry into the hepatocytes[59-61]. Various receptors involved in lipoprotein-viral particle entry into hepatocytes are posited, including the scavenger receptor B1 (SR-B1) and LDLc receptor[62-65]. IFN therapy leads to down-regulation of SR-B1 expression[66]. Finally, statin use in HCV-1 has been associated with increased SVR to dual PEG-IFN/RBV therapy[55] and in boceprevir-based triple therapy[30]. Taken together the above studies support the notion that decreased lipoprotein expression may in turn impact serum lipoproteins and lipids profile measures affecting SVR. However, these associations need to be interpreted with caution, because the favourable IL28B CC genotype is also associated with increased LDLc[67-69].
The presence of advanced liver fibrosis and cirrhosis has long been recognized to be associated with lower SVR rates to IFN-based treatment[33,34,70]. Actually, advanced fibrosis and cirrhosis have been shown to be major independent predictors of non-response to SOC[34,44,70]. Furthermore, in treatment-naïve patients, triple therapies with boceprevir and telaprevir (SPRINT-2[30] and ADVANCE[29] studies, respectively), showed that the severity of liver fibrosis together with IL28B genotype affect treatment outcome. Similar results were found when treatment-experienced patients were taken into account[57,71], although in these patients other factors such as IL28B genotype and pattern of previous response seemed to be the best predictors of response to triple therapy[38].
Host genetic factors have long been suggested that might play an important role for the observed differences in HCV clearance and response to treatment among different ethnic groups. Over 40 genes have been linked to modulation of anti-HCV therapy affecting either response to treatment or side effects to drugs[72,73]. However, only after sequencing of the entire human genome in 2001, major advances have been made in genotyping technologies, with the large-scale discovery of SNPs and the implementation of GWAS.
Initial studies investigated candidate genes to identify differences or SNPs between two populations. Several SNPs were identified in genes that are involved in the innate defense against HCV including MxA, OAS1, EIF2AK2, IFNAR1, IL-6, MHC, MAPKAPK3 and KIR receptors[74-82]. In addition, pretreatment hepatic gene expression such as an 8-gene subset (GIP2/IFI15/ISG15, ATF5, IFIT1, MX1, USP18/UBP43, DUSP1, CEB1, and RPS28)[83] and a two-gene signature (IFI27 and CXLC9)[84] predicted the response to treatment with a sufficient predictive accuracy in the majority of patients studied. Interestingly, in a recent paper by Chen et al[85], treatment response was linked to cell-specific activation patterns: interferon-stimulated gene 15 (ISG 15) protein up-regulation was more pronounced in hepatocytes of non-responders but also in Kupffer cells of responders. Even more recently, the same group showed that the gene expression pattern, at either the mRNA or the protein level, is more predictive of treatment outcome than the host (IL28B) or viral genotype[86]. Furthermore, the level of expression of intrahepatic mir-122 has been associated with HCV treatment response[87]. Expression of mir-122 was significantly lower in primary non-responders compared to early responders[86], while 8 other microRNAs (mir-34b, mir-145, mir-143, mir-652, mir-18a, mir-27b, mir-422b, and mir-378) were also found to be differentially expressed in responders and non-responders[88].
However, the validity of the reported associations varies, since a small number of them have repeatedly been reported from several independent large cohorts and have been verified to apply to different ethnicities. For this reason, it is till now IL28B polymorphisms that are considered as ‘‘state of the art’’ in HCV pharmacogenetics, which can potentially tailor future therapies[89].
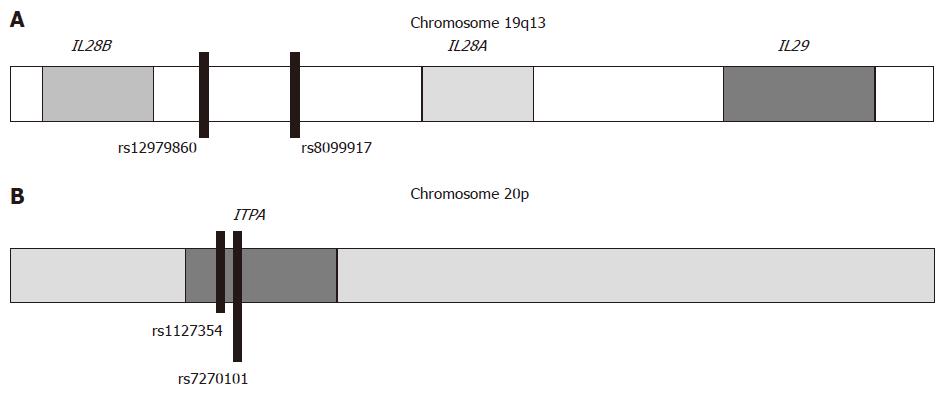
Independent GWAS have identified genetic variants near the IL28B gene that are strongly associated with treatment response[16-18] and spontaneous HCV clearance[32,90,91]. Two bi-allelic SNPs, in linkage disequilibrium[92], were most strongly associated with favourable response; SNP rs12979860 located 3 kb upstream of the IL28B gene (favourable response CC genotype, and unfavourable CT/TT genotypes) and rs8099917 located 8 kb downstream of the IL28B gene and 16 kb upstream of the IL28A gene (favourable response TT genotype, and unfavourable GT/GG genotypes) (Figure 2A). Subsequently, these treatment response findings were confirmed by many studies in different populations such as HCV-1 patients[27,93-97], HCV-4 patients[98-101], patients with recent HCV infection[102], adults and children with spontaneous HCV clearance[103-105], HCV/HIV co-infected patients[106-108], and patients with recurrent HCV infection after orthotopic liver transplantation[109]. The results were reproduced in five recent meta-analyses in Asian, Caucasian and African patients[110-114].
The influence of IL28B on treatment response in genotype 2 or genotype 3 infections is less clinically relevant. Several studies from Europe and Japan have demonstrated an association between IL28B and SVR rates[115,116], while other studies failed to show a clear effect[117,118] although some studies showed that viral elimination within the first weeks of treatment and the achievement of rapid virological response (RVR) was significantly faster in CCrs12979860 and TTrs8099917 patients[118,119]. Thus, determination of IL28B genotype may be more relevant in slow responders to treatment (e.g., non-RVR patients)[120]. Recently, a meta-analysis of 23 studies involving 3042 patients showed that in HCV-2 and HCV-3 patients with favourable IL28B polymorphisms (either CCrs12979860 or TTrs8099917 patients), RVR rates were increased by 13%-15% and SVR rates by 5%[121].
The mechanisms behind these associations are related to the host innate immune response[122,123]. IL28B gene along with IL28A and IL29 belong to the type III IFN family, also named IFN-λ and they are located on the human chromosome 19. In particular, IL28A corresponds to IFN-λ2, IL28B to IFN-λ3 and IL29 to IFN-λ1[124]. The corresponding cytokines are induced by viral infection and their antiviral activity is mediated by triggering the Janus kinase-Signal Transducer and Activator of Transcription (JAK-STAT) pathway, following to their interaction with a heterodimeric class II cytokine receptor that consists of IL-10 and IL-28 receptors[125-127]. The JAK-STAT pathway activates ISGs which are known to cause apoptosis, growth inhibition, and inhibition of viral replication[128]. IL28B genotype has strongly been associated with intrahepatic ISGs expression, with the unfavourable genotypes expressing higher baseline ISGs levels compared with the favourable genotype[129,130]. This finding could indicate an exhaustion of innate immunity prior to treatment in patients with unfavourable IL28B genotype. Raglow et al[131] showed that this relationship is reversed in normal liver tissue, where TTrs12979860 genotype expresses lower levels of ISGs than CCrs12979860 genotype. Recently, two independent studies identified dysregulation in several pathways of innate immunity and natural killer cells activity in patients with unfavourable IL28B genotype, resulting in a muted response to IFN therapy[132,133].
However, it is not clear so far, if there is a direct correlation between IL28B polymorphisms and IL28B expression level. IL28 mRNA in the peripheral blood mononuclear cells (PBMCs) of patients with favourable IL28B genotype was significantly higher, whereas the presence of the risk alleles CT/CCrs8099917 were associated with lower expression of IFN-λ[17,18]. Shi et al[134] demonstrated that mRNA and serum levels of IL28B were lower in patients with CT/TTrs12979860 unfavourable genotypes. Moreover, in the era of liver transplantation, these genetic variations were significantly associated with IL28B mRNA expression in both the resected liver derived from the recipients and the donated liver[109]. On the contrary, other studies have not found such a correlation between IL28B genotype and IL28B gene expression[129,135]. Abe et al[136] showed that the expression levels of IL28B in the liver were lower in patients with the favorable IL28B genotype. This is in discordance with what happens in the periphery (PBMCs).
Knowledge about the impact of IL28B SNPs on the different phases of viral elimination shed light on the functional implications of IL28B variability. The favourable IL28B genotypes enhance the reduction of HCV RNA during the early phases (“first” and “second”) of viral kinetics. This is translated into increased rates of RVR, complete early virological response (EVR) on-treatment and finally SVR[27,133,137-143]. In fact, most RVR patients carry the good-response IL28B genotype. Furthermore, IL28B genotype can be combined with a chemotactic chemokine, the interferon-gamma inducible protein 10 kDa (IP-10), which strongly predicts the HCV RNA decline during the first days of therapy for all HCV genotypes[144]. In fact, it has been shown that the combination of increased baseline plasma IP-10 levels in association with IL28B non-favourable genotypes can accurately predict the first-phase decline of HCV RNA during treatment[135,145,146]. Besides, in acute HCV infection, IL28B genotype together with serum IP-10 levels can identify patients who are most likely to undergo spontaneous clearance and those in need of early antiviral therapy[147].
