Published online Dec 21, 2013. doi: 10.3748/wjg.v19.i47.8850
Revised: November 13, 2013
Accepted: November 28, 2013
Published online: December 21, 2013
Hepatocellular carcinoma (HCC) is one of the most common cancers, and is also the leading cause of death worldwide. Studies have shown that cellular reprogramming contributes to chemotherapy and/or radiotherapy resistance and the recurrence of cancers. In this article, we summarize and discuss the latest findings in the area of cellular reprogramming in HCC. The aberrant expression of transcription factors OCT4, KLF4, SOX2, c-MYC, NANOG, and LIN28 have been also observed, and the expression of these transcription factors is associated with unfavorable clinical outcomes in HCC. Studies indicate that cellular reprogramming may play a critical role in the occurrence and recurrence of HCC. Recent reports have shown that DNA methylation, miRNAs, tumor microenvironment, and signaling pathways can induce the expression of stemness transcription factors, which leads to cellular reprogramming in HCC. Furthermore, studies indicate that therapies based on cellular reprogramming could revolutionize HCC treatment. Finally, a novel therapeutic concept is discussed: reprogramming control therapy. A potential reprogramming control therapy method could be developed based on the reprogramming demonstrated in HCC studies and applied at two opposing levels: differentiation and reprogramming. Our increasing understanding and control of cellular programming should facilitate the exploitation of this novel therapeutic concept and its application in clinical HCC treatment, which may represent a promising strategy in the future that is not restricted to liver cancer.
Core tip: Cellular reprogramming contributes to chemoresistance and radioresistance and cancer recurrence in hepatocellular carcinoma (HCC). Recent findings on cellular reprogramming in HCC are summarized and discussed, including stemness transcription factors, DNA methylation, miRNAs, tumor microenvironments, and signaling pathways. The novel therapeutic concept of reprogramming control therapy is also described, which may be a promising strategy for HCC therapy in the future.
- Citation: Zheng YW, Nie YZ, Taniguchi H. Cellular reprogramming and hepatocellular carcinoma development. World J Gastroenterol 2013; 19(47): 8850-8860
- URL: https://www.wjgnet.com/1007-9327/full/v19/i47/8850.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i47.8850
Liver cancer is one of the most common tumors worldwide. An estimated 749000 new liver cancer cases and 695000 cancer deaths occurred worldwide in 2008[1]. Half of these cases and deaths were estimated to have occurred in sub-Saharan Africa and Southeast Asia. Among primary liver cancers, hepatocellular carcinoma (HCC) represents the major histological subtype, which accounts for 70%-85% of the total liver cancer burden worldwide[2].
Reports have shown that tumor recurrence[3] and patient survival[4,5] are correlated with HCC differentiation. Based on the Edmondson-Steiner’s classification, HCC can be graded from I to IV: well-differentiated (grade I), moderately differentiated (grade II), poorly differentiated (grade III), and undifferentiated (grade IV) HCC[6]. The prognosis of poorly differentiated carcinoma is worse than that of well-differentiated carcinoma[4], and the five-year survival of patients with poorly differentiated HCC is significantly worse than that of patients with moderately or well-differentiated HCC[7]. Ample evidence demonstrates that the poor prognosis and low five-year survival with poorly differentiated carcinoma are correlated with the expression of specific genes[4,8,9] and signal pathway activation[10,11], which can increase the resistance to chemotherapeutic drugs and the frequency of HCC recurrence.
Evidence shows that aggressive poorly differentiated human cancers express high levels of embryonic stem cell-like genes, suggesting that reprogramming to a more dedifferentiated state occurs during tumor progression[12]. Moreover, if different reprogramming factors are activated, cancer cells can form well-differentiated and poorly differentiated sarcomas[13]. Poorly differentiated cancers have a higher content of prospectively isolated cancer stem cells than well-differentiated cancers[14]. These data support the view that cancer is a reprogramming-like disease and that cancer stem cells (CSC) may arise through a reprogramming-like mechanism before initiating tumor formation and progression in HCC. Therefore, understanding the role of cellular reprogramming may facilitate the development of new therapeutic strategies for HCC.
