Published online Feb 21, 2013. doi: 10.3748/wjg.v19.i7.1056
Revised: September 13, 2012
Accepted: September 22, 2012
Published online: February 21, 2013
AIM: To characterize oxidase- and urease-producing bacterial isolates, grown aerobically, that originated from antral biopsies of patients suffering from acid peptic diseases.
METHODS: A total of 258 antral biopsy specimens were subjected to isolation of bacteria followed by tests for oxidase and urease production, acid tolerance and aerobic growth. The selected isolates were further characterized by molecular techniques viz. amplifications for 16S rRNA using universal eubacterial and HSP60 gene specific primers. The amplicons were subjected to restriction analysis and partial sequencing. A phylogenetic tree was generated using unweighted pair group method with arithmetic mean (UPGMA) from evolutionary distance computed with bootstrap test of phylogeny. Assessment of acidity tolerance of bacteria isolated from antrum was performed using hydrochloric acid from 10-7 mol/L to 10-1 mol/L.
RESULTS: Of the 258 antral biopsy specimens collected from patients, 179 (69.4%) were positive for urease production by rapid urease test and 31% (80/258) yielded typical Helicobacter pylori (H. pylori) after 5-7 d of incubation under a microaerophilic environment. A total of 240 (93%) antral biopsies yielded homogeneous semi-translucent and small colonies after overnight incubation. The partial 16S rRNA sequences revealed that the isolates had 99% similarity with Pseudomonas species. A phylogenetic tree on the basis of 16S rRNA sequences denoted that JQ927226 and JQ927227 were likely to be related to Pseudomonas fluorescens (P. fluorescens). On the basis of HSP60 sequences applied to the UPGMA phylogenetic tree, it was observed that isolated strains in an aerobic environment were likely to be P. fluorescens, and HSP60 sequences had more discriminatory potential rather than 16S rRNA sequences. Interestingly, this bacterium was acid tolerant for hours at low pH. Further, a total of 250 (96.9%) genomic DNA samples of 258 biopsy specimens and DNA from 240 bacterial isolates were positive for the 613 bp amplicons by targeting P. fluorescens-specific conserved putative outer membrane protein gene sequences.
CONCLUSION: This study indicates that bacterial isolates from antral biopsies grown aerobically were P. fluorescens, and thus acid-tolerant bacteria other than H. pylori can also colonize the stomach and may be implicated in pathogenesis/protection.
-
Citation: Patel SK, Pratap CB, Verma AK, Jain AK, Dixit VK, Nath G.
Pseudomonas fluorescens -like bacteria from the stomach: A microbiological and molecular study. World J Gastroenterol 2013; 19(7): 1056-1067 - URL: https://www.wjgnet.com/1007-9327/full/v19/i7/1056.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i7.1056
Helicobacter pylori (H. pylori) is a gram-negative, microaerophilic bacterium found primarily in the stomach. It has been implicated in chronic gastritis, gastric ulcers, duodenal ulcers and stomach cancers that were previously believed to be of non-microbial origin[1-3]. There was a misconception that no bacteria could live in the stomach because of its highly acidic environment. For the first time, Steer and Colin-Jones[4] published their results regarding the presence of gram-negative, oxidase- and urease-producing bacteria, but they proposed that it was Pseudomonas (P.), a contaminant and not related to peptic ulcer. In an effort to grow H. pylori from an antral biopsy, we could see that a peculiar type of bacterial colony was growing consistently after overnight incubation while H. pylori was taking 3-5 d to grow. These colonies were also oxidase and urease producers, but growing in an aerobic environment. There are reports that show the presence of a variety of bacteria in the stomach by isolation, DNA profiling and polymerase chain reaction (PCR)-based analysis, and some of them are urease producers hindering the specificity of the urea breath test[5-7]. Therefore, we aimed to characterize this type of bacterial isolate and to analyze whether they are colonizers or contaminants.
The study subjects were patients attending inpatient services of the Department of Gastroenterology, University Hospital of Banaras Hindu University, Varanasi, Uttar Pradesh, India. This hospital provides tertiary-level health services for the eastern part of Northern India. The culture isolation, phenotypic and molecular characterizations were carried out in the Department of Microbiology, Institute of Medical Sciences.
A total of 258 patients suffering from upper gastrointestinal (UGI) diseases like non-ulcer dyspepsia (NUD), peptic ulcer diseases (PUD) including gastric ulcer and duodenal ulcer, and gastric carcinoma were enrolled during a period of 3 years (2007-2010) and three antral biopsy pieces from each patient were collected. Before taking a biopsy, the endoscope was rinsed with detergent followed by water, and disinfected with 2% alkaline glutaraldehyde for 30 min then rinsed with sterile water. In a similar way, biopsy forceps were washed and sterilized and one biopsy forceps was used for one patient exclusively. The biopsy specimens were collected by endoscopic forceps from each individual with full aseptic precautions after taking well-informed consent. The work was approved by the Ethics Committee of the Institute of Medical Sciences, Banaras Hindu University. Patients with mucosal breaks greater than 5 mm in size with apparent depth were diagnosed as having ulcer and those with ulcerato-infiltrative lesions with positive histology/brush cytology were considered as having stomach carcinoma. The patients not having ulcerative lesions but suffering from dyspeptic symptoms were diagnosed as NUD. Individuals with normal endoscopic findings without gastroduodenal symptoms but having other gut problems were treated as healthy controls. In the present study, those patients were excluded who had a history of previous gastric surgery, active UGI bleeding, chronic alcoholism, intake of antibiotics and proton pump inhibitors during the last 4 wk or those taking non-steroidal anti-inflammatory drugs. Further individuals less than 18 years of age, pregnant or lactating mothers or those having illnesses like cirrhosis, chronic renal failure or ischemic heart disease were also excluded.
The three biopsy pieces were pooled and homogenized into phosphate saline buffer together in an all glass disposable homogenizer and were divided into three aliquots. The first aliquot of the tissue homogenate was transferred immediately into a rapid urease test (RUT) medium and the second was plated within 30 min of collection onto the media used for bacterial culture [Mueller Hinton agar without supplement and media containing brain heart infusion agar (Difco, Becton Dickinson, Sparks, MD, United States), supplemented with 7% sheep blood, 0.4% IsoVitaleX, and Skirrow selective supplement (vancomycin 10 μg/mL; polymixin B sulfate 2.5 IU/mL; trimethoprim lactate 5 μg/mL) (Difco, Becton Dickinson, Sparks, MD, United States)]. The non-enriched plate was incubated at 37 °C in an aerobic atmosphere while the other was incubated in the presence of 5% O2, 10% CO2, and 85% N2 for 3-7 d. Several small colonies could be seen after overnight incubation in the first plate. The other plate was examined every alternate day after 3-7 d to see if colonies other than those observed after overnight incubation developed. Small translucent colonies developed after 5-7 d of incubation other than those originally after observed overnight incubation on non-enriched nonselective medium. These colonies were further subjected to morphological and biochemical tests viz. motility, oxidase, catalase, and urease. All the isolates were divided into two groups on the basis of the incubation period, i.e., those isolates obtained after overnight incubation designated as group A and the other group B possessing those strains isolated after 5-7 d of incubation. The third aliquot was subjected to genomic DNA extraction.
For the RUT, biopsy specimens were inoculated into 1 mL of 10% urea in deionized water (pH 6.8), to which two drops of 1% phenol red solution was added, and incubated at 37 °C for 24 h. A positive result was recorded when the color changed from yellow to pink within 30 min. If there was mild color change within 30 min, the RUT tubes were incubated for a further 24 h.