The new DAAs along with PEG-IFN/RBV combination are the new SOC in HCV-1 patients. Thus, it is necessary to redefine the role of the IL28B genotype in the decision to treat and how to treat these patients. SNPs in the IL28B region are the best baseline predictors of SVR and could be used as independent factors for treatment decision-making. In particular, treatment-naïve HCV-1 patients with genotype CCrs12979860 have the same probability (more than 80%) in achieving SVR either with dual therapy or triple therapy with telaprevir or boceprevir[27,29,30]. So, in these patients, the best therapeutic choice would be the standard PEG-IFN/RBV therapy, in order to avoid the higher costs and significant side effects of triple therapy[148,149]. However, during treatment, monitoring of HCV kinetics with milestones such as RVR and/or EVR, has been proved stronger predictor of treatment outcome[150,151]. Therefore, treatment-naïve HCV-1 patients with favorable IL28B genotype who do not achieve RVR should be considered candidates for more effective therapy with DAAs.
Response-guided therapy has become standard practice for HCV-1 patients and guidelines recommend shortening or prolongation of treatment duration, based on-treatment virological response. However, since the discovery of its significance, the incorporation of IL28B genotype in HCV management algorithms is intensively investigated. Huang et al[152] showed that IL28B genotype combined with baseline viral load might help in identifying HCV-1 patients who will or will not benefit from a shortened 24-wk regimen. The positive predictive value of these two factors (TTrs8099917 and lower baseline viral load) was 80% and the negative predictive value 91%. In the same line, Liu et al[153] showed that HCV-1 patients with the TTrs8099917 genotype and low baseline viral load (HCV RNA < 600000 IU/mL) could benefit from a shorter duration of combination therapy. Sarrazin et al[154] in a following study showed that the CCrs12979860 polymorphism was significantly associated with SVR, but in patients with on-treatment virologic response, SVR rates were similar for different IL28B genotypes. This finding indicates that virological response during therapy seems to determine the chance to achieve an SVR in an IL28B-independent way. In addition, two cohorts[155,156] have analyzed the efficacy of prolongation of treatment duration in HCV-1 slow-responders up to 72 wk and demonstrated that the benefits of extended therapy were restricted only to patients with CT/TTrs12979860 genotype.
Despite the fact that DAAs clearly attenuates the association between IL28B genotype and HCV treatment response, there are studies suggesting that the favourable IL28B alleles could also predict response to triple therapy in treatment-naïve and treatment-experienced HCV-1 patients[157-160]. In SPRINT-2[30] and RESPOND-2[37], where treatment-naïve or prior relapsers/non-responders were included respectively, CCrs12979860 genotype was significantly associated with increased SVR rates and could be used as a predictor for shortening therapy[161]. However, the sub-analysis of the REALISE study[57] failed to prove a significant association[162]. Further studies with DAAs are needed to identify whether the predictive role of IL28B genotype remains.
For the time being, studies with new DAAs and/or IFN-free therapy regimens are being conducted. In INFORM-1 study, where 83 patients were treated with mericitabine plus danoprevir or placebo, CCrs12979860 was associated with an increased second phase decline of HCV RNA[163]. In parallel, PROPEL[164] and JUMP-C[165] studies showed higher SVR rates in IL28B CCrs12979860 genotype patients receiving mericitabine plus PEG-IFN/RBV. The efficacy of interferon-free combination of faldaprevir and deleobuvir with RBV for the treatment of HCV-1 patients was also associated with IL28B genotype[166] though in other IFN-free regimens[167,168] the effect of IL28B genotype was unclear. For example, the ATOMIC study[169], where the efficacy of treatment with sofosbuvir along with PEG-IFN/RBV was investigated, showed promising results in shortening the duration of treatment to 12 wk, independently of the IL28B genotype. The importance of IL28B genotype on response to future interferon-free combination DAA regimens remains to be determined with larger and longer duration studies.
RBV is a main component in the currently available antiviral regimens for the treatment of CHC. One of the most common side effects of RBV therapy is anemia that may result in dose reduction or discontinuation in up to 15% of patients[4,7,10], which may have deleterious impact on SVR.
The mechanisms of hemolytic anemia induced by RBV administration are complex. RBV causes a relative deficiency of ATP in human erythrocytes by depleting guanosine triphosphate (GTP) and subsequently leading to inhibition of the ATP-dependent oxidative metabolism. This causes oxidative damage to the erythrocyte membranes and leads to extravascular hemolysis by the reticuloendothelial system[170-172]. ITPA gene, which encodes a protein that hydrolyses inosine triphosphate (ITP), has been found to play a significant role in RBV-induced anemia. A reduced ITPA activity leads to the accumulation of ITP in erythrocytes, which allows for the substitution of ITP for GTP in ATP biosynthesis. This substitution reduces ATP depletion and protects against hemolytic anemia[173-177].
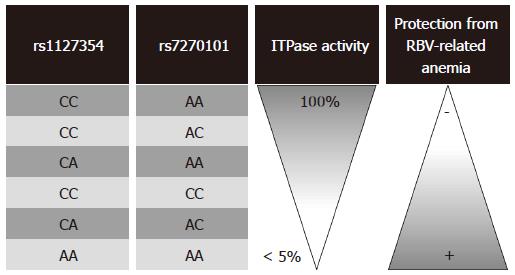
In two large GWAS[19,20], two functional SNPs, rs1127354 and rs7270101, within the ITPA gene were strongly associated with protection from RBV treatment-related hemolytic anemia and decreased the need for RBV dose reduction. The rs1127354 SNP is a missense variant in exon 2 of ITPA gene, while rs7270101 is located in ITPA intron 2 and alters splicing (Figure 2B). The AA/ACrs1127354 protective genotypes, as well as the CC/CArs7270101 protective genotypes, are associated with decreased ITPase activity and as a consequence decreased hemolytic side effects from RBV therapy (Figure 3). Following studies steadily reproduces the finding that polymorphic variation of the ITPA gene leads to enzymatic deficiency, which in turn is a major determinant of RBV-induced hemolytic anemia in HCV-1 patients[178-191]. However, ITPA SNPs effect on therapeutic outcome is unclear. Some studies have shown no association[19,178,180,186,191], and others have reported a possible association with treatment outcomes in CHC patients[20,181,182,192]. These discrepancies could be attributed to the geographic variance of ITPA SNPs and different inclusion/exclusion criteria used for patient recruitment. Taking into account that anemia is one of the main adverse events leading to premature termination of therapy, any marker able to predict the risk of severe anemia before treatment would be of outmost importance. For this purpose, Tsubota et al[193] and Kurosaki et al[192] constructed predictions models incorporating ITPA genotype along with baseline hemoglobin, creatinine clearance and quantitative hemoglobin decline at week 2 of treatment. We need of course, further validation before entering these algorithms into clinical practice
Prediction of anemia remains important in the era of DAAs, because these newer therapies still require RBV and PEG-IFN in combination, while in addition increases the frequency as well as the severity, and hence, clinical relevance of this adverse event[30,194-197]. Suzuki et al[198] studied 61 patients receiving triple telaprevir-based therapy and showed that decreases in haemoglobin levels were greater in patients with unfavourable (CCrs1127354) than favourable (CA/AArs1127354) genotypes in the ITPA gene, especially during the first 12 wk of treatment. Mean RBV dose during the first 12 wk was lower in patients with CCrs1127354 than CA/AArs1127354 genotypes, but the total dose of RBV and SVR rates were not different. Similar results obtained by Chayama et al[158] in a study of 94 Japanese HCV-1 patients treated with PEG-IFN/RBV and telaprevir. Patients with the anemia-susceptible ITPA SNP CCrs1127354 typically required ribavirin dose reduction earlier than did patients with other genotypes. However, this polymorphism was not proved to be a predictive marker of SVR. Recently, Ogawa et al[199] in a prospective multicenter study with 292 Japanese patients showed that CCrs1127354 genotype along with baseline hemoglobin less than 13.5 g/dL and estimated glomerular filtration rate < 80 mL/min per 1.73 m2 were independent pretreatment predictors for developing severe anemia (Hb < 8.5 g/dL) during the treatment period. No effect on treatment outcome was proved.
In sum, ITPA genotyping is able to detect HCV-infected patients who are at higher risk of developing anaemia. These patients should be monitored more closely in order to avoid premature withdrawals from treatment.
Prediction of SVR, which is translated in stopping disease progression, is the main requirement in CHC. Factors that predict SVR in turn have impact on treatment decision making, treatment duration and planning for the individual patient. Major advances in genetics during the last decade allow the identification of specific markers associated with viral response. Among them, IL-28B has been strongly associated with response to current treatment for HCV-1, in all reports, including large cohorts of patients. However, for the single patient, the use of IL-28B alone as a predictive factor of treatment outcome and decision to treat is not sufficient. An important question arising and needing an urgent answer is how host and viral factors could be integrated in clinical practice. However, since in the same patient various host and viral factors interact, predictive analysis should be cautious, considering all these factors in combination. In the upcoming years of personalized medicine, the comprehension of HCV pathogenesis based on the knowledge of both host and viral genotypes could lead to individually tailored HCV therapies.