Classical tumor formation theory, i.e., clonal evolution theory, suggests that each cell in a tumor is biological homogeneous[15], whereas the alternative theory considers that the cells within a tumor are not identical, which is also known as tumor heterogeneity[16]. In the alternative theory, all of cell types can arise from a signal cell, known as a CSC, which has the potential for self-renewal and differentiation[17]. Ample evidence supports a major role for the CSC model in tumor heterogeneity. Lapidot et al[18] first demonstrated a critical role for CSC in human acute myeloid leukemia, where leukemic stem cells (LSC) initiated human acute myeloid leukemia after transplantation into SCID mice. The existence of LSC prompted further research into other types of cancer. CSC have recently been identified in several solid tumors, including breast, brain, colorectal, pancreas, liver, melanoma, and prostate cancers[19]. CSC possess the properties of normal stem cells, i.e., self-renewal and differentiation. Self-renewal enables CSC to produce another CSC with essentially the same developmental and replication potential, which can increase the capacity for self-protection against drugs, toxins, and radiation. Differentiation involves the production of different types of cancer cells that trigger tumor initiation, maintain tumor growth, and finally form a bulk tumor.
Studies have shown that reprogramming factors have specific expression signatures in human tumors (Table 1) and that the expression levels of these factors are correlated with the differentiation grades of tumor. Ben-Porath et al[12] found that poorly differentiated tumors preferentially overexpressed embryonic stem cell (ESC) genes. Moreover, the activation targets of reprogramming factors, such as NANOG, OCT4, SOX2 and c-MYC, are more frequently overexpressed in poorly differentiated tumors than well-differentiated tumors[12]. Chiou et al[20] reported that the expression levels of NANOG, OCT4 and CD133 were correlated with a poor survival prognosis in patients with oral squamous cell carcinoma. Reprogramming factors also play essential roles in maintaining the properties of CSC in tumors. Silencing the expression of Oct-4 in CD133+ lung cancer can significantly inhibit the capacity for self-renewal, enhance CD133+ cell differentiation into CD133- cells, and reverse the effects of chemotherapy or radiotherapy[21]. These data suggest that reprogramming factors play critical roles in the origin and development of CSC.
| Type of cancer | Transcription factors |
| Breast cancer | NANOG[22], SOX2[23], OCT4[24] and KLF4[22] |
| Colorectal cancer | NANOG[25], SOX2[26] and OCT4[26] |
| Gastric cancer | NANOG[27], SOX2[27] and OCT3/4[27] |
| Hepatic cancer | NANOG[28], SOX2[29], OCT4[29] and KLF4[30] |
| Lung cancer | NANOG[31], SOX2[32] and OCT4[33] |
| Esophageal cancer | NANOG[34], SOX2[35], OCT3/4[35] and LIN28[36] |
| Ovarian cancer | OCT4[37] and LIN 28[38] |
Studies have shown that the occurrence of CSC is related to cellular reprogramming, but the origin of CSC remains a conundrum. However, important new evidence has demonstrated that there are two possible routes for CSC emergence.
First, CSC may arise from normal stem cells (SC) that lose the ability to regulate proliferation. Kim et al[39] showed that SC are more readily reprogrammed into induced pluripotent stem cells (iPS) compared with somatic cells. OCT4 and either KLF4 or c-MYC are sufficient to generate iPS from neural SC[39], which suggests that SC can be reprogrammed, and the process may be much easier than reprogramming mature cells. Riggi et al[40] successfully reprogrammed mesenchymal SC (MSC) into Ewing sarcoma cancer SC by inducing the expression of the ESC genes OCT4, SOX2 and NANOG using the EWS-FLI1 fusion gene. Chiba et al[41] reported that normal SC can be transformed into CSC after overexpressing the BMI-1 gene, which had the potential for tumor formation.