The organisms which appeared after overnight incubation were tested for growth at 4 °C, 35 °C and 42 °C onto Muller Hinton agar; incubation was maintained for 14 d to distinguish Pseudomonas spp.[8,9]. Those organisms showing growth at 4 °C were sub cultured twice and incubated at the appropriate temperature for more than 10 d each time.
Five isolates were suspended into different molar concentrations of hydrochloric acid (10-1 to 10-7 mol/L corresponding to pH 1.0 to 7.0) and a CFU was maintained as 106 CFU/mL. After the intervals of 0 min, 5 min, 10 min, 20 min, 30 min and 60 min, 100 μL acidic suspension (105 CFU) were transferred into 3 mL BHI broth. After overnight incubation optical density was recorded with the help of a spectrophotometer at 600 nm wavelength and bacterial count was expressed in CFU/mL. This experiment was repeated twice.
Extraction of genomic DNA from both types of bacterial isolates (A total of 320 strains including groups A + B) as well as from tissue homogenate was performed using a standard proteinase K and phenol-chloroform method[10]. To exclude the possibility of cross contamination of DNA during DNA extraction, one set of double distilled was included in each batch of DNA extraction.
Confirmation of H. pylori was done at a molecular level by nested PCR targeting the conserved HSP60 gene and its restriction fragment length polymorphism. The reaction was performed in 25 μL final volume containing 10 ng of DNA, 1 U of Taq polymerase (Bangalore Genie, India), 200 mmol/L (each) deoxynucleotide triphosphate (MBI, Fermentas) and 1.5 mmol/L MgCl2 in standard PCR buffer and 10 pmol of each primer as described by Singh et al[11]. Primer sequences and PCR conditions are displayed in Table 1. For the internal amplification, the PCR product from the primary cycle was diluted 1/50 and 1 μL was used as the template in the nested PCR. The conditions for the PCR amplification, first and nested reactions were the same. DNA from H. pylori reference J99 and a tube containing water in place of DNA were assayed in each PCR run as positive and negative controls, respectively.
| Target gene | Primer name | Primer sequence (5’-3’) | PCR condition (number of cycles, size of product) |
| 16S rRNA | 16S F | TTGGAGAGTTTGATCCTGGCC | 94 °C, 30 s; 59 °C, 30 s; 72 °C, 30 s (30, 1155 bp) |
| 16S R | ACGTCATCCCACCTTCTC | ||
| HSP60 | |||
| Primary | HSP1 | AAGGCATGCAATTTGATAGAGGCT | 94 °C, 30 s; 56 °C, 30 s; 72 °C, 30 s (35, 594 bp) |
| HSP2 | CTTTTTTCTCTTTCATTTCCACTT | ||
| Nested | HSPN1 | TTGATAGAGGCTACCTCTCC | 94 °C, 30 s; 56 °C, 30 s; 72 °C, 30 s (35, 501 bp) |
| HSPN2 | TGTCATAATCGCTTGTCGTGC | ||
| Putative membrane-bound protein (PFMP) | |||
| Primary | PFMPF | TCTKRYCRMGAATCRARACWRYC | 94 °C, 1 min; 50 °C, 1 min; 72 °C, 1 min (35, 704 bp) |
| PFMPR | GKTWYTGCKCRWWKCSYTSMMC | ||
| Nested | PFMPNF | TGCGYWMMWCCYWRWCCWTGA | 94 °C, 1 min; 50 °C, 1 min; 72 °C, 1 min (35, 613 bp) |
| PFMPNR | AKCABGGTSCWGCMVRCCRBGC | ||
| mupV | |||
| Primary | mupVF | TGAGTTCGATGTGACCTGCCTG | 94 °C, 1 min; 55 °C, 1 min; 72 °C, 1 min (35 cycles, 722 bp) |
| mupVR | AACTCGCCAGATTGTCGTACAC | ||
| Nested | mupVnF | CAGCATTATCCTGCCACTGAC | 94 °C, 1 min; 55 °C, 1 min; 72 °C, 1 min (35, 611 bp) |
| mupVnR | ATGATGTCCTGGCACACCTGATC |
After amplification, the PCR products (501 bp) were precipitated with 2.5 volumes of ethanol. The pellets were washed twice with 70% ethanol and dissolved in Tris-EDTA buffer (pH 8.0). A 10 μL precipitated amplified DNA sample was then digested with 10 U of restriction enzyme HindIII in appropriate buffered solution recommended by the manufacturer (Bangalore Genie, India) and incubated for 3 h at 37 °C. The digested DNA fragments were analyzed by electrophoresis on 2% agarose gels (Bangalore Genie, India) containing 0.5 μg of ethidium bromide per mL. The gel was run at 70 V with TBE (Tris Boric acid EDTA) buffer for 3 h and was examined by a transilluminator and photographed. The sizes of digested DNA fragments were estimated from distances of molecular weight standards and compared with in silico restriction digestion.
For each group of isolates, the 16S rRNA from 8 randomly selected isolates was amplified using forward primer-16SF and reverse primer-16SR[12]. PCR amplification was performed in a thermocycler (Biometra, Germany) according to standard procedures (Table 1). After amplification by universal eubacterial primers, the PCR products (1155 bp) were precipitated with 2.5 volumes of ethanol. The pellets were washed twice with 70% ethanol and dissolved in Tris-EDTA buffer (pH 8.0). A 10 μL precipitated amplified DNA sample was then digested with 10 U of restriction enzyme EcoR1, in appropriate buffered solution recommended by the manufacturer (Bangalore Genie) and incubated for 3 h at 37 °C. The visualization of the restriction fragment was done by the same method as described in the previous paragraph.
The amplified PCR products were purified from salts and primers using QIA quick PCR purification kit (Qiagen, United States). A total of 8 purified amplicons generated targeting HSP60 and 2 amplicon 16S rRNA genes were outsourced for partial sequencing to Bangalore Genei, India. Sequences were analyzed using BLASTN (http://www.ncbi.nlm.nih.gov/BLAST) to verify the identity of the sequences: whether H. pylori or some other microorganism.
Reference sequences of Pseudomonas group and other enteric pathogens used for phylogenetic analyses were retrieved from Genbank. The partial 16S rRNA sequences and HSP60 sequence for the strains were aligned with reference sequences using Clustal X version 1.81, with default parameters[13]. Phylogenetic and molecular evolutionary analyses were performed using MEGA version 4[14]. The phylogenetic tree was generated using the unweighted pair group method with arithmetic mean (UPGMA) from evolutionary distance computed with bootstrap test of phylogeny. The degree of statistical support for branches was determined with 500 bootstrap replicates.
The putative membrane-bound protein coding gene present in all the strains of P. fluorescens is available in NCBI Genbank. Due to a similarity of sequences of about 55% in P. fluorescens, we therefore planned to design nested degenerate primers to amplify the partial sequence of putative membrane bound protein so that it was able to amplify all strains of P. fluorescens. Forward and reverse oligo-nucleotide degenerate primers derived from the region located between bases 92072 and 92775 of the Pseudomonas fluorescens (Pseudomonas fluorescens Pf0-1; GenBank Accession number CP000094.2 and GI: 253992019) were synthesized. Internal primers were derived from the region between bases 92114-92726 for nested PCR. DNA extracted from P. putida and P. aeruginosa was used to monitor sensitivity against P. putida and P. aeruginosa. During the PCR assay in each batch, Mili Q water was used as a template to ensure that there was no contamination by water and PCR reagent[15].