P- Reviewers: Arain SA, Muratori L S- Editor: Zhai HH L- Editor: A E- Editor: Ma S
| 1. | Shepard CW, Finelli L, Alter MJ. Global epidemiology of hepatitis C virus infection. Lancet Infect Dis. 2005;5:558-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1927] [Cited by in RCA: 1931] [Article Influence: 96.6] [Reference Citation Analysis (0)] |
| 2. | Manesis EK, Papatheodoridis GV, Touloumi G, Karafoulidou A, Ketikoglou J, Kitis GE, Antoniou A, Kanatakis S, Koutsounas SJ, Vafiadis I. Natural course of treated and untreated chronic HCV infection: results of the nationwide Hepnet.Greece cohort study. Aliment Pharmacol Ther. 2009;29:1121-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 3. | Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45:529-538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1764] [Cited by in RCA: 1843] [Article Influence: 97.0] [Reference Citation Analysis (0)] |
| 4. | Fried MW, Shiffman ML, Reddy KR, Smith C, Marinos G, Gonçales FL, Häussinger D, Diago M, Carosi G, Dhumeaux D. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N Engl J Med. 2002;347:975-982. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4847] [Cited by in RCA: 4747] [Article Influence: 206.4] [Reference Citation Analysis (0)] |
| 5. | Furusyo N, Katoh M, Tanabe Y, Kajiwara E, Maruyama T, Shimono J, Sakai H, Nakamuta M, Nomura H, Masumoto A. Interferon alpha plus ribavirin combination treatment of Japanese chronic hepatitis C patients with HCV genotype 2: a project of the Kyushu University Liver Disease Study Group. World J Gastroenterol. 2006;12:784-790. [PubMed] |
| 6. | Manns MP, McHutchison JG, Gordon SC, Rustgi VK, Shiffman M, Reindollar R, Goodman ZD, Koury K, Ling M, Albrecht JK. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet. 2001;358:958-965. [PubMed] |
| 7. | Hadziyannis SJ, Sette H, Morgan TR, Balan V, Diago M, Marcellin P, Ramadori G, Bodenheimer H, Bernstein D, Rizzetto M. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann Intern Med. 2004;140:346-355. [PubMed] |
| 8. | European Association for the Study of the Liver. EASL Clinical Practice Guidelines: management of hepatitis C virus infection. J Hepatol. 2011;55:245-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 889] [Cited by in RCA: 919] [Article Influence: 65.6] [Reference Citation Analysis (0)] |
| 9. | Ghany MG, Nelson DR, Strader DB, Thomas DL, Seeff LB, American Association for Study of Liver Diseases. An update on treatment of genotype 1 chronic hepatitis C virus infection: 2011 practice guideline by the American Association for the Study of Liver Diseases. Hepatology. 2011;54:1433-1444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 803] [Cited by in RCA: 844] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 10. | Fried MW. Side effects of therapy of hepatitis C and their management. Hepatology. 2002;36:S237-S244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 208] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 11. | Jang JY, Chung RT. New treatments for chronic hepatitis C. Korean J Hepatol. 2010;16:263-277. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Welsch C, Jesudian A, Zeuzem S, Jacobson I. New direct-acting antiviral agents for the treatment of hepatitis C virus infection and perspectives. Gut. 2012;61 Suppl 1:i36-i46. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 144] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 13. | Zeisel MB, Lupberger J, Fofana I, Baumert TF. Host-targeting agents for prevention and treatment of chronic hepatitis C - perspectives and challenges. J Hepatol. 2013;58:375-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 14. | Chuang WL, Yu ML. Host factors determining the efficacy of hepatitis C treatment. J Gastroenterol. 2013;48:22-30. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 15. | Asselah T, Estrabaud E, Bieche I, Lapalus M, De Muynck S, Vidaud M, Saadoun D, Soumelis V, Marcellin P. Hepatitis C: viral and host factors associated with non-response to pegylated interferon plus ribavirin. Liver Int. 2010;30:1259-1269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 117] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 16. | Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2776] [Cited by in RCA: 2723] [Article Influence: 170.2] [Reference Citation Analysis (0)] |
| 17. | Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105-1109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1779] [Cited by in RCA: 1775] [Article Influence: 110.9] [Reference Citation Analysis (0)] |
| 18. | Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M, Abate ML, Bassendine M, Spengler U, Dore GJ, Powell E. IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet. 2009;41:1100-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1505] [Cited by in RCA: 1504] [Article Influence: 94.0] [Reference Citation Analysis (0)] |
| 19. | Fellay J, Thompson AJ, Ge D, Gumbs CE, Urban TJ, Shianna KV, Little LD, Qiu P, Bertelsen AH, Watson M. ITPA gene variants protect against anaemia in patients treated for chronic hepatitis C. Nature. 2010;464:405-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 371] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 20. | Ochi H, Maekawa T, Abe H, Hayashida Y, Nakano R, Kubo M, Tsunoda T, Hayes CN, Kumada H, Nakamura Y. ITPA polymorphism affects ribavirin-induced anemia and outcomes of therapy--a genome-wide study of Japanese HCV virus patients. Gastroenterology. 2010;139:1190-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 143] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 21. | Hepburn MJ, Hepburn LM, Cantu NS, Lapeer MG, Lawitz EJ. Differences in treatment outcome for hepatitis C among ethnic groups. Am J Med. 2004;117:163-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 83] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 22. | Jeffers LJ, Cassidy W, Howell CD, Hu S, Reddy KR. Peginterferon alfa-2a (40 kd) and ribavirin for black American patients with chronic HCV genotype 1. Hepatology. 2004;39:1702-1708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 195] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 23. | Muir AJ, Bornstein JD, Killenberg PG, Atlantic Coast Hepatitis Treatment Group. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N Engl J Med. 2004;350:2265-2271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 431] [Cited by in RCA: 401] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 24. | Cheung RC, Currie S, Shen H, Ho SB, Bini EJ, Anand BS, Bräu N, Wright TL. Chronic hepatitis C in Latinos: natural history, treatment eligibility, acceptance, and outcomes. Am J Gastroenterol. 2005;100:2186-2193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 49] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 25. | Conjeevaram HS, Fried MW, Jeffers LJ, Terrault NA, Wiley-Lucas TE, Afdhal N, Brown RS, Belle SH, Hoofnagle JH, Kleiner DE. Peginterferon and ribavirin treatment in African American and Caucasian American patients with hepatitis C genotype 1. Gastroenterology. 2006;131:470-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 366] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 26. | Missiha S, Heathcote J, Arenovich T, Khan K. Impact of asian race on response to combination therapy with peginterferon alfa-2a and ribavirin in chronic hepatitis C. Am J Gastroenterol. 2007;102:2181-2188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 27. | Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, Urban T, Afdhal NH, Jacobson IM, Esteban R. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology. 2010;139:120-129.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 535] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 28. | Clark PJ, Thompson AJ, McHutchison JG. IL28B genomic-based treatment paradigms for patients with chronic hepatitis C infection: the future of personalized HCV therapies. Am J Gastroenterol. 2011;106:38-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 29. | Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R. Telaprevir for previously untreated chronic hepatitis C virus infection. N Engl J Med. 2011;364:2405-2416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1905] [Cited by in RCA: 1861] [Article Influence: 132.9] [Reference Citation Analysis (0)] |
| 30. | Poordad F, McCone J, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195-1206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1996] [Cited by in RCA: 1980] [Article Influence: 141.4] [Reference Citation Analysis (0)] |
| 31. | Nguyen LH, Nguyen MH. Systematic review: Asian patients with chronic hepatitis C infection. Aliment Pharmacol Ther. 2013;37:921-936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 32. | Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, O’Huigin C, Kidd J, Kidd K, Khakoo SI, Alexander G. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798-801. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1706] [Cited by in RCA: 1687] [Article Influence: 105.4] [Reference Citation Analysis (0)] |
| 33. | Poynard T, McHutchison J, Goodman Z, Ling MH, Albrecht J. Is an “a la carte” combination interferon alfa-2b plus ribavirin regimen possible for the first line treatment in patients with chronic hepatitis C? The ALGOVIRC Project Group. Hepatology. 2000;31:211-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 250] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 34. | Shiffman ML, Suter F, Bacon BR, Nelson D, Harley H, Solá R, Shafran SD, Barange K, Lin A, Soman A. Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2007;357:124-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 410] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 35. | Dalgard O, Bjøro K, Ring-Larsen H, Bjornsson E, Holberg-Petersen M, Skovlund E, Reichard O, Myrvang B, Sundelöf B, Ritland S, Hellum K, Frydén A, Florholmen J, Verbaan H, North-C Group. Pegylated interferon alfa and ribavirin for 14 versus 24 weeks in patients with hepatitis C virus genotype 2 or 3 and rapid virological response. Hepatology. 2008;47:35-42. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 151] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 36. | Lagging M, Langeland N, Pedersen C, Färkkilä M, Buhl MR, Mørch K, Dhillon AP, Alsiö A, Hellstrand K, Westin J, Norkrans G, NORDynamIC Study Group. Randomized comparison of 12 or 24 weeks of peginterferon alpha-2a and ribavirin in chronic hepatitis C virus genotype 2/3 infection. Hepatology. 2008;47:1837-1845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 126] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 37. | Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, Poordad F, Goodman ZD, Sings HL, Boparai N. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1207-1217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1306] [Cited by in RCA: 1308] [Article Influence: 93.4] [Reference Citation Analysis (0)] |