The alternative theory hypothesizes that CSC may be reprogrammed from somatic cells, which acquire the capacities for self-renewal and tumor initiation after genetic lesions. After forcing the expression of exogenous OSKM (OCT4, SOX2, KLF4, MYC) in the human somatic fibroblast line TIG1, Nagata et al[42] isolated induced cancer SC (iCSC) from cell populations with the capacity for self-renewal. The lack of a functional RB1 can also trigger reprogramming, which generates cells with the properties of CSC from mouse fibroblasts[43]. Therefore, studies suggest that CSC can be reprogrammed from somatic cells. Moreover, the dedifferentiation of tumor cells may also lead to stemness property of cells. Recent studies suggest that tumor cells could also be a source of CSC. The expression of the reprogramming factors, OCT4 and NANOG, was detected in poorly differentiated lung adenocarcinoma, whereas ectopic expression of OCT4 and NANOG increased the proportion of the CD133-expressing subpopulation, sphere formation, and enhanced drug resistance in lung adenocarcinoma[44]. Similar results were also observed in melanoma and colon cancer[45,46]. For example, exogenous expression of the OCT4 gene or the transmembrane delivery of OCT4 protein promoted the dedifferentiation of melanoma cells into CSC-like cells by the induced expression of endogenous OCT4, NANOG and KLF4[45]. Su et al[46] showed that HT29/CD44- cells can be reprogrammed into CSC with significantly increased expression levels of c-MYC, STAT3, SOX2 and OCT4 by the CD44-SRC-integrin axis.
Transcription factors: Recently, it was demonstrated that forced expression of combinations of four transcription factors, i.e., OCT4, KLF4, SOX2, and c-MYC or OCT4, SOX2, NANOG and LIN28, can reprogram somatic cells into iPS that closely resemble ESC[47-50]. Increasing evidence has demonstrated that aberrant expression of reprogramming factors may confer primitive and aggressive traits, which are associated with unfavorable clinical outcomes in HCC. OCT4, NANOG and SOX2 have been detected in HCC cell lines and in tumor specimens from patients with HCC, and Oct4 could play a significant role in activating the Wnt/β-catenin and transforming growth factor-β (TGF-β) signaling pathways[51]. Huang et al[29] demonstrated that SOX2- and OCT4A-positive expression were significantly associated with an aggressive phenotype in HCC. SOX2 or OCT4A are independent prognostic factors for HCC, but the coexpression of SOX2/OCT4A has the poorest prognosis in HCC[29]. Increased expression of Nanog is also correlated with a poorer clinical outcome in HCC, whereas the overexpression of NANOG in NANOG- cells increases the capacity for self-renewal by the insulin-like growth factor receptor (IGF1R) signaling pathway in HCC[28]. Of interest, expression of the pluripotent transcription factor KLF4 is decreased or lost in primary HCC[30]. The loss of KLF4 expression is also significantly associated with poor survival in HCC[30]. Evidence suggests that KLF4 is a putative tumor suppressor gene. The enforced restoration of KLF4 expression markedly inhibits cell migration, invasion, and growth in vitro, and significantly attenuates tumor growth and metastasis in HCC animal models[30,52]. Reprogramming factors are expressed preferentially in hepatocellular carcinoma SC (HCSC). Expression levels of CD44, OCT4 and BMI1 were specifically upregulated in CD45-CD90+ cells isolated from the tumor tissues and blood samples of patients with HCC compared with those in CD45-CD90+ cells isolated from normal livers[53]. Ma et al[54] found that CD133+ HCC cells expressed consistently higher mRNA levels of β-catenin, OCT-3/4, BMI, SMO, and NOTCH-1 than CD133− HCC cells.