Forward and reverse oligonucleotides were derived from the conserved region located between bases 68101 and 68822 of the mupirocin biosynthetic gene cluster of Pseudomonas fluorescens (NCIMB 10586). An internal primer was derived from the region between bases 68796 and 68796 (GenBank Accession number AF318063.2; gene GI: 20150006). PCR amplification was similar to amplification of HSP60 gene and conditions are described in Table 1.
Fingerprinting of 71 randomly selected strains from group A was performed based on randomly amplified polymorphic DNA (RAPD) PCR methods by using primers RAPD3 5’-TACAGCTCG-3’and RAPD5 5’AGCACTGCCT-3’ (this study). PCR was carried out in 25 μL volume using 10 ng of genomic DNA, 1 U of Taq polymerase (Bangalore Genie, India), and 15 pmol of each primer (Bangalore Genie), 200 mmol/L (each) deoxynucleotide triphosphate (Bangalore Genie, India) and 2 mmol/L MgCl2 in standard PCR buffer. Amplification reactions were carried out in a thermal cycler (Biometra, Goettingen, Germany).
The gel images were analyzed under ultraviolet light in a gel documentation system (Alpha Innotech, United States). Cluster analysis of all the isolates was done on the basis of the fingerprint generated. Based on the presence or absence of different DNA fragments in the fingerprints of the P. fluorescens strains, a binary data matrix was created. Overall similarity between the pair of strains was calculated from the binary data matrix using the simple matching-dice coefficient. The resulting similarity matrix was used as the input data for cluster analysis by NTSYS pc2.0 programme of UPGMA[16].
The level of significance between the two proportions, i.e., culture rates and molecular detection rates, was calculated by Fischer’s Exact Probability test.
Of the 258 antral biopsy specimens collected from patients, 69.4% (179/258) were found to be positive by RUT and 31% (80/258) by culture for typical H. pylori, after 5-7 d of incubation under a microaerophilic environment; these were gram-negative curved rods and were positive for oxidase, catalase and urease. However, 258 antral biopsies yielded 240 (93%) homogeneous semi-translucent and small colonies after overnight incubation. These isolates also grew aerobically but the colonies had an opaque, small character contrary to the translucent one which is typical for H. pylori (Table 2).
| Diseases | Antral biopsies | RUT, positivity | H. pylori HSP60, positivity | P value1 | P. fluorescens putative outer membrane protein, positivity | P value2 | Isolation of different types of bacteria, positivity | P value3 | |
| Group A | Group B | ||||||||
| PUD | 65 | 51 (78.5) | 59 (90.8) | < 0.001 | 63 (96.9) | < 0.010 | 61 (93.8) | 23 (35.4) | < 0.001 |
| NUD | 123 | 92 (74.8) | 109 (88.6) | < 0.001 | 121 (98.4) | < 0.001 | 119 (96.7) | 39 (31.7) | < 0.001 |
| CA | 49 | 23 (46.9) | 24 (48.9) | < 0.050 | 46 (93.9) | < 0.001 | 43 (87.5) | 12 (24.5) | < 0.001 |
| Normal | 21 | 13 (61.9) | 19 (90.4) | < 0.001 | 20 (95.2) | 0.001 | 17 (80.9) | 6 (28.6) | < 0.001 |
| Total | 258 | 179 (69.4) | 211 (81.8) | < 0.001 | 250 (96.9) | < 0.001 | 240 (93.0) | 80 (31.0) | < 0.001 |
A total of 100 isolates randomly selected from group A were subjected to extensive phenotypic characterization. All of them were gram-negative, oxidase-, catalase- and urease-positive. All were non-fermenters, showed variable nitrate reduction and failed to utilize simple sugars (glucose, lactose, sucrose, mannitol and maltose). Citrate was utilized by all of them. All the tested strains were Methyl Red negative and Voges-Praskauer negative or equivocal. All the strains were oxidase- and catalase-positive. Indole test was negative but on mixing with the Kovac’s reagent a typical greenish color developed. All these isolates were able to multiply at 4 °C. These findings intimated that those isolates which appeared after overnight incubation were P. fluorescens.
The bacterial count of P. fluorescens was 7.3 × 108, 6.9 × 108, 7 × 108, 6.1 × 108, 7.6 × 108, 6.5 × 108 and 9.8 × 108 CFU/mL for 0 min, 5 min, 10 min, 15 min, 20 min, 30 min and 60 min after acid exposure (pH 1.0). Average bacterial counts were 9.3 × 108, 9.8 × 108, 1.0 × 109, 9.5 × 108 and 9.8 × 108 CFU/mL for acid tolerance of low pH 2, pH 3, pH 4, pH 5, pH 6 and pH 7, respectively, for different time intervals (0 min, 5 min, 10 min, 15 min, 20 min, 30 min and 60 min). Similarly, bacterial growth was approximately the same in control experiments where acidic solution was replaced by LB Broth. The exposure to acidic pH showed that P. fluorescens growth was not killed by exposure to lower pH 1.0 for an hour.
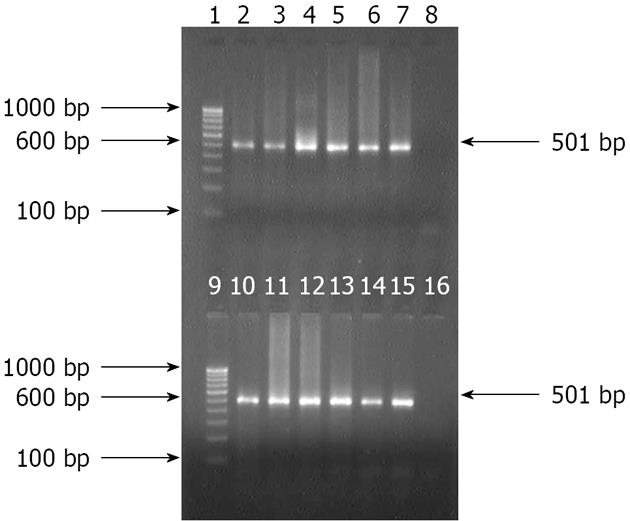
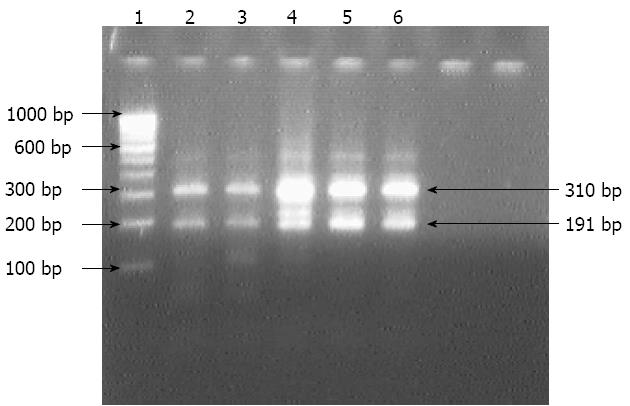
Isolates from group B, which grew under microaerophilic environment, were subjected to amplification by primers specific for HSP60 gene of H. pylori. All the 80 isolates from 258 patients were positive for the 501 bp of amplicon for the corresponding gene. However, 211 (81.8%) of 258 antral biopsies were positive for H. pylori DNA as the 501 bp amplicon was produced by nested PCR. All nested amplicons of 501 bp were restricted into two fragments of 310 bp and 191 bp by the HindIII restriction enzyme (Figures 1 and 2). Although colonies grown after overnight incubation could give amplification with first round of PCR primers, nested PCR did not generate 501 bp amplicon specific to H. pylori. Similarly, 240 aerobic isolates could not amplify in second round PCR.