| 38. | Poordad F, Bronowicki JP, Gordon SC, Zeuzem S, Jacobson IM, Sulkowski MS, Poynard T, Morgan TR, Molony C, Pedicone LD. Factors that predict response of patients with hepatitis C virus infection to boceprevir. Gastroenterology. 2012;143:608-618.e1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 150] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 39. | Palmer C, Corpuz T, Guirguis M, O’Toole S, Yan K, Bu Y, Jorgenson J, Talbot M, Loi K, Lloyd A. The effect of obesity on intrahepatic cytokine and chemokine expression in chronic hepatitis C infection. Gut. 2010;59:397-404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 40. | Ortiz V, Berenguer M, Rayón JM, Carrasco D, Berenguer J. Contribution of obesity to hepatitis C-related fibrosis progression. Am J Gastroenterol. 2002;97:2408-2414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 160] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 41. | Bressler BL, Guindi M, Tomlinson G, Heathcote J. High body mass index is an independent risk factor for nonresponse to antiviral treatment in chronic hepatitis C. Hepatology. 2003;38:639-644. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 268] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 42. | Berg T, von Wagner M, Nasser S, Sarrazin C, Heintges T, Gerlach T, Buggisch P, Goeser T, Rasenack J, Pape GR. Extended treatment duration for hepatitis C virus type 1: comparing 48 versus 72 weeks of peginterferon-alfa-2a plus ribavirin. Gastroenterology. 2006;130:1086-1097. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 366] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 43. | Zeuzem S, Hultcrantz R, Bourliere M, Goeser T, Marcellin P, Sanchez-Tapias J, Sarrazin C, Harvey J, Brass C, Albrecht J. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C in previously untreated patients infected with HCV genotypes 2 or 3. J Hepatol. 2004;40:993-999. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 254] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 44. | Jacobson IM, Brown RS, Freilich B, Afdhal N, Kwo PY, Santoro J, Becker S, Wakil AE, Pound D, Godofsky E. Peginterferon alfa-2b and weight-based or flat-dose ribavirin in chronic hepatitis C patients: a randomized trial. Hepatology. 2007;46:971-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 210] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 45. | Deltenre P, Louvet A, Lemoine M, Mourad A, Fartoux L, Moreno C, Henrion J, Mathurin P, Serfaty L. Impact of insulin resistance on sustained response in HCV patients treated with pegylated interferon and ribavirin: a meta-analysis. J Hepatol. 2011;55:1187-1194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (1)] |
| 46. | Eslam M, Aparcero R, Kawaguchi T, Del Campo JA, Sata M, Khattab MA, Romero-Gomez M. Meta-analysis: insulin resistance and sustained virological response in hepatitis C. Aliment Pharmacol Ther. 2011;34:297-305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 83] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 47. | Del Campo JA, Ampuero J, Rojas L, Conde M, Rojas A, Maraver M, Millán R, García-Valdecasas M, García-Lozano JR, González-Escribano MF. Insulin resistance predicts sustained virological response to treatment of chronic hepatitis C independently of the IL28b rs12979860 polymorphism. Aliment Pharmacol Ther. 2013;37:74-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 48. | Ogawa E, Furusyo N, Murata M, Ikezaki H, Ihara T, Hayashi T, Toyoda K, Taniai H, Okada K, Kainuma M. Insulin resistance undermines the advantages of IL28B polymorphism in the pegylated interferon alpha-2b and ribavirin treatment of chronic hepatitis C patients with genotype 1. J Hepatol. 2012;57:534-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 36] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 49. | Serfaty L, Forns X, Goeser T, Ferenci P, Nevens F, Carosi G, Drenth JP, Lonjon-Domanec I, DeMasi R, Picchio G. Insulin resistance and response to telaprevir plus peginterferon α and ribavirin in treatment-naive patients infected with HCV genotype 1. Gut. 2012;61:1473-1480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 60] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 50. | Gopal K, Johnson TC, Gopal S, Walfish A, Bang CT, Suwandhi P, Pena-Sahdala HN, Clain DJ, Bodenheimer HC, Min AD. Correlation between beta-lipoprotein levels and outcome of hepatitis C treatment. Hepatology. 2006;44:335-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 90] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 51. | Akuta N, Suzuki F, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M, Arase Y. Predictive factors of early and sustained responses to peginterferon plus ribavirin combination therapy in Japanese patients infected with hepatitis C virus genotype 1b: amino acid substitutions in the core region and low-density lipoprotein cholesterol levels. J Hepatol. 2007;46:403-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 218] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 52. | Backus LI, Boothroyd DB, Phillips BR, Mole LA. Predictors of response of US veterans to treatment for the hepatitis C virus. Hepatology. 2007;46:37-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 183] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 53. | Economou M, Milionis H, Filis S, Baltayiannis G, Christou L, Elisaf M, Tsianos E. Baseline cholesterol is associated with the response to antiviral therapy in chronic hepatitis C. J Gastroenterol Hepatol. 2008;23:586-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 33] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 54. | Sheridan DA, Price DA, Schmid ML, Toms GL, Donaldson P, Neely D, Bassendine MF. Apolipoprotein B-associated cholesterol is a determinant of treatment outcome in patients with chronic hepatitis C virus infection receiving anti-viral agents interferon-alpha and ribavirin. Aliment Pharmacol Ther. 2009;29:1282-1290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 55. | Harrison SA, Rossaro L, Hu KQ, Patel K, Tillmann H, Dhaliwal S, Torres DM, Koury K, Goteti VS, Noviello S. Serum cholesterol and statin use predict virological response to peginterferon and ribavirin therapy. Hepatology. 2010;52:864-874. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 56. | Ramcharran D, Wahed AS, Conjeevaram HS, Evans RW, Wang T, Belle SH, Yee LJ, Virahep-C Study Group. Associations between serum lipids and hepatitis C antiviral treatment efficacy. Hepatology. 2010;52:854-863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 68] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 57. | Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, Focaccia R, Younossi Z, Foster GR, Horban A. Telaprevir for retreatment of HCV infection. N Engl J Med. 2011;364:2417-2428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1230] [Cited by in RCA: 1214] [Article Influence: 86.7] [Reference Citation Analysis (0)] |
| 58. | Berg T, Andreone P, Pol S, Roberts S, Younossi Z, Diago M, Lawitz E, Focaccia R, Foster GR, Horban A. Predictors of virologic response with telaprevir-based combination treatment in HCV genotype 1-infected patients with prior peginterferon/ribavirin treatment failure: post hoc analysis of the phase III REALIZE study. Hepatology. 2011;54:S24. [DOI] [Full Text] |
| 59. | André P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Bréchot C, Paranhos-Baccalà G, Lotteau V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J Virol. 2002;76:6919-6928. [PubMed] |
| 60. | André P, Perlemuter G, Budkowska A, Bréchot C, Lotteau V. Hepatitis C virus particles and lipoprotein metabolism. Semin Liver Dis. 2005;25:93-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 134] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 61. | Dreux M, Cosset FL. HCV and lipoproteins: is oxLDL an Achilles’ heel of the Trojan horse? Hepatology. 2006;43:903-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 62. | Monazahian M, Böhme I, Bonk S, Koch A, Scholz C, Grethe S, Thomssen R. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J Med Virol. 1999;57:223-229. [PubMed] |
| 63. | Carrière M, Rosenberg AR, Conti F, Chouzenoux S, Terris B, Sogni P, Soubrane O, Calmus Y, Podevin P. Low density lipoprotein receptor transcripts correlates with liver hepatitis C virus RNA in patients with alcohol consumption. J Viral Hepat. 2006;13:633-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 64. | Molina S, Castet V, Fournier-Wirth C, Pichard-Garcia L, Avner R, Harats D, Roitelman J, Barbaras R, Graber P, Ghersa P. The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J Hepatol. 2007;46:411-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 219] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 65. | Petit JM, Minello A, Duvillard L, Jooste V, Monier S, Texier V, Bour JB, Poussier A, Gambert P, Verges B. Cell surface expression of LDL receptor in chronic hepatitis C: correlation with viral load. Am J Physiol Endocrinol Metab. 2007;293:E416-E420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 66. | Murao K, Imachi H, Yu X, Cao WM, Nishiuchi T, Chen K, Li J, Ahmed RA, Wong NC, Ishida T. Interferon alpha decreases expression of human scavenger receptor class BI, a possible HCV receptor in hepatocytes. Gut. 2008;57:664-671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 67. | Li JH, Lao XQ, Tillmann HL, Rowell J, Patel K, Thompson A, Suchindran S, Muir AJ, Guyton JR, Gardner SD. Interferon-lambda genotype and low serum low-density lipoprotein cholesterol levels in patients with chronic hepatitis C infection. Hepatology. 2010;51:1904-1911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 68. | Clark PJ, Thompson AJ, Zhu M, Vock DM, Zhu Q, Ge D, Patel K, Harrison SA, Urban TJ, Naggie S. Interleukin 28B polymorphisms are the only common genetic variants associated with low-density lipoprotein cholesterol (LDL-C) in genotype-1 chronic hepatitis C and determine the association between LDL-C and treatment response. J Viral Hepat. 2012;19:332-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 69. | Sheridan DA, Bridge SH, Felmlee DJ, Crossey MM, Thomas HC, Taylor-Robinson SD, Toms GL, Neely RD, Bassendine MF. Apolipoprotein-E and hepatitis C lipoviral particles in genotype 1 infection: evidence for an association with interferon sensitivity. J Hepatol. 2012;57:32-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 70. | Everson GT, Hoefs JC, Seeff LB, Bonkovsky HL, Naishadham D, Shiffman ML, Kahn JA, Lok AS, Di Bisceglie AM, Lee WM, Dienstag JL, Ghany MG, Morishima C, HALT-C Trial Group. Impact of disease severity on outcome of antiviral therapy for chronic hepatitis C: Lessons from the HALT-C trial. Hepatology. 2006;44:1675-1684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 105] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 71. | Jacobson IM, Marcellin P, Zeuzem S, Sulkowski MS, Esteban R, Poordad F, Bruno S, Burroughs MH, Pedicone LD, Boparai N. Refinement of stopping rules during treatment of hepatitis C genotype 1 infection with boceprevir and peginterferon/ribavirin. Hepatology. 2012;56:567-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 72. | Schlecker C, Ultsch A, Geisslinger G, Lötsch J. The pharmacogenetic background of hepatitis C treatment. Mutat Res. 2012;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 73. | Estrabaud E, Vidaud M, Marcellin P, Asselah T. Genomics and HCV infection: progression of fibrosis and treatment response. J Hepatol. 2012;57:1110-1125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 83] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 74. | Knapp S, Yee LJ, Frodsham AJ, Hennig BJ, Hellier S, Zhang L, Wright M, Chiaramonte M, Graves M, Thomas HC. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun. 2003;4:411-419. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 173] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 75. | Matsuyama N, Mishiro S, Sugimoto M, Furuichi Y, Hashimoto M, Hijikata M, Ohta Y. The dinucleotide microsatellite polymorphism of the IFNAR1 gene promoter correlates with responsiveness of hepatitis C patients to interferon. Hepatol Res. 2003;25:221-225. [PubMed] |