DNA methylation: Epigenetic studies have demonstrated that specific DNA methylation patterns, including global hypomethylation and promoter hypermethylation, may be early events in HCC[55]. A genome-wide DNA methylation microarray analysis showed that side population (SP) cells had a different DNA methylation status compared with non-SP cells in HCC[56]. Recent discoveries have shown that DNA methylation is an essential epigenetic mechanism during iPS reprogramming[57]. Demethylating agents and demethylase proteins may activate pluripotent gene promoters, thereby facilitating cellular reprogramming and ultimately enhancing the efficiency of iPS generation. Wang et al[58] found that chemoresistant cells exhibited increased expression levels of OCT4 in HCC, whereas the expression of OCT4 was regulated by DNA methylation. More recent reports have shown that the expression of OCT4 is associated with the protein level of lipid storage droplet (LSD) in pluripotent cancer cells and human testicular seminoma tissues[59]. CD133 expression is also regulated by DNA methylation in HCC[60]. The elevated expression of CD133 is associated with the demethylation of Line-1 in HCC[60]. Moreover, TGF-β-1 can inhibit the expression of DNA methyltransferases (DNMT)1 and DNMT3β, thereby leading to significant demethylation in the CD133 promoter-1 in CD133- Huh7 cells[61]. Studies of MSC have shown that methylation of the tumor suppressor genes, HIC1 and RASSF1A, is sufficient to successfully reprogram the MSC into cancer stem/initiating cells[62]. These studies suggest that the demethylation of reprogramming factors and/or methylation of tumor suppressor genes contribute to reprogramming in HCC and to the origination of HCSC.
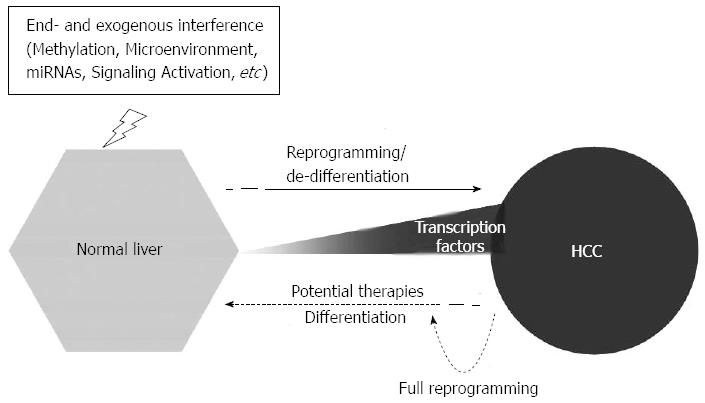
MicroRNAs: MicroRNAs (miRNAs) are well-characterized regulators of development and differentiation[63]. Studies have demonstrated that specific miRNAs have high expression levels in ESC and that they play a critical role in the control of pluripotency-related genes[64,65]. The clusters of miRNA-302s/367s[66] or miRNA-302s/369s/200c[67] can directly reprogram mouse and human somatic cells to pluripotency and increase the expression levels of OCT4 and SOX2. Studies have shown that miRNA-302 is a direct target of OCT4 and SOX2 in human ESC[68], whereas miRNA-302 and OCT4/SOX2 may work as a positive feedback system in cellular reprogramming. Moreover, the reprogramming miRNA-302 is highly expressed in a rare subpopulation of glioma cell lines. miR-302 expression causes tumorsphere formation and significant upregulation of pluripotent genes[69]. Results indicate that miRNAs participate in the neoplastic transformation of HCSC in HCC. In total, 68 miRNAs have been found to be overexpressed, whereas 10 miRNAs were underexpressed in a SP of HCC cells compared with fetal liver cells[70]. miRNA can also regulate the expression of cancer SC markers in HCC. OCT4 was regulated by miRNA-145 in T3A-A3, which are CSC-like cells[71], whereas miRNA-148 attenuated the expression of CD90 and CD44 in HCC[72]. miRNA-181 family members were highly expressed in (epithelial cell adhesion molecule+ (EpCAM+AFP+) HCC cells, and the inhibition of miRNA-181 led to a reduction in the quantity of EpCAM+ HCC cells and their tumor-initiating ability[73]. These reports suggest that miRNAs are potential factors in the reprogramming of HCC (Figure 1).