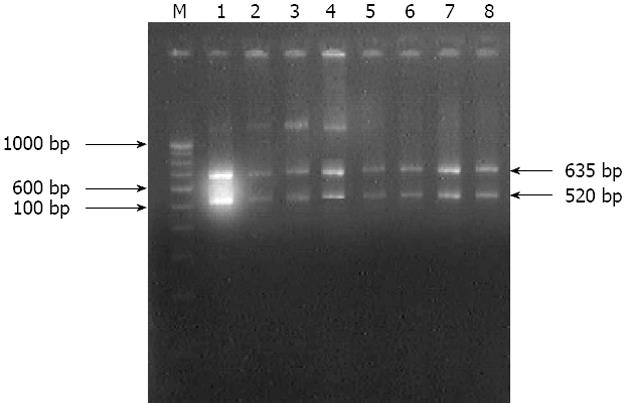
The isolates analyzed by using EcoR1 restriction endonuclease enzyme on amplicons generated by 16S rRNA specific primers fell into two groups: group A isolates could not be restricted, while group B isolates which were restricted into two fragments of 635 bp and 520 bp which were similar to in silico restriction of H. pylori J99 (Figure 3).
The partial nucleotide sequence of 16S rRNA of 2 isolates (GenBank accession number JQ927226 and JQ927227) from group A represented no restriction site for EcoR1 restriction enzyme, and comparison of the nucleotide sequences with the NCBI database showed similarity of 99% with P. fluorescens. Similarly, partial nucleotide sequence of unrestricted amplified HSP60 gene (590 bp) of the isolates that grew after overnight incubation aerobically also showed 96% similarity with P. fluorescens.
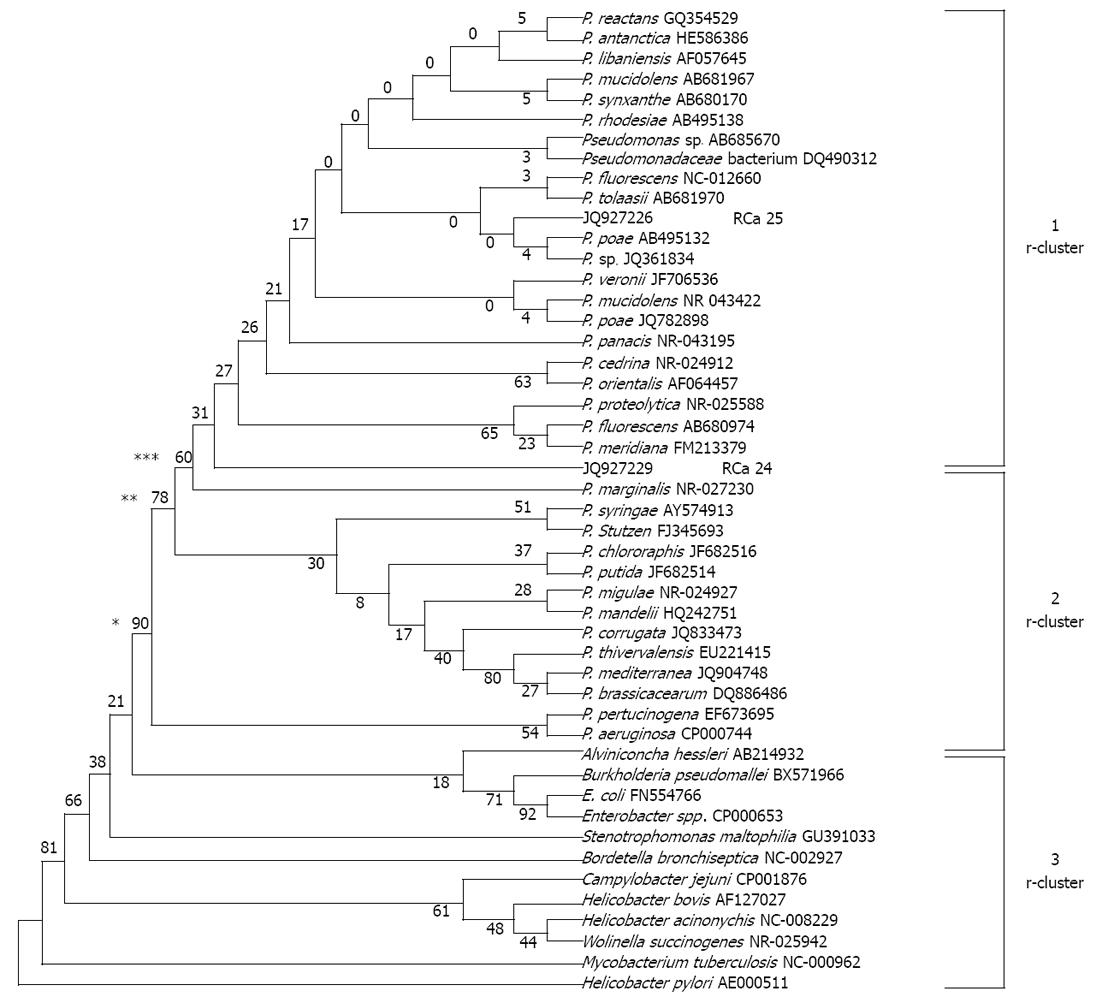
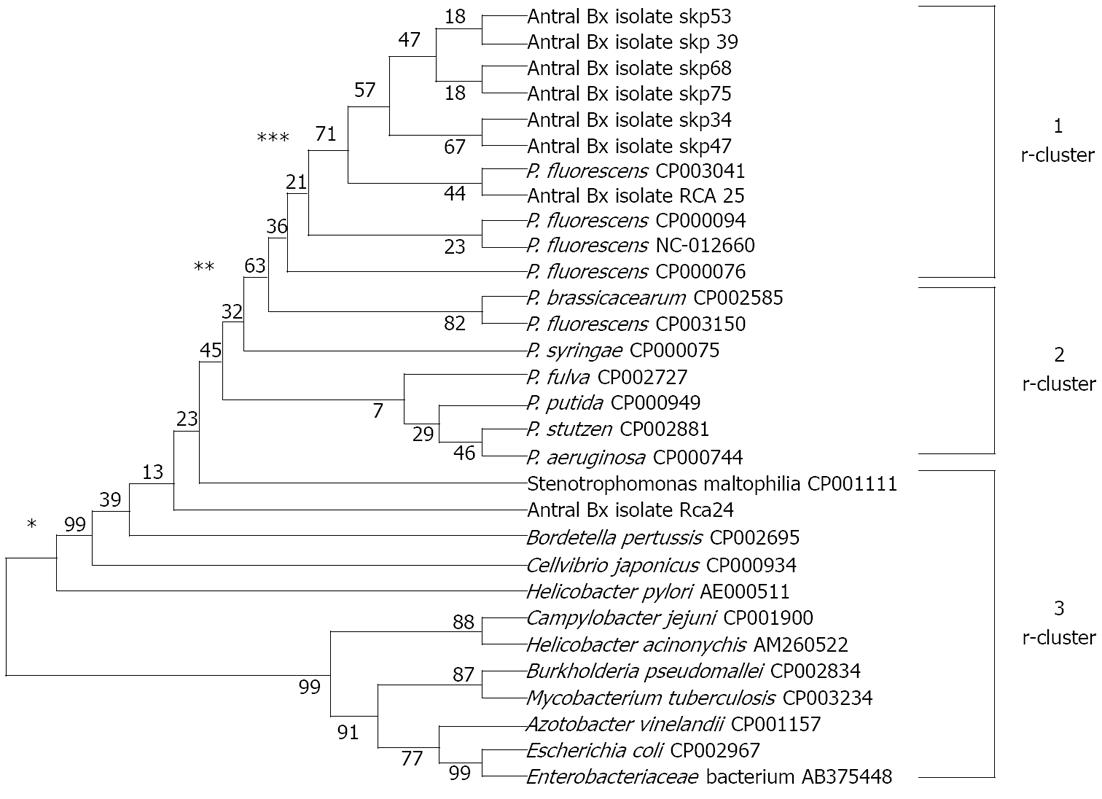
The partial sequence of the 16S rRNA gene of two P. fluorescens-like isolated strains and sequences of 16S rRNA gene from 34 reference strains representative of the principal Pseudomonas phyla and 12 other bacterial pathogens were used for comparison of a phylogenetic tree. Similarly, a partial sequence of HSP60 gene of 8 P. fluorescens-like isolates and sequence of HSP60 gene of 5 P. fluorescens reference strains and 6 other reference strains representative of the principal Pseudomonas phyla and 11 other clinical pathogens were used to prepare a phylogenetic tree. To simplify the comparisons for the resulting phylogenetic tree by the UPGMA method, we named clusters based on 16S rRNA gene data, r-clusters, and those based on HSP60 data, h-clusters. The tree showed, on the basis of 16S rRNA sequences, it can be grouped clearly into three r-clusters on the basis of number of bootstraps i.e., 90, 78 and 60. Cluster r-1 represents a fluorescens group along with two sequences submitted to gene bank (accession number as JQ927226 and JQ927227 for RCa25 and RCa24 respectively). Cluster r-2 represents Pseudomonas aeruginosa, Pseudomonas syringae, Pseudomonas stuzeri, Pseudomonas chlororaphis, Pseudomonas putida, Pseudomonas pertucinogena group and 4 P. fluorescens reference strains, and r-3 cluster represents other microbes and enteric pathogens rather than non-Pseudomonas spp. From cluster analysis it is clear that JQ927226 and JQ927227 are related to P. fluorescens (Figure 4).