| 76. | Naito M, Matsui A, Inao M, Nagoshi S, Nagano M, Ito N, Egashira T, Hashimoto M, Mishiro S, Mochida S. SNPs in the promoter region of the osteopontin gene as a marker predicting the efficacy of interferon-based therapies in patients with chronic hepatitis C. J Gastroenterol. 2005;40:381-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 55] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 77. | Huang Y, Yang H, Borg BB, Su X, Rhodes SL, Yang K, Tong X, Tang G, Howell CD, Rosen HR. A functional SNP of interferon-gamma gene is important for interferon-alpha-induced and spontaneous recovery from hepatitis C virus infection. Proc Natl Acad Sci USA. 2007;104:985-990. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 91] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 78. | Rhodes SL, Erlich H, Im KA, Wang J, Li J, Bugawan T, Jeffers L, Tong X, Su X, Rosen HR, Yee LJ, Liang TJ, Yang H, Virahep-C Study Group. Associations between the human MHC and sustained virologic response in the treatment of chronic hepatitis C virus infection. Genes Immun. 2008;9:328-333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 79. | Tsukada H, Ochi H, Maekawa T, Abe H, Fujimoto Y, Tsuge M, Takahashi H, Kumada H, Kamatani N, Nakamura Y. A polymorphism in MAPKAPK3 affects response to interferon therapy for chronic hepatitis C. Gastroenterology. 2009;136:1796-1805.e6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 80. | Yee LJ, Im K, Borg B, Yang H, Liang TJ. Interleukin-6 haplotypes and the response to therapy of chronic hepatitis C virus infection. Genes Immun. 2009;10:365-372. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 81. | Knapp S, Warshow U, Hegazy D, Brackenbury L, Guha IN, Fowell A, Little AM, Alexander GJ, Rosenberg WM, Cramp ME. Consistent beneficial effects of killer cell immunoglobulin-like receptor 2DL3 and group 1 human leukocyte antigen-C following exposure to hepatitis C virus. Hepatology. 2010;51:1168-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 127] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 82. | Vidal-Castiñeira JR, López-Vázquez A, Díaz-Peña R, Alonso-Arias R, Martínez-Borra J, Pérez R, Fernández-Suárez J, Melón S, Prieto J, Rodrigo L. Effect of killer immunoglobulin-like receptors in the response to combined treatment in patients with chronic hepatitis C virus infection. J Virol. 2010;84:475-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 83. | Chen L, Borozan I, Feld J, Sun J, Tannis LL, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology. 2005;128:1437-1444. [PubMed] |
| 84. | Asselah T, Bieche I, Narguet S, Sabbagh A, Laurendeau I, Ripault MP, Boyer N, Martinot-Peignoux M, Valla D, Vidaud M. Liver gene expression signature to predict response to pegylated interferon plus ribavirin combination therapy in patients with chronic hepatitis C. Gut. 2008;57:516-524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 152] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 85. | Chen L, Borozan I, Sun J, Guindi M, Fischer S, Feld J, Anand N, Heathcote J, Edwards AM, McGilvray ID. Cell-type specific gene expression signature in liver underlies response to interferon therapy in chronic hepatitis C infection. Gastroenterology. 2010;138:1123-1133.e1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 87] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 86. | McGilvray I, Feld JJ, Chen L, Pattullo V, Guindi M, Fischer S, Borozan I, Xie G, Selzner N, Heathcote EJ. Hepatic cell-type specific gene expression better predicts HCV treatment outcome than IL28B genotype. Gastroenterology. 2012;142:1122-1131.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 87. | Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat Med. 2009;15:31-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 252] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 88. | Murakami Y, Tanaka M, Toyoda H, Hayashi K, Kuroda M, Tajima A, Shimotohno K. Hepatic microRNA expression is associated with the response to interferon treatment of chronic hepatitis C. BMC Med Genomics. 2010;3:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 89. | Afdhal NH, McHutchison JG, Zeuzem S, Mangia A, Pawlotsky JM, Murray JS, Shianna KV, Tanaka Y, Thomas DL, Booth DR. Hepatitis C pharmacogenetics: state of the art in 2010. Hepatology. 2011;53:336-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 119] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 90. | Tillmann HL, Thompson AJ, Patel K, Wiese M, Tenckhoff H, Nischalke HD, Lokhnygina Y, Kullig U, Göbel U, Capka E, Wiegand J, Schiefke I, Güthoff W, Grüngreiff K, König I, Spengler U, McCarthy J, Shianna KV, Goldstein DB, McHutchison JG, Timm J, Nattermann J, German Anti-D Study Group. A polymorphism near IL28B is associated with spontaneous clearance of acute hepatitis C virus and jaundice. Gastroenterology. 2010;139:1586-1592, 1592.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 241] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 91. | Duggal P, Thio CL, Wojcik GL, Goedert JJ, Mangia A, Latanich R, Kim AY, Lauer GM, Chung RT, Peters MG. Genome-wide association study of spontaneous resolution of hepatitis C virus infection: data from multiple cohorts. Ann Intern Med. 2013;158:235-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 173] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 92. | Kobayashi M, Suzuki F, Akuta N, Sezaki H, Suzuki Y, Hosaka T, Kawamura Y, Kobayashi M, Saitoh S, Arase Y. Association of two polymorphisms of the IL28B gene with viral factors and treatment response in 1,518 patients infected with hepatitis C virus. J Gastroenterol. 2012;47:596-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 93. | McCarthy JJ, Li JH, Thompson A, Suchindran S, Lao XQ, Patel K, Tillmann HL, Muir AJ, McHutchison JG. Replicated association between an IL28B gene variant and a sustained response to pegylated interferon and ribavirin. Gastroenterology. 2010;138:2307-2314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 246] [Cited by in RCA: 261] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 94. | Montes-Cano MA, García-Lozano JR, Abad-Molina C, Romero-Gómez M, Barroso N, Aguilar-Reina J, Núñez-Roldán A, González-Escribano MF. Interleukin-28B genetic variants and hepatitis virus infection by different viral genotypes. Hepatology. 2010;52:33-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 166] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 95. | Hayes CN, Kobayashi M, Akuta N, Suzuki F, Kumada H, Abe H, Miki D, Imamura M, Ochi H, Kamatani N. HCV substitutions and IL28B polymorphisms on outcome of peg-interferon plus ribavirin combination therapy. Gut. 2011;60:261-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 85] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 96. | Venegas M, Villanueva RA, González K, Brahm J. IL28B polymorphisms associated with therapy response in Chilean chronic hepatitis C patients. World J Gastroenterol. 2011;17:3636-3639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 35] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 97. | Cieśla A, Bociąga-Jasik M, Sobczyk-Krupiarz I, Głowacki MK, Owczarek D, Cibor D, Sanak M, Mach T. IL28B polymorphism as a predictor of antiviral response in chronic hepatitis C. World J Gastroenterol. 2012;18:4892-4897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 98. | De Nicola S, Aghemo A, Rumi MG, Galmozzi E, Valenti L, Soffredini R, De Francesco R, Prati GM, D’Ambrosio R, Cheroni C. Interleukin 28B polymorphism predicts pegylated interferon plus ribavirin treatment outcome in chronic hepatitis C genotype 4. Hepatology. 2012;55:336-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 99. | Asselah T, De Muynck S, Broët P, Masliah-Planchon J, Blanluet M, Bièche I, Lapalus M, Martinot-Peignoux M, Lada O, Estrabaud E. IL28B polymorphism is associated with treatment response in patients with genotype 4 chronic hepatitis C. J Hepatol. 2012;56:527-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 146] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 100. | Derbala M, Rizk N, Shebl F, Alkaabi S, Eldweik N, John A, Sharma M, Yaqoob R, Almohanadi M, Butt M. Interleukin-28 and hepatitis C virus genotype-4: treatment-induced clearance and liver fibrosis. World J Gastroenterol. 2012;18:7003-7008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 14] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 101. | Antaki N, Bibert S, Kebbewar K, Asaad F, Baroudi O, Alideeb S, Hadad M, Abboud D, Sabah H, Bochud PY. IL28B polymorphisms predict response to therapy among chronic hepatitis C patients with HCV genotype 4. J Viral Hepat. 2013;20:59-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 102. | Grebely J, Petoumenos K, Hellard M, Matthews GV, Suppiah V, Applegate T, Yeung B, Marks P, Rawlinson W, Lloyd AR, Booth D, Kaldor JM, George J, Dore GJ, ATAHC Study Group. Potential role for interleukin-28B genotype in treatment decision-making in recent hepatitis C virus infection. Hepatology. 2010;52:1216-1224. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 136] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 103. | di Iulio J, Ciuffi A, Fitzmaurice K, Kelleher D, Rotger M, Fellay J, Martinez R, Pulit S, Furrer H, Günthard HF. Estimating the net contribution of interleukin-28B variation to spontaneous hepatitis C virus clearance. Hepatology. 2011;53:1446-1454. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 104. | Ruiz-Extremera A, Muñoz-Gámez JA, Salmerón-Ruiz MA, de Rueda PM, Quiles-Pérez R, Gila-Medina A, Casado J, Belén Martín A, Sanjuan-Nuñez L, Carazo A. Genetic variation in interleukin 28B with respect to vertical transmission of hepatitis C virus and spontaneous clearance in HCV-infected children. Hepatology. 