Microenvironment: Microenvironment plays a role in HCC, although its role during cellular reprogramming remains unclear. Hypoxia is a well-known characteristic of the tumor microenvironment, including HCC. In the emerging field of induced pluripotency, Yoshida et al[74] have shown that hypoxia can significantly improve the generation of iPS colonies following reprogramming. Seven hypoxia-related prognostic genes, i.e., CCNG2, EGLN3, ERO1L, WDR45L, FGF21, MAT1A and RCL1, which were dysregulated in HCC, were associated with chronic hypoxia, and were correlated with a poor prognosis in HCC[75]. CCNG2[76] and EGLN3[77] were upregulated in CSC, whereas MAT1A deficiency increases the expression of CD133+ HCSC[78]. Mathieu et al[79] showed that hypoxia by hypoxia-inducible factor (HIF) could induce a hESC-like transcriptional program, including induction of the reprogramming factors, OCT4, NANOG, SOX2, KLF4, cMYC and miRNA-302, in 11 cancer cell types, including HCC. Haraguchi et al[80] reported that CD13 is a marker for semiquiescent CSC in human liver cancer cell lines, where the expression of CD13 is accompanied by the expression of carbonic anhydrase 9 (CA9), a hypoxia marker in HCC.
The tumor environment is always characterized by inflammation. Interleukin (IL)-6, an inflammatory cytokine, led to HCC from an IL-6-driven transformed SC with inactivated TGF-β signaling[81]. Moreover, a subset of highly chemoresistant and invasive HSC were screened that had aberrant expression levels of cytokine IL-6 and TWIST. The secretion of IL-6 and TWIST can significantly increase the expression levels of let-7 and miR-181, which contribute to chemoresistance and cell invasion in HCC[82].
Both of Hepatitis B virus (HBV) and Hepatitis C virus (HCV) are the major etiological agents of chronic liver disease and HCC. In vitro and in vivo studies have shown that OCT4, NANOG, KLF-4, β-catenin and (EpCAM) are activated by HBx, and the upregulated expression of multiple stem genes demonstrates that HBx contributes to hepatocarcinogenesis, at least partly, by promoting changes in gene expression, which are characteristics of CSC[83]. Moreover, HCV can also induce the cancer stem cell-like signatures in cell culture and mouse model.
Reprogramming is likely to induce drastic molecular changes that involve the upregulation of pluripotent genes and the repression of differentiation genes. Thus, signaling pathways have profound effects on the reprogramming of somatic cells into iPS[84]. A class comparison analysis showed that 793 genes were differentially expressed in hepatic stem cell-like HCC (HpSC-HCC) and mature hepatocyte-like HCC (MH-HCC)[85]. A pathway analysis indicated that differentially expressed genes were significantly associated with SC signaling pathways, including Wnt/β-catenin, TGF-β and ERK/MAPK signaling[85]. These results suggest that signaling pathways have significant effects on cell reprogramming in HCC.
Wnt/β-catenin: It is well-known that Wnt/β-catenin signaling can control ESC self-renewal and the maintenance of stemness[86], and it also regulates the expression of ESC genes[87]. Furthermore, it may contribute to the reprogramming of somatic cells in pluripotent cells[88]. Yamashita et al[89] identified a novel prognostic HCC subtype based on EpCAM expression, which resembled hepatic progenitor cells with activated stem cell markers and Wnt/β-catenin signaling. The expression of EpCAM was associated with the activation of Wnt/β-catenin signaling[89]. Similar results were reported by Yang et al[90] who found that OV6+ cancer cells could endogenously activate Wnt/β-catenin signaling in HCC. Expression of OV6 increases after the activation of Wnt/β-catenin signaling, whereas inhibition of Wnt/β-catenin signaling leads to a decrease in the proportion of OV6+ cells in HCC[90]. Moreover, the activation of Wnt/β-catenin signaling could be inhibited by silencing the expression of OCT4, with a reduction in WNT-10b and β-catenin and an increase in TCF3[51]. These results indicate that Wnt/β-catenin signaling may be an essential part of cellular reprogramming and the maintenance of stem-like characteristics in HCC.