The tree shows that, on the basis of HSP60 sequences, three h-clusters could be observed (bootstraps number, 99, 63 and 71): the sequences of seven isolated strains from the gastric niche could be grouped in h-cluster I along with P. fluorescens strains while cluster II represents non-fluorescens pseudomonas species with the exception of P. fluorescens F113 (CP003150). The h-cluster III grouped non-Pseudomonas sp. including enteric pathogens along with H. pylori. Interestingly, one strain of P. fluorescens RCa 24 fell into this III cluster with closeness to Stenotrophomonas maltophilia and Bordetella pertusis (Figure 5).
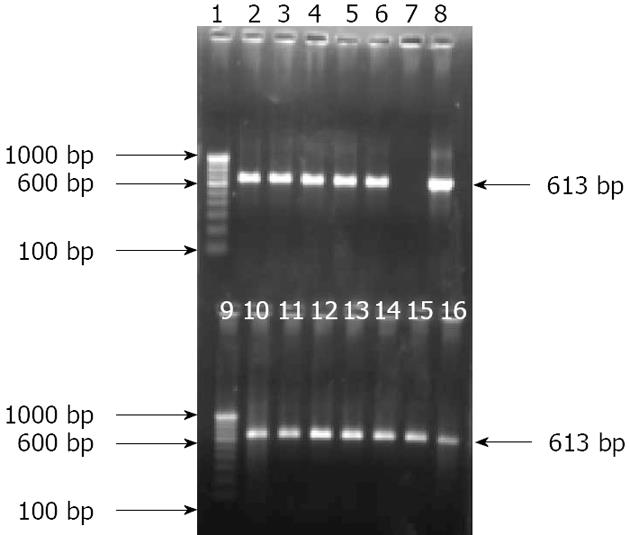
One specific pair of primers targeting putative outer membrane protein was used to identify P. fluorescens. The other specific primers targeting mupirocin biosynthetic gene were used to screened out whether any P. fluorescens-like isolate was producing mupirocin. Genomic DNA extracted from group-A isolates and all biopsies was used as template for PCR amplification. A total of 250 (96.9%) out of 258 biopsy specimen genomic DNA samples and 240 bacterial isolates were positive for the amplification of the corresponding gene, i.e., the putative outer membrane protein gene sequences (Table 2 and Figure 6). Although putative outer membrane protein gene targeting primers were degenerate, they were unable to produce the 613 bp amplicon from Pseudomonas aeruginosa and Pseudomonas putida at similar PCR conditions. The mupirocin biosynthetic gene targeting primers were unable to produce amplification of the 722 bp or 611 bp amplicon either in primary or secondary round PCR, respectively, from any of the isolates (data not shown).
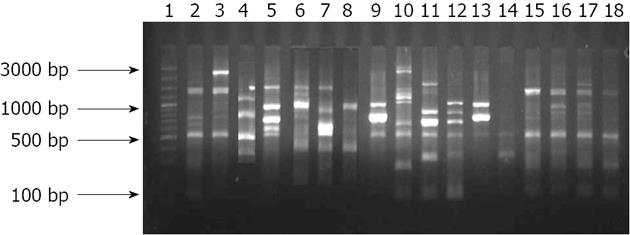
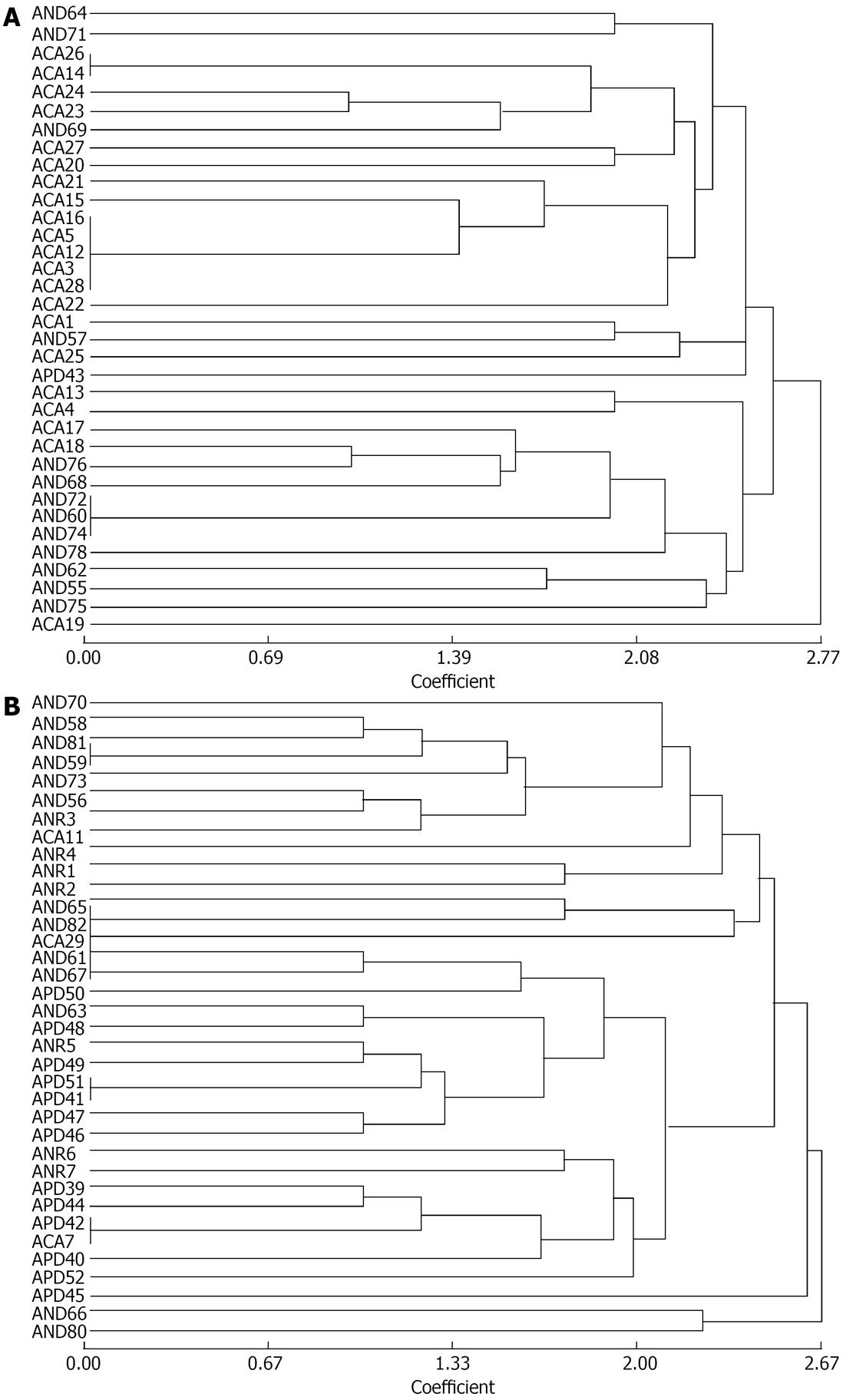
All the 71 strains tested from group-A yielded significant PCR products with RAPD primers. The strains generated approximately 3-13 well-defined bands between 150 bp to 2.5 kb sizes (9 bands on average) with each isolate yielding a unique profile of products. Cluster analysis with a RAPD-PCR based method showed that only a few isolates exhibited an identical profile. Nearly all the isolates appeared as dissimilar from each strain, but 5 strains isolated from cancer patients showed similar banding pattern at 0.0 coefficient (Figures 7 and 8).