2011;53:1830-1838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 95] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 105. | Grebely J, Page K, Sacks-Davis R, van der Loeff MS, Rice TM, Bruneau J, Morris MD, Hajarizadeh B, Amin J, Cox AL, Kim AY, McGovern BH, Schinkel J, George J, Shoukry NH, Lauer GM, Maher L, Lloyd AR, Hellard M, Dore GJ, Prins M, InC3 Study Group. The effects of female sex, viral genotype, and IL28B genotype on spontaneous clearance of acute hepatitis C virus infection. Hepatology. 2014;59:109-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 298] [Cited by in RCA: 302] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 106. | Rallón NI, Naggie S, Benito JM, Medrano J, Restrepo C, Goldstein D, Shianna KV, Vispo E, Thompson A, McHutchison J. Association of a single nucleotide polymorphism near the interleukin-28B gene with response to hepatitis C therapy in HIV/hepatitis C virus-coinfected patients. AIDS. 2010;24:F23-F29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 163] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 107. | Pineda JA, Caruz A, Rivero A, Neukam K, Salas I, Camacho A, Palomares JC, Mira JA, Martínez A, Roldán C. Prediction of response to pegylated interferon plus ribavirin by IL28B gene variation in patients coinfected with HIV and hepatitis C virus. Clin Infect Dis. 2010;51:788-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 108. | Aparicio E, Parera M, Franco S, Pérez-Alvarez N, Tural C, Clotet B, Martínez MA. IL28B SNP rs8099917 is strongly associated with pegylated interferon-α and ribavirin therapy treatment failure in HCV/HIV-1 coinfected patients. PLoS One. 2010;5:e13771. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 109. | Fukuhara T, Taketomi A, Motomura T, Okano S, Ninomiya A, Abe T, Uchiyama H, Soejima Y, Shirabe K, Matsuura Y. Variants in IL28B in liver recipients and donors correlate with response to peg-interferon and ribavirin therapy for recurrent hepatitis C. Gastroenterology. 2010;139:1577-1585, 1585.e1-3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 168] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 110. | Chen Y, Xu HX, Wang LJ, Liu XX, Mahato RI, Zhao YR. Meta-analysis: IL28B polymorphisms predict sustained viral response in HCV patients treated with pegylated interferon-α and ribavirin. Aliment Pharmacol Ther. 2012;36:91-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 111. | Rangnekar AS, Fontana RJ. Meta-analysis: IL-28B genotype and sustained viral clearance in HCV genotype 1 patients. Aliment Pharmacol Ther. 2012;36:104-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 112. | Jia Z, Ding Y, Tian S, Niu J, Jiang J. Test of IL28B polymorphisms in chronic hepatitis C patients treated with PegIFN and ribavirin depends on HCV genotypes: results from a meta-analysis. PLoS One. 2012;7:e45698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 113. | Luo Y, Jin C, Ling Z, Mou X, Zhang Q, Xiang C. Association study of IL28B: rs12979860 and rs8099917 polymorphisms with SVR in patients infected with chronic HCV genotype 1 to PEG-INF/RBV therapy using systematic meta-analysis. Gene. 2013;513:292-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 114. | Jiménez-Sousa MA, Fernández-Rodríguez A, Guzmán-Fulgencio M, García-Álvarez M, Resino S. Meta-analysis: implications of interleukin-28B polymorphisms in spontaneous and treatment-related clearance for patients with hepatitis C. BMC Med. 2013;11:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 72] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 115. | Sarrazin C, Susser S, Doehring A, Lange CM, Müller T, Schlecker C, Herrmann E, Lötsch J, Berg T. Importance of IL28B gene polymorphisms in hepatitis C virus genotype 2 and 3 infected patients. J Hepatol. 2011;54:415-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 180] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 116. | Kawaoka T, Hayes CN, Ohishi W, Ochi H, Maekawa T, Abe H, Tsuge M, Mitsui F, Hiraga N, Imamura M. Predictive value of the IL28B polymorphism on the effect of interferon therapy in chronic hepatitis C patients with genotypes 2a and 2b. J Hepatol. 2011;54:408-414. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 117. | Rauch A, Kutalik Z, Descombes P, Cai T, Di Iulio J, Mueller T, Bochud M, Battegay M, Bernasconi E, Borovicka J. Genetic variation in IL28B is associated with chronic hepatitis C and treatment failure: a genome-wide association study. Gastroenterology. 2010;138:1338-1345, 1345.e1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 850] [Cited by in RCA: 867] [Article Influence: 57.8] [Reference Citation Analysis (0)] |
| 118. | Yu ML, Huang CF, Huang JF, Chang NC, Yang JF, Lin ZY, Chen SC, Hsieh MY, Wang LY, Chang WY. Role of interleukin-28B polymorphisms in the treatment of hepatitis C virus genotype 2 infection in Asian patients. Hepatology. 2011;53:7-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 157] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 119. | Moghaddam A, Melum E, Reinton N, Ring-Larsen H, Verbaan H, Bjøro K, Dalgard O. IL28B genetic variation and treatment response in patients with hepatitis C virus genotype 3 infection. Hepatology. 2011;53:746-754. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 103] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 120. | Mangia A, Thompson AJ, Santoro R, Piazzolla V, Tillmann HL, Patel K, Shianna KV, Mottola L, Petruzzellis D, Bacca D. An IL28B polymorphism determines treatment response of hepatitis C virus genotype 2 or 3 patients who do not achieve a rapid virologic response. Gastroenterology. 2010;139:821-827, 827.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 235] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 121. | Schreiber J, Moreno C, Garcia BG, Louvet A, Trepo E, Henrion J, Thabut D, Mathurin P, Deltenre P. Meta-analysis: the impact of IL28B polymorphisms on rapid and sustained virological response in HCV-2 and -3 patients. Aliment Pharmacol Ther. 2012;36:353-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 122. | Asahina Y, Tsuchiya K, Muraoka M, Tanaka K, Suzuki Y, Tamaki N, Hoshioka Y, Yasui Y, Katoh T, Hosokawa T. Association of gene expression involving innate immunity and genetic variation in interleukin 28B with antiviral response. Hepatology. 2012;55:20-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 123. | Heim MH. Innate immunity and HCV. J Hepatol. 2013;58:564-574. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 124. | Meager A, Visvalingam K, Dilger P, Bryan D, Wadhwa M. Biological activity of interleukins-28 and -29: comparison with type I interferons. Cytokine. 2005;31:109-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 175] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 125. | Kotenko SV, Gallagher G, Baurin VV, Lewis-Antes A, Shen M, Shah NK, Langer JA, Sheikh F, Dickensheets H, Donnelly RP. IFN-lambdas mediate antiviral protection through a distinct class II cytokine receptor complex. Nat Immunol. 2003;4:69-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1416] [Cited by in RCA: 1482] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 126. | Sheppard P, Kindsvogel W, Xu W, Henderson K, Schlutsmeyer S, Whitmore TE, Kuestner R, Garrigues U, Birks C, Roraback J. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nat Immunol. 2003;4:63-68. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1221] [Cited by in RCA: 1215] [Article Influence: 55.2] [Reference Citation Analysis (0)] |
| 127. | Zhang L, Jilg N, Shao RX, Lin W, Fusco DN, Zhao H, Goto K, Peng LF, Chen WC, Chung RT. IL28B inhibits hepatitis C virus replication through the JAK-STAT pathway. J Hepatol. 2011;55:289-298. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 104] [Cited by in RCA: 108] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 128. | Maher SG, Sheikh F, Scarzello AJ, Romero-Weaver AL, Baker DP, Donnelly RP, Gamero AM. IFNalpha and IFNlambda differ in their antiproliferative effects and duration of JAK/STAT signaling activity. Cancer Biol Ther. 2008;7:1109-1115. [PubMed] |
| 129. | Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, McKenzie A, Patel K, Shianna KV. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology. 2010;52:1888-1896. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 303] [Cited by in RCA: 314] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 130. | Dill MT, Duong FH, Vogt JE, Bibert S, Bochud PY, Terracciano L, Papassotiropoulos A, Roth V, Heim MH. Interferon-induced gene expression is a stronger predictor of treatment response than IL28B genotype in patients with hepatitis C. Gastroenterology. 2011;140:1021-1031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 208] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 131. | Raglow Z, Thoma-Perry C, Gilroy R, Wan YJ. IL28B genotype and the expression of ISGs in normal liver. Liver Int. 2013;33:991-998. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 132. | Golden-Mason L, Bambha KM, Cheng L, Howell CD, Taylor MW, Clark PJ, Afdhal N, Rosen HR, Virahep-C Study Group. Natural killer inhibitory receptor expression associated with treatment failure and interleukin-28B genotype in patients with chronic hepatitis C. Hepatology. 2011;54:1559-1569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 57] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 133. | Naggie S, Osinusi A, Katsounas A, Lempicki R, Herrmann E, Thompson AJ, Clark PJ, Patel K, Muir AJ, McHutchison JG. Dysregulation of innate immunity in hepatitis C virus genotype 1 IL28B-unfavorable genotype patients: impaired viral kinetics and therapeutic response. Hepatology. 2012;56:444-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 134. | Shi X, Pan Y, Wang M, Wang D, Li W, Jiang T, Zhang P, Chi X, Jiang Y, Gao Y. IL28B genetic variation is associated with spontaneous clearance of hepatitis C virus, treatment response, serum IL-28B levels in Chinese population. PLoS One. 2012;7:e37054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 78] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 135. | Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y, Yamashita T, Nakamura M, Shirasaki T, Horimoto K, Tanaka Y, Tokunaga K, Mizokami M, Kaneko S, Hokuriku Liver Study Group. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 344] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 136. | Abe H, Hayes CN, Ochi H, Maekawa T, Tsuge M, Miki D, Mitsui F, Hiraga N, Imamura M, Takahashi S. IL28 variation affects expression of interferon stimulated genes and peg-interferon and ribavirin therapy. J Hepatol. 