TGF-β: TGF-β signaling pathway has been reported in many cellular processes in adult organisms and the developing embryo, including cell growth, differentiation, apoptosis, and homeostasis. Ichida et al[91] demonstrated that TGF-β signaling is involved with cellular reprogramming. The inhibition of TGF-β signaling can promote the completion of reprogramming by the induction of Nanog[91]. Recent studies have shown that the TGF-β signaling pathway can regulate cellular reprogramming in HCC. HCSC exhibit the unexpected loss of Transforming growth factor beta receptor II, which could lead to inactivation of the TGF-β signaling pathway[81]. Toll-like receptor 4/NANOG-dependent tumor-initiating stem-like cells (TICs) were also detected with an inactivated TGF-β signaling pathway. Restoration of the TGF-β signaling pathway can inhibit the expression of pluripotent genes, including NANOG, CD133, OCT4 and SOX2, as well as tumorigenesis and abrogate the chemoresistance of TICs[92].
Mitogen-activated protein kinase/ERK kinase: The mitogen-activated protein kinase/ERK kinase (MAPK/ERK) signaling pathway has been detected in mouse ESC[93]. During reprogramming, the inhibition of MAPK/ERK could promote the transformation of pre-iPS into ground state pluripotent SC, which are cells associated with inhibition of the glycogen synthase kinase-3 (GSK3) signaling pathway[94]. It has been reported that CD133+ HCC exhibit a substantial increase in MAPK/ERK pathway activation[95,96] and that activation of the MAPK/ERK pathway can enhance proliferation, tumor angiogenesis, and initiate tumors in CD133+ HCC. Moreover, MAPK inhibition using the MAPK kinase 1 (MEK1) inhibitor PD98059 leads to a significant increase in TGF-β-induced apoptosis in CD133+ HCC[97].
In addition to these signaling pathways, the BMI-1 and Insulin-like growth factor-1 signal pathways also play key roles during cellular reprogramming in HCC. BMI-1 expression was highly correlated with the CSC phenotype in CD133+ HCC cells, and a modification in BMI-1 expression resulted in a similar change in the maintenance of a CD133 subpopulation in HCC[98]. Insulin-like growth factor (IGF2) and IGF1R can be upregulated in NANOG+ CSC, and a specific inhibitor of IGF1R signaling may significantly inhibit self-renewal and NANOG expression in HCSC, thereby indicating that IGF1R signaling participates in NANOG-mediated cellular reprogramming in HCC[28].
The detection and treatment of HCC have greatly improved with the advances in medicine; however, HCC remains largely incurable due to tumor recurrence. Conventional anticancer approaches, surgical resection, chemotherapy, and radiotherapy are primarily directed at bulk tumor populations. However, these strategies are frequently ineffective because of resistance to drugs and/or radiation[99]. Increasing evidence indicates that cellular reprogramming is involved with self-renewal, drug and/or radiation resistance, and tumorigenicity in HCC, and the concept of using precancerous cells and their progeny, CSC, in cancer therapy could provide unique insights into early cancer diagnosis, treatment, and preventive therapy[100]. Cellular reprogramming could also be a potentially useful therapeutic target in HCC.
Methylation: Given the essential role of DNA methylation during cellular reprogramming in HCC, DNA methylation may be a therapeutic target in HCC. Enhancer of zeste homolog 2 (EZH2) is a histone methyltransferase that catalyzes the addition of methyl groups to H3K27, and the blocking of H3K27 methylation leads to a significant reduction in TF-induced reprogramming[101]. 3-deazaneplanocin A, an S-adenosylhomocysteine hydrolase inhibitor, is an efficient inhibitor of the function of EZH2, which reduces the levels of H3K27 me3 in HCC cells, thereby reducing the number of EpCAM+ cells and the self-renewal capacity of these cells[102]. Lysine-specific histone demethylase 1 (LSD1) is a histone demethylase, and specific small bioactive inhibitors of LSD1 can enhance H3K4 methylation, derepress epigenetically suppressed genes, and inhibit the proliferation of pluripotent cancer cells, including teratocarcinoma, embryonic carcinoma, seminoma, and ESC[59]. All these studies suggest that methylation of histone 3 may be a potential target in HCC therapy.