Traditionally, the human stomach has been viewed as an inhospitable environment for microorganisms because of its acidic environment along with several other antimicrobial factors. With the discovery of H. pylori and other gastric helicobacters, and subsequent insight into the mechanisms by which these organisms adapt to the gastric environment[17], the existence of a bacterial community adapted to this human niche seems quite plausible. This is the first report of its kind showing the presence of a P. fluorescens-like bacterium in the human stomach. We isolated the bacteria from NUD, gastric ulcer, duodenal ulcer and gastric carcinoma patients with the belief that only H. pylori is associated with acid peptic diseases. However, P. fluorescens grew on a simple medium like Mueller-Hinton agar without an antibiotic supplement. The small white colonies had Gram-negative slightly curved rods and produced oxidase, urease and catalase enzymes. These colonies appeared after overnight incubation in an aerobic environment. These isolates exhibited growth at 4 °C, which is one of the key characteristics of P. fluorescens[9]. When these isolates were exposed to acidic pH as low as pH 1.0 for 1 h, the subsequent growth of the bacteria was not affected, indicating that they were acid tolerant. However, due to deviation from the classical H. pylori growth characteristics, we were prompted to consider them as non-H. pylori and submit them for further characterization. For the purpose, H. pylori specific primers for amplifying HSP60 gene-specific sequences were applied[11]. However, an approximate 600 bp sized amplicon was produced by the first round of PCR, but a restriction site specific for H. pylori for HindIII enzyme was absent in these amplicons. Further, none of these isolates could yield a 501 bp amplicon for H. pylori by nested PCR primers. Thus, the possibility of these isolates being H. pylori was excluded. On the other hand, the typical H. pylori isolates which grew after 5-7 d of incubation yielded 501 bp amplicon by nested PCR protocol targeting the HSP60 gene.
Identification based on partial nucleotide sequencing of 16S rRNA of the representative isolates showed 99% sequence similarity with γ-protobacteria. On the basis of enzyme restriction analysis as well as partial nucleotide sequencing of 16S rRNA, the strains isolated from the acidic environment of the stomach either could be grouped in the genus Pseudomonas or with a closely related new genus. Further, on the basis of blasting of partial nucleotide sequences of HSP60 gene sequences on the NCBI gene data bank, isolated strains could be grouped with P. fluorescens. On the basis of h cluster based on the UPGMA phylogenetic tree, it may be concluded that the HSP60 gene sequence has better discriminatory power than 16S rRNA. Further, as one of the isolates could be grouped with Stenotrophomonas maltophilia and Bordetella pertusis, the possibility of the presence of bacteria other than P. fluorescens may not be denied.
The relatively conserved 16S rRNA gene of P. fluorescens has been targeted for PCR-based amplification in culture isolates, but with variable specificity[18]. Therefore, we decided to target a conserved putative membrane-bound protein gene of P. fluorescens. The putative membrane-bound protein specific primer screened in the present study was found to be specific for P. fluorescens and it yielded an amplicon of 613 bp, confirming the isolates as being P. fluorescens. Mupirocin is known for its antibacterial activity and has been reported in only one strain of P. fluorescens (NCIMB 10586). A mupirocin-producing gene cluster could not be traced in any of the P. fluorescens isolates despite confirmation by 3 genes, i.e., 16S rRNA, HSP60 and putative membrane bound protein genes. None of our isolates showed the presence of mupV (mupirocin). It is quite possible that this gene might be either absent in all the strains or have too much polymorphism at the loci of primer annealing[19]. We could isolate the P. fluorescens strain from 93% of antral biopsies of the patients suffering from gastric diseases. These isolates were further confirmed by PCR-based amplification targeting 3 conserved genes. In DNA of antral biopsy tissue also, 98.4% of NUD and 93.9% of gastric cancer patients were found to be positive for the P. fluorescens DNA by nested PCR, while 95.2% of normal stomach had P. fluorescens and 96.9% of patients with PUD were found to have the bacterium. Thus our observation indicates a high prevalence and density of these non-H. pylori bacteria in the gastric mucosa of patients. Further, to ascertain that the isolated P. fluorescens were not contaminants, we carried out whole genome fingerprinting of the 71 randomly selected isolates by using RAPD[20,21] with two RAPD primers. Cluster analysis indicated that almost all the strains had different banding patterns with only a few exceptions, confirming that the isolates were of different clones and thus ruling out the possibility of cross contamination. Moreover, from time to time, we did specific PCR amplification for the P. fluorescens on DNA extracted from samples of tap water and washouts of the endoscope, which never yielded the required amplicon even by nested protocol for putative outer membrane protein.
It is interesting to mention that before the implication of H. pylori in acid peptic diseases and stomach cancer, Steer and Collin-Jones[4] reported that 80% of antral biopsies have Pseudomonas spp. There are reports showing the presence of non-Helicobacter bacteria in gastric biopsies of patients suffering from gastric atrophy[22,23]. In cases of reduced gastric acidity due to antacids, it also has been reported that there is presence of several other bacteria[24-26]. Two more studies based on profiling of bacterial flora by temperature gradient gel electrophoresis, 16S rRNA sequence analysis[7] and molecular analysis of the bacterial microbiota[27] have shown presence of several bacterial species including Pseudomonas in the stomach. This bacterium seems to be able to colonize the stomach due to its ability to produce urease enzyme. Similarly, it was reported that a non-H. pylori bacterium, Ochrobacrum anthropi, could be implicated in causation of gastritis in the Squirrel monkey[6].