2011;54:1094-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 137. | Scott J, Holte S, Urban T, Burgess C, Coppel E, Wang C, Corey L, McHutchison J, Goldstein D. IL28B genotype effects during early treatment with peginterferon and ribavirin in difficult-to-treat hepatitis C virus infection. J Infect Dis. 2011;204:419-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 138. | Lindh M, Lagging M, Arnholm B, Eilard A, Nilsson S, Norkrans G, Söderholm J, Wahlberg T, Wejstål R, Westin J. IL28B polymorphisms determine early viral kinetics and treatment outcome in patients receiving peginterferon/ribavirin for chronic hepatitis C genotype 1. J Viral Hepat. 2011;18:e325-e331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 139. | Bochud PY, Bibert S, Negro F, Haagmans B, Soulier A, Ferrari C, Missale G, Zeuzem S, Pawlotsky JM, Schalm S. IL28B polymorphisms predict reduction of HCV RNA from the first day of therapy in chronic hepatitis C. J Hepatol. 2011;55:980-988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 89] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 140. | Arends JE, Fransen JH, Hoepelman AI, van Baarle D. Association between IL28B polymorphisms and first-phase viral load decrease in chronic hepatitis C virus-infected patients treated with peginterferon alfa-2b/ribavirin. Int J Antimicrob Agents. 2011;38:538-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 141. | Hsu CS, Hsu SJ, Chen HC, Tseng TC, Liu CH, Niu WF, Jeng J, Liu CJ, Lai MY, Chen PJ. Association of IL28B gene variations with mathematical modeling of viral kinetics in chronic hepatitis C patients with IFN plus ribavirin therapy. Proc Natl Acad Sci USA. 2011;108:3719-3724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 142. | Lindh M, Lagging M, Färkkilä M, Langeland N, Mørch K, Nilsson S, Norkrans G, Pedersen C, Buhl MR, Westin J. Interleukin 28B gene variation at rs12979860 determines early viral kinetics during treatment in patients carrying genotypes 2 or 3 of hepatitis C virus. J Infect Dis. 2011;203:1748-1752. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 143. | Howell CD, Gorden A, Ryan KA, Thompson AJ, Ibrahim C, Fried M, Afdhal NH, McHutchison JG, Shianna KV, Goldstein DB. Single nucleotide polymorphism upstream of interleukin 28B associated with phase 1 and phase 2 of early viral kinetics in patients infected with HCV genotype 1. J Hepatol. 2012;56:557-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 144. | Askarieh G, Alsiö A, Pugnale P, Negro F, Ferrari C, Neumann AU, Pawlotsky JM, Schalm SW, Zeuzem S, Norkrans G. Systemic and intrahepatic interferon-gamma-inducible protein 10 kDa predicts the first-phase decline in hepatitis C virus RNA and overall viral response to therapy in chronic hepatitis C. Hepatology. 2010;51:1523-1530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 99] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 145. | Fattovich G, Covolo L, Bibert S, Askarieh G, Lagging M, Clément S, Malerba G, Pasino M, Guido M, Puoti M, Gaeta GB, Santantonio T, Raimondo G, Bruno R, Bochud PY, Donato F, Negro F, ITAHEC Study Group. IL28B polymorphisms, IP-10 and viral load predict virological response to therapy in chronic hepatitis C. Aliment Pharmacol Ther. 2011;33:1162-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 146. | Lagging M, Askarieh G, Negro F, Bibert S, Söderholm J, Westin J, Lindh M, Romero A, Missale G, Ferrari C, Neumann AU, Pawlotsky JM, Haagmans BL, Zeuzem S, Bochud PY, Hellstrand K, DITTO-HCV Study Group. Response prediction in chronic hepatitis C by assessment of IP-10 and IL28B-related single nucleotide polymorphisms. PLoS One. 2011;6:e17232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 110] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 147. | Beinhardt S, Aberle JH, Strasser M, Dulic-Lakovic E, Maieron A, Kreil A, Rutter K, Staettermayer AF, Datz C, Scherzer TM. Serum level of IP-10 increases predictive value of IL28B polymorphisms for spontaneous clearance of acute HCV infection. Gastroenterology. 2012;142:78-85.e2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 148. | Cammà C, Petta S, Enea M, Bruno R, Bronte F, Capursi V, Cicchetti A, Colombo GL, Di Marco V, Gasbarrini A, Craxì A, WEF Study Group. Cost-effectiveness of boceprevir or telaprevir for untreated patients with genotype 1 chronic hepatitis C. Hepatology. 2012;56:850-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 149. | Liu S, Cipriano LE, Holodniy M, Owens DK, Goldhaber-Fiebert JD. New protease inhibitors for the treatment of chronic hepatitis C: a cost-effectiveness analysis. Ann Intern Med. 2012;156:279-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 150. | Mangia A, Thompson AJ, Santoro R, Piazzolla V, Copetti M, Minerva N, Petruzzellis D, Mottola L, Bacca D, McHutchison JG. Limited use of interleukin 28B in the setting of response-guided treatment with detailed on-treatment virological monitoring. Hepatology. 2011;54:772-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 151. | Liu CH, Liang CC, Liu CJ, Tseng TC, Lin CL, Yang SS, Su TH, Lin JW, Chen JH, Chen PJ. Interleukin 28B genetic polymorphisms play a minor role in identifying optimal treatment duration in HCV genotype 1 slow responders to pegylated interferon plus ribavirin. Antivir Ther. 2012;17:1059-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 152. | Huang CF, Huang JF, Yang JF, Hsieh MY, Lin ZY, Chen SC, Wang LY, Juo SH, Chen KC, Chuang WL. Interleukin-28B genetic variants in identification of hepatitis C virus genotype 1 patients responding to 24 weeks peginterferon/ribavirin. J Hepatol. 2012;56:34-40. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 153. | Liu CH, Liang CC, Liu CJ, Tseng TC, Lin CL, Yang SS, Su TH, Hsu SJ, Lin JW, Chen JH. Interleukin 28B genetic polymorphisms and viral factors help identify HCV genotype-1 patients who benefit from 24-week pegylated interferon plus ribavirin therapy. Antivir Ther. 2012;17:477-484. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 154. | Sarrazin C, Schwendy S, Möller B, Dikopoulos N, Buggisch P, Encke J, Teuber G, Goeser T, Thimme R, Klinker H. Improved responses to pegylated interferon alfa-2b and ribavirin by individualizing treatment for 24-72 weeks. Gastroenterology. 2011;141:1656-1664. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 155. | Pearlman BL, Ehleben C. The IL-28B genotype predicts which slow-responding hepatitis C-infected patients will benefit from treatment extension. Am J Gastroenterol. 2011;106:1370-1371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 156. | Scherzer TM, Stättermayer AF, Strasser M, Laferl H, Maieron A, Stauber R, Datz C, Dulic-Lakovic E, Steindl-Munda P, Hofer H. Impact of IL28B on treatment outcome in hepatitis C virus G1/4 patients receiving response-guided therapy with peginterferon alpha-2a (40KD)/ribavirin. Hepatology. 2011;54:1518-1526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 157. | Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Kobayashi M. Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology. 2010;52:421-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 201] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 158. | Chayama K, Hayes CN, Abe H, Miki D, Ochi H, Karino Y, Toyota J, Nakamura Y, Kamatani N, Sezaki H. IL28B but not ITPA polymorphism is predictive of response to pegylated interferon, ribavirin, and telaprevir triple therapy in patients with genotype 1 hepatitis C. J Infect Dis. 2011;204:84-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 111] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 159. | Furusyo N, Ogawa E, Nakamuta M, Kajiwara E, Nomura H, Dohmen K, Takahashi K, Satoh T, Azuma K, Kawano A, Tanabe Y, Kotoh K, Shimoda S, Hayashi J, Kyushu University Liver Disease Study (KULDS) Group. Telaprevir can be successfully and safely used to treat older patients with genotype 1b chronic hepatitis C. J Hepatol. 2013;59:205-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 160. | Akuta N, Suzuki F, Fukushima T, Kawamura Y, Sezaki H, Suzuki Y, Hosaka T, Kobayashi M, Hara T, Kobayashi M. Prediction of treatment efficacy and telaprevir-resistant variants after triple therapy in patients infected with hepatitis C virus genotype 1. J Clin Microbiol. 2013;51:2862-2868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 161. | Poordad F, Bronowicki JP, Gordon SC, Zeuzem S, Jacobson I, Sulkowski M, Poynard T, Morgan TR, Burroughs M, Sniukiene V. IL28B Polymorphism Predicts Virologic Response in Patients with Hepatitis C Genotype 1 Treated with Boceprevir Combination Therapy. J Hepatol. 2011;54:S6. [DOI] [Full Text] |
| 162. | Pol S, Aerssens J, Zeuzem S, Andreone P, Lawitz EJ, Roberts S, Younossi Z, Foster GR, Focaccia R, Horban A. Limited impact of IL28B genotype on response rates in telaprevir-treated patients with prior treatment failure. J Hepatol. 2013;58:883-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 163. | Chu TW, Kulkarni R, Gane EJ, Roberts SK, Stedman C, Angus PW, Ritchie B, Lu XY, Ipe D, Lopatin U. Effect of IL28B genotype on early viral kinetics during interferon-free treatment of patients with chronic hepatitis C. Gastroenterology. 2012;142:790-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 76] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 164. | Wedemeyer H, Jensen D, Herring R, Ferenci P, Ma MM, Zeuzem S, Rodriguez-Torres M, Bzowej N, Pockros P, Vierling J. PROPEL: a randomized trial of mericitabine plus peginterferon alpha-2a/ribavirin therapy in treatment-naïve HCV genotype 1/4 patients. Hepatology. 2013;58:524-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 165. | Pockros PJ, Jensen D, Tsai N, Taylor R, Ramji A, Cooper C, Dickson R, Tice A, Kulkarni R, Vierling JM. JUMP-C: a randomized trial of mericitabine plus pegylated interferon alpha-2a/ribavirin for 24 weeks in treatment-naïve HCV genotype 1/4 patients. Hepatology. 2013;58:514-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 166. | Zeuzem S, Soriano V, Asselah T, Bronowicki JP, Lohse AW, Müllhaupt B, Schuchmann M, Bourlière M, Buti M, Roberts SK. Faldaprevir and deleobuvir for HCV genotype 1 infection. N Engl J Med. 2013;369:630-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 183] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 167. | Poordad F, Lawitz E, Kowdley KV, Everson GT, Freilich B, Cohen CJ, Siggelkow S, Heckaman M, Menon H, Pilot-Matias T. 12-week interferon-free regimen of ABT-450/r ABT-333 ribavirin achieved SVR12 in more than 90% of treatment-naïve HCV genotype-1-infected subjects and 47% of previous non-responders. J Hepatol. 2012;56 Suppl 2:S549-S550. [DOI] [Full Text] |
| 168. | Sulkowski M, Gardiner D, Lawitz E, Hinestrosa F, Nelson DR, Thuluvath P, Rodriguez-Torres M, Lok A, Schwartz H, Reddy KR. Potent viral suppression with all-oral combination of daclatasvir (NS5A inhibitor) an GS-7977 (NS5B inhibitor), /− ribavirin, in treatment-naive patients with chronic HCV GT1, 2, or 3. J Hepatol. 2012;56:S560. [DOI] [Full Text] |