Such a high prevalence of P. fluorescens in human stomach raises many questions: Is it prevalent in the stomach of patients of other subcontinents and continents? Does it have any pathogenic role? Does it have some protective role? Is it simply a part of commensal flora of human stomach? There is a study from Venezuela[5] reporting that Pseudomonas strains may interfere with the identification of H. pylori. They suggested that one should not rely on rapid urease, catalase and oxidase tests for identification of H. pylori. However, studies are needed to observe and identify the presence of P. fluorescens in other parts of the world also. With regard to its pathogenic potential, P. fluorescens is known as an unusual pathogen of humans. It has been reported as causing septicemia in humans, especially associated with transfusion and cancer[28-30]. Moreover, there is a report indicating that P. fluorescens encodes the Crohn’s disease-associated I2 sequence and T cell super antigen, thus implicating it in the pathogenesis of Crohn’s disease[31].
However, its commensal state in the stomach may be speculated strongly due to its prevalence in stomach at such a high level and density (it could be isolated in the majority of the antral biopsy). Assumptions may be made that P. fluorescens might be producing some antibacterial substances, such as is produced by P. aeruginosa, P. aeruginosa is known for producing 4-hydroxy-2-alkylquinoline which is inhibitory in vitro to H. pylori[32]. Moreover, one of the P. fluorescens strains is already known for production of mupirocin which is very effective against methicillin-resistant Staphylococcus aureus. This bacterium has been stated to have a probiotic role in the gills of fish[33]. Furthermore, low isolation of H. pylori and fewer incidences of acid peptic diseases including gastric cancer in North Indians may also be speculated on the basis of the probiotic activity of P. fluorescens in the stomach. In addition, there are reports of the unique property of P. fluorescens to inhibit the growth of other bacteria, fungi and nematodes causing plant pathology[34-40].
In view of the suggestions made by previous studies[41-43], there is a strong need to explore the exact role of H. pylori in stomach diseases because the commensal role of H. pylori cannot be rejected outright. In a similar way, the observations made in the present study strongly indicate that further exploration of the different aspects of associations of P. fluorescens with human disease and health should be carried out. The pathogenic potential may be explored in animal models like gerbils.
This study concludes that P. fluorescens is as common as H. pylori in the stomach of humans. Colonies that appeared on enriched BHI agar after 72 h of strictly microaerophilic incubation were H. pylori while those growing faster in an aerobic atmosphere were different. These different growths could be identified as P. fluorescens. The activity of P. fluorescens in the stomach may be speculated to be either pathogenic or probiotic.
Although Helicobacter pylori (H. pylori) has been implicated in acid peptic diseases along with stomach cancer, there are reports indicating the presence of several other bacterial species in the stomach. Antral biopsies from North Indian subjects frequently yielded a bacterial growth on selective, non-enriched simple medium in an aerobic environment at 37 °C of small, low convex, and pinhead-size translucent colonies. Presence of these types of growth provoked questions about its characterization and its status: whether it was a contaminant from the environment during antral biopsy collection.
The big question to be answered is further characterization of these isolates and to ensure that they are actual colonizers of the stomach. Do these acid tolerant isolates have pathogenic potential if they are real colonizers?
Pseudomonas fluorescens (P. fluorescens)-like bacteria colonize the stomach quite frequently of North Indian patients at high density, as polymerase chain reaction (PCR)-based detection and isolation rates were both comparable. In contrast, the density of H. pylori seems to be quite low as nested PCR-based detection is significantly high as compared to the isolation rate of the bacterium. P. fluorescens isolates are urease producers and acid tolerant. Although the mupirocin gene could not be detected in any of the P. fluorescens isolates, the probiotic (inhibitory to H. pylori) role of the bacterium in the stomach may be speculated.
The potential of P. fluorescens as a probiotic may be explored because despite very high prevalence of H.pylori in India the incidences of acid peptic diseases and stomach cancer are quite low.
Nested polymerase chain reaction is a modified technique of PCR intended to increase sensitivity and specificity of primer and to reduce the contamination in amplicons due to the amplification of undesired primer annealing sites. Phylogenetics is the study of evolutionary relations among groups of organisms such as strains or species, which are based on molecular sequencing data matrices. Probiotics are live microorganisms which when administered in adequate amounts confer a health benefit on the host, e.g., lactic acid bacteria.
The study is well carried out from the methodological perspective. This manuscript characterized the bacterial isolates growing aerobically from antral biopsies were P. fluorescens, which was the acid tolerant bacteria other than H. pylori. On the basis of 16S rRNA and HSP60 sequence and from phylogenetic sequence analysis these organisms were closely related to P. fluorescens. The authors also reconfirmed this unknown bacterium as P. fluorescens by PCR positivity of P. fluorescens specific conserved putative outer membrane protein gene. Finally, the authors concluded that P. fluorescens is as common as H. pylori in the human stomach.
P- Reviewers Slomiany BL, Wang JT S- Editor Gou SX L- Editor Logan S E- Editor Lu YJ
| 1. | Marshall BJ, Warren JR. Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet. 1984;1:1311-1315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3302] [Cited by in RCA: 3265] [Article Influence: 79.6] [Reference Citation Analysis (1)] |
| 2. | Cover TL, Blaser MJ. Helicobacter pylori and gastroduodenal disease. Annu Rev Med. 1992;43:135-145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 154] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 3. | McGowan CC, Cover TL, Blaser MJ. Helicobacter pylori and gastric acid: biological and therapeutic implications. Gastroenterology. 1996;110:926-938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 139] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 4. | Steer HW, Colin-Jones DG. Mucosal changes in gastric ulceration and their response to carbenoxolone sodium. Gut. 1975;16:590-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 80] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 5. | Domínguez-Bello MG, Reyes N, Teppa-Garrán A, Romero R. Interference of Pseudomonas strains in the identification of Helicobacter pylori. J Clin Microbiol. 2000;38:937. [PubMed] |
| 6. | Khanolkar-Gaitonde SS, Reubish GK, Lee CK, Stadtländer CT. Isolation of bacteria other than Helicobacter pylori from stomachs of squirrel monkeys (Saimiri spp.) with gastritis. Dig Dis Sci. 2000;45:272-280. [PubMed] |
| 7. | Monstein HJ, Tiveljung A, Kraft CH, Borch K, Jonasson J. Profiling of bacterial flora in gastric biopsies from patients with Helicobacter pylori-associated gastritis and histologically normal control individuals by temperature gradient gel electrophoresis and 16S rDNA sequence analysis. J Med Microbiol. 2000;49:817-822. [PubMed] |
| 8. | Blazevic DJ, Koepcke MH, Matsen JM. Incidence and identification of Pseudomonas fluorescens and Pseudomonas putida in the clinical laboratory. Appl Microbiol. 1973;25:107-110. [PubMed] |
| 9. | Gilligan PH. Pseudomonas and Burkholderia. 6th ed. Manual of clinical microbiology. Washington: ASM Press 1995; 509-519. |