| 169. | Kowdley KV, Lawitz E, Crespo I, Hassanein T, Davis MN, DeMicco M, Bernstein DE, Afdhal N, Vierling JM, Gordon SC. Sofosbuvir with pegylated interferon alfa-2a and ribavirin for treatment-naive patients with hepatitis C genotype-1 infection (ATOMIC): an open-label, randomised, multicentre phase 2 trial. Lancet. 2013;381:2100-2107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 223] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 170. | De Franceschi L, Fattovich G, Turrini F, Ayi K, Brugnara C, Manzato F, Noventa F, Stanzial AM, Solero P, Corrocher R. Hemolytic anemia induced by ribavirin therapy in patients with chronic hepatitis C virus infection: role of membrane oxidative damage. Hepatology. 2000;31:997-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 328] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 171. | Russmann S, Grattagliano I, Portincasa P, Palmieri VO, Palasciano G. Ribavirin-induced anemia: mechanisms, risk factors and related targets for future research. Curr Med Chem. 2006;13:3351-3357. [PubMed] |
| 172. | Homma M, Hosono H, Hasegawa Y, Kohda Y. Morphological transformation and phosphatidylserine exposure in erythrocytes treated with ribavirin. Biol Pharm Bull. 2009;32:1940-1942. [PubMed] |
| 173. | Sumi S, Marinaki AM, Arenas M, Fairbanks L, Shobowale-Bakre M, Rees DC, Thein SL, Ansari A, Sanderson J, De Abreu RA. Genetic basis of inosine triphosphate pyrophosphohydrolase deficiency. Hum Genet. 2002;111:360-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 199] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 174. | Cao H, Hegele RA. DNA polymorphisms in ITPA including basis of inosine triphosphatase deficiency. J Hum Genet. 2002;47:620-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 79] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 175. | Arenas M, Duley J, Sumi S, Sanderson J, Marinaki A. The ITPA c.94C>A and g.IVS2+21A>C sequence variants contribute to missplicing of the ITPA gene. Biochim Biophys Acta. 2007;1772:96-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 62] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 176. | Stepchenkova EI, Tarakhovskaya ER, Spitler K, Frahm C, Menezes MR, Simone PD, Kolar C, Marky LA, Borgstahl GE, Pavlov YI. Functional study of the P32T ITPA variant associated with drug sensitivity in humans. J Mol Biol. 2009;392:602-613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 177. | Hitomi Y, Cirulli ET, Fellay J, McHutchison JG, Thompson AJ, Gumbs CE, Shianna KV, Urban TJ, Goldstein DB. Inosine triphosphate protects against ribavirin-induced adenosine triphosphate loss by adenylosuccinate synthase function. Gastroenterology. 2011;140:1314-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 101] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 178. | Thompson AJ, Fellay J, Patel K, Tillmann HL, Naggie S, Ge D, Urban TJ, Shianna KV, Muir AJ, Fried MW. Variants in the ITPA gene protect against ribavirin-induced hemolytic anemia and decrease the need for ribavirin dose reduction. Gastroenterology. 2010;139:1181-1189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 161] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 179. | Sakamoto N, Tanaka Y, Nakagawa M, Yatsuhashi H, Nishiguchi S, Enomoto N, Azuma S, Nishimura-Sakurai Y, Kakinuma S, Nishida N. ITPA gene variant protects against anemia induced by pegylated interferon-α and ribavirin therapy for Japanese patients with chronic hepatitis C. Hepatol Res. 2010;40:1063-1071. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 180. | Thompson AJ, Santoro R, Piazzolla V, Clark PJ, Naggie S, Tillmann HL, Patel K, Muir AJ, Shianna KV, Mottola L. Inosine triphosphatase genetic variants are protective against anemia during antiviral therapy for HCV2/3 but do not decrease dose reductions of RBV or increase SVR. Hepatology. 2011;53:389-395. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 181. | Azakami T, Hayes CN, Sezaki H, Kobayashi M, Akuta N, Suzuki F, Kumada H, Abe H, Miki D, Tsuge M. Common genetic polymorphism of ITPA gene affects ribavirin-induced anemia and effect of peg-interferon plus ribavirin therapy. J Med Virol. 2011;83:1048-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 182. | Kurosaki M, Tanaka Y, Tanaka K, Suzuki Y, Hoshioka Y, Tamaki N, Kato T, Yasui Y, Hosokawa T, Ueda K. Relationship between polymorphisms of the inosine triphosphatase gene and anaemia or outcome after treatment with pegylated interferon and ribavirin. Antivir Ther. 2011;16:685-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 183. | Domingo P, Guardiola JM, Salazar J, Torres F, Mateo MG, Pacho C, Del Mar Gutierrez M, Lamarca K, Fontanet A, Martin J. Association of ITPA gene polymorphisms and the risk of ribavirin-induced anemia in HIV/hepatitis C virus (HCV)-coinfected patients receiving HCV combination therapy. Antimicrob Agents Chemother. 2012;56:2987-2993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 184. | Nishimura T, Osaki R, Shioya M, Imaeda H, Aomatsu T, Takeuchi T, Okumura Y, Fujiyama Y, Andoh A. Polymorphism of the inosine triphosphate pyrophosphatase gene predicts ribavirin-induced anemia in chronic hepatitis C patients. Mol Med Rep. 2012;5:517-520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 185. | Miyamura T, Kanda T, Nakamoto S, Wu S, Jiang X, Arai M, Fujiwara K, Imazeki F, Yokosuka O. Roles of ITPA and IL28B genotypes in chronic hepatitis C patients treated with peginterferon plus ribavirin. Viruses. 2012;4:1264-1278. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 186. | Eskesen AN, Melum E, Moghaddam A, Bjøro K, Verbaan H, Ring-Larsen H, Dalgard O. Genetic variants at the ITPA locus protect against ribavirin-induced hemolytic anemia and dose reduction in an HCV G2/G3 cohort. Eur J Gastroenterol Hepatol. 2012;24:890-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 187. | Ahmed WH, Furusyo N, Zaky S, Eldin AS, Aboalam H, Ogawa E, Murata M, Hayashi J. Pre-treatment role of inosine triphosphate pyrophosphatase polymorphism for predicting anemia in Egyptian hepatitis C virus patients. World J Gastroenterol. 2013;19:1387-1395. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 18] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 188. | Scherzer TM, Stättermayer AF, Stauber R, Maieron A, Strasser M, Laferl H, Schwarzer R, Datz C, Rutter K, Beinhardt S. Effect of gender and ITPA polymorphisms on ribavirin-induced anemia in chronic hepatitis C patients. J Hepatol. 2013;59:964-971. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 189. | Rau M, Stickel F, Russmann S, Manser CN, Becker PP, Weisskopf M, Schmitt J, Dill MT, Dufour JF, Moradpour D. Impact of genetic SLC28 transporter and ITPA variants on ribavirin serum level, hemoglobin drop and therapeutic response in patients with HCV infection. J Hepatol. 2013;58:669-675. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 190. | Fujino T, Aoyagi Y, Takahashi M, Yada R, Yamamoto N, Ohishi Y, Nishiura A, Kohjima M, Yoshimoto T, Fukuizumi K. Association of ITPA polymorphism with outcomes of peginterferon-α plus ribavirin combination therapy. World J Gastrointest Pharmacol Ther. 2013;4:54-60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 3] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 191. | Di Marco V, Calvaruso V, Grimaudo S, Ferraro D, Pipitone RM, Di Stefano R, Craxì A. Role of IL-28B and inosine triphosphatase polymorphisms in efficacy and safety of Peg-Interferon and ribavirin in chronic hepatitis C compensated cirrhosis with and without oesophageal varices. J Viral Hepat. 2013;20:113-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 192. | Kurosaki M, Tanaka Y, Nishida N, Sakamoto N, Enomoto N, Matsuura K, Asahina Y, Nakagawa M, Watanabe M, Sakamoto M. Model incorporating the ITPA genotype identifies patients at high risk of anemia and treatment failure with pegylated-interferon plus ribavirin therapy for chronic hepatitis C. J Med Virol. 2013;85:449-458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 193. | Tsubota A, Shimada N, Abe H, Yoshizawa K, Agata R, Yumoto Y, Ika M, Namiki Y, Nagatsuma K, Matsudaira H. Several factors including ITPA polymorphism influence ribavirin-induced anemia in chronic hepatitis C. World J Gastroenterol. 2012;18:5879-5888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 13] [Cited by in RCA: 13] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 194. | Hézode C, Forestier N, Dusheiko G, Ferenci P, Pol S, Goeser T, Bronowicki JP, Bourlière M, Gharakhanian S, Bengtsson L, McNair L, George S, Kieffer T, Kwong A, Kauffman RS, Alam J, Pawlotsky JM, Zeuzem S, PROVE2 Study Team. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N Engl J Med. 2009;360:1839-1850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 849] [Cited by in RCA: 793] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 195. | McHutchison JG, Everson GT, Gordon SC, Jacobson IM, Sulkowski M, Kauffman R, McNair L, Alam J, Muir AJ, PROVE1 Study Team. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827-1838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 851] [Cited by in RCA: 809] [Article Influence: 50.6] [Reference Citation Analysis (0)] |
| 196. | Kwo PY, Lawitz EJ, McCone J, Schiff ER, Vierling JM, Pound D, Davis MN, Galati JS, Gordon SC, Ravendhran N. Efficacy of boceprevir, an NS3 protease inhibitor, in combination with peginterferon alfa-2b and ribavirin in treatment-naive patients with genotype 1 hepatitis C infection (SPRINT-1): an open-label, randomised, multicentre phase 2 trial. Lancet. 2010;376:705-716. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 519] [Cited by in RCA: 517] [Article Influence: 34.5] [Reference Citation Analysis (0)] |
| 197. | McHutchison JG, Manns MP, Muir AJ, Terrault NA, Jacobson IM, Afdhal NH, Heathcote EJ, Zeuzem S, Reesink HW, Garg J, Bsharat M, George S, Kauffman RS, Adda N, Di Bisceglie AM, PROVE3 Study Team. Telaprevir for previously treated chronic HCV infection. N Engl J Med. 2010;362:1292-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 570] [Cited by in RCA: 535] [Article Influence: 35.7] [Reference Citation Analysis (0)] |
| 198. | Suzuki F, Suzuki Y, Akuta N, Sezaki H, Hirakawa M, Kawamura Y, Hosaka T, Kobayashi M, Saito S, Arase Y. Influence of ITPA polymorphisms on decreases of hemoglobin during treatment with pegylated interferon, ribavirin, and telaprevir. Hepatology. 2011;53:415-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 87] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 199. | Ogawa E, Furusyo N, Nakamuta M, Kajiwara E, Nomura H, Dohmen K, Takahashi K, Satoh T, Azuma K, Kawano A, Tanabe Y, Kotoh K, Shimoda S, Hayashi J, Kyushu University Liver Disease Study (KULDS) Group. Clinical milestones for the prediction of severe anemia by chronic hepatitis C patients receiving telaprevir-based triple therapy. J Hepatol. 2013;59:667-674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 46] [Article Influence: 3.8] [Reference Citation Analysis (0)] |