| 10. | Ho SA, Hoyle JA, Lewis FA, Secker AD, Cross D, Mapstone NP, Dixon MF, Wyatt JI, Tompkins DS, Taylor GR. Direct polymerase chain reaction test for detection of Helicobacter pylori in humans and animals. J Clin Microbiol. 1991;29:2543-2549. [PubMed] |
| 11. | Singh V, Mishra S, Rao GR, Jain AK, Dixit VK, Gulati AK, Mahajan D, McClelland M, Nath G. Evaluation of nested PCR in detection of Helicobacter pylori targeting a highly conserved gene: HSP60. Helicobacter. 2008;13:30-34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 12. | Carroll NM, Jaeger EE, Choudhury S, Dunlop AA, Matheson MM, Adamson P, Okhravi N, Lightman S. Detection of and discrimination between gram-positive and gram-negative bacteria in intraocular samples by using nested PCR. J Clin Microbiol. 2000;38:1753-1757. [PubMed] |
| 13. | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876-4882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29832] [Cited by in RCA: 27640] [Article Influence: 987.1] [Reference Citation Analysis (0)] |
| 14. | Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596-1599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22436] [Cited by in RCA: 19670] [Article Influence: 1092.8] [Reference Citation Analysis (0)] |
| 15. | Grahn N, Olofsson M, Ellnebo-Svedlund K, Monstein HJ, Jonasson J. Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol Lett. 2003;219:87-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 16. | Romesbrug HC. Cluster Analysis for Researchers. Belmont (CA): Lifetime Learning Publications 1984; . |
| 17. | Merrell DS, Goodrich ML, Otto G, Tompkins LS, Falkow S. pH-regulated gene expression of the gastric pathogen Helicobacter pylori. Infect Immun. 2003;71:3529-3539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 181] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 18. | Kumar NR, Thirumalai V Arasu, Gunasekaran P. Genotyping of antifungal compounds producing plant growth promoting rhizobacteria, Pseudomonas fluorescens. Curr Sci. 2002;82:1463-1468. |
| 19. | Fuller AT, Mellows G, Woolford M, Banks GT, Barrow KD, Chain EB. Pseudomonic acid: an antibiotic produced by Pseudomonas fluorescens. Nature. 1971;234:416-417. [PubMed] |
| 20. | Charan AR, Reddy VP, Reddy PN, Reddy SS. Assessment of genetic diversity in Pseudomonas fluorescens using PCR-based methods. Biorem Biodiv Bioavail. 2011;5:10–16. |
| 21. | Manceau C, Horvais A. Assessment of genetic diversity among strains of Pseudomonas syringae by PCR-restriction fragment length polymorphism analysis of rRNA operons with special emphasis on P. syringae pv. tomato. Appl Environ Microbiol. 1997;63:498-505. [PubMed] |
| 22. | Dolby JM, Webster AD, Borriello SP, Barclay FE, Bartholomew BA, Hill MJ. Bacterial colonization and nitrite concentration in the achlorhydric stomachs of patients with primary hypogammaglobulinaemia or classical pernicious anaemia. Scand J Gastroenterol. 1984;19:105-110. [PubMed] |
| 23. | Jonkers D, van den Broek E, van Dooren I, Thijs C, Dorant E, Hageman G, Stobberingh E. Antibacterial effect of garlic and omeprazole on Helicobacter pylori. J Antimicrob Chemother. 1999;43:837-839. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 53] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 24. | Mauritz W, Graninger W, Schindler I, Karner J, Zadrobilek E, Sporn P. [Pathogenic flora in the gastric juice and bronchial secretion of long-term ventilated intensive-care patients]. Anaesthesist. 1985;34:203-207. [PubMed] |
| 25. | Howden CW, Hunt RH. Relationship between gastric secretion and infection. Gut. 1987;28:96-107. [PubMed] |
| 26. | Houben GM, Hooi J, Hameeteman W, Stockbrügger RW. Twenty-four-hour intragastric acidity: 300 mg ranitidine b.d., 20 mg omeprazole o.m., 40 mg omeprazole o.m. vs. placebo. Aliment Pharmacol Ther. 1995;9:649-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 27. | Bik EM, Eckburg PB, Gill SR, Nelson KE, Purdom EA, Francois F, Perez-Perez G, Blaser MJ, Relman DA. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA. 2006;103:732-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 730] [Cited by in RCA: 780] [Article Influence: 41.1] [Reference Citation Analysis (0)] |
| 28. | Murray AE, Bartzokas CA, Shepherd AJ, Roberts FM. Blood transfusion-associated Pseudomonas fluorescens septicaemia: is this an increasing problem? J Hosp Infect. 1987;9:243-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 29. | Hsueh PR, Teng LJ, Pan HJ, Chen YC, Sun CC, Ho SW, Luh KT. Outbreak of Pseudomonas fluorescens bacteremia among oncology patients. J Clin Microbiol. 1998;36:2914-2917. [PubMed] |
| 30. | Gershman MD, Kennedy DJ, Noble-Wang J, Kim C, Gullion J, Kacica M, Jensen B, Pascoe N, Saiman L, McHale J. Multistate outbreak of Pseudomonas fluorescens bloodstream infection after exposure to contaminated heparinized saline flush prepared by a compounding pharmacy. Clin Infect Dis. 2008;47:1372-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 31. | Wei B, Huang T, Dalwadi H, Sutton CL, Bruckner D, Braun J. Pseudomonas fluorescens encodes the Crohn’s disease-associated I2 sequence and T-cell superantigen. Infect Immun. 2002;70:6567-6575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 107] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 32. | Lacey SL, Mehmet S, Taylor GW. Inhibition of Helicobacter pylori growth by 4-hydroxy-2-alkyl-quinolines produced by Pseudomonas aeruginosa. J Antimicrob Chemother. 1995;36:827-831. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 33. | Gram L, Melchiorsen J, Spanggaard B, Huber I, Nielsen TF. Inhibition of vibrio anguillarum by Pseudomonas fluorescens AH2, a possible probiotic treatment of fish. Appl Environ Microbiol. 1999;65:969-973. [PubMed] |
| 34. | Haas D, Défago G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol. 2005;3:307-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1651] [Cited by in RCA: 1131] [Article Influence: 56.6] [Reference Citation Analysis (0)] |
| 35. | Haas D, Keel C. Regulation of antibiotic production in root-colonizing Peudomonas spp. and relevance for biological control of plant disease. Annu Rev Phytopathol. 2003;41:117-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 592] [Cited by in RCA: 384] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 36. | Sharifi-Tehrani A, Zala M, Natsch A, Loccoz YM, Defago G. Biocontrol of soil-borne fungal plant diseases by 2,4-diacetylphloroglucinol-producing fluorescent pseudomonads with different restriction profiles of amplified 16S rDNA. Er J Plant Pathol. 1998;104:631–643. |
| 37. | Thomashow LS, Weller D; Current concepts in the use of introduced bacteria for biological disease control: mechanisms and antifungal metabolites. In: Stacey G, Keen NT, editors. Plant-microbe interactions 1996. United Kingdom: Chapman and Hall, Ltd.: 187-236. . [DOI] [Full Text] |
| 38. | Toyoda H, Katsuragi K, Tamari T and Ouchi S. DNA sequence of genes for detoxification of fusaric acid a wilt-inducing agent produced by Fusarium spp. Journal of Phytopathology. 1991;133:265-277. [DOI] [Full Text] |
| 39. | Loper JE. Role of fluorescent siderophore production in biological control of Pythium ultimum by a Pseudomonas fluorescens strain. Phytopathology. 1988;78:166-172. [DOI] [Full Text] |
| 40. | Neilands JB, Leong SA. Siderophores in relation to plant growth and disease. Ann Rev Plant Physiol. 1986;37:187-208. [RCA] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 165] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 41. | Blaser MJ. Not all Helicobacter pylori strains are created equal: should all be eliminated? Lancet. 1997;349:1020-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 103] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 42. | Blaser MJ. Helicobacters are indigenous to the human stomach: duodenal ulceration is due to changes in gastric microecology in the modern era. Gut. 1998;43:721-727. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 43. | Blaser MJ. Hypothesis: the changing relationships of Helicobacter pylori and humans: implications for health and disease. J Infect Dis. 1999;179:1523-1530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 226] [Cited by in RCA: 210] [Article Influence: 8.1] [Reference Citation Analysis (1)] |