miRNA: It is known that miRNAs are involved with the reprogramming of HCC and that they directly regulate the expression of reprogramming factors; however, miRNA can also act as a barrier during reprogramming. Evidence suggests that miRNA-34 is a reprogramming suppression miRNA, which can repress the expression of pluripotent genes, including NANOG, SOX2 and MYCN[103]. The expression of pluripotent genes in HCC can also be downregulated by miRNAs. miRNA-145 can directly target OCT4 to arrest the cell cycle and inhibit the tumor growth of T3A-A3[71]. Moreover, miRNAs can regulate self-renewal, differentiation, and chemoresistance in HCSC. The inhibition of let-7 increases the chemosensitivity to sorafenib and doxorubicin by directly targeting SOCS-1 and Caspase-3, whereas silencing of miR-181 expression leads to a reduction in the motility and invasion by directly targeting RASSF1A, TIMP3, and nemo-like kinase in CD133+ HCC[82]. Zhang et al[104] demonstrated that overexpression of miR-150 downregulates c-Myb protein levels and leads to a significant reduction in CD133+ cells, which is accompanied with significant inhibition of cell growth and tumorsphere formation. Ma et al[105] reported that antagonizing miR-130b reduces the resistance to chemotherapeutic agents, leads in the loss of in vivo tumorigenicity, and inhibits self-renewal in CD133+ TICs through TP53INP1 silencing.
Using chemotherapeutic drugs to select chemoresistant cancer cells in HCC, Wang et al[58] showed that chemoresistant cells exhibit CSC features with dramatically increased Oct4 levels and a highly activated OCT4-TCL1-AKT-ABCG2 pathway. OCT4 knockdown and/or AKT pathway inhibition can reduce the resistance to chemotherapy both in vitro and in vivo[58]. Oikawa et al[106] focused on Sal-like protein 4 (SALL4) and found that elevated expression of SALL4 in tumors is associated with poor survival in HCC. The silencing of SALL4 expression significantly inhibits in vitro and in vivo tumor growth with increased differentiation[106]. Yamashita et al[85] suggested that RNAi-mediated knockdown of EpCAM can reduce self-renewal, tumorigenicity, migration, and drug resistance in HCC cells. Haraguchi et al[80] demonstrated that CD13 could ROS-induced DNA damage after genotoxic chemotherapy or radiation stress and protect cells from apoptosis. The combination of a CD13 inhibitor and the genotoxic chemotherapeutic agent fluorouracil (5-FU) drastically reduces the tumor volume in mouse xenograft models[80].
Reports have shown that the abnormal activation and/or inhibition of signaling pathways in CSC, as well as the regulation of signal pathways, may be effective approaches to HCC therapy. Yamashita et al[89] found that TCF/β-catenin binding inhibitors were much more sensitive to EpCAM+ HCC than EpCAM− HCC, and they significantly inhibited the growth of EpCAM+ HCC. CD133+ HCC cells that survived chemotherapy had increased preferential expression levels of proteins involved with the AKT/PKB and BCL-2 pathways. AKT/PKB pathway-related cell survival proteins significantly reduce after treatment with an AKT1 inhibitor. Coincubation of an AKT1 inhibitor with DOX or 5-FU almost completely inhibits the preferential survival effect induced by CD133+ cells in HCC[107]. HCSC also exhibit an inactivated TGF-β signaling pathway[81]. A CD133+ population demonstrated significant resistance to TGF-β induced apoptosis compared with CD133− cells in HCC, whereas the MEK1 inhibitor PD98059 leads to a significant increase in TGF-β-induced apoptosis in CD133+ cells[97].
Given that the formation of tumors involves various cancer cells that differentiate from CSC, it is expected that CSC will become less malignant if forced to differentiate into mature cells. Tang et al[81] demonstrated that IL-6 can drive the differentiation of HCC from hepatic stem/progenitor cells with inactivated TGF-β signaling. Chow et al[108] found that MYC-driven tumors contains a subset of cells (SP cells), which are characterized by Hoechst 33342 efflux. SP tumor cells exhibit markers of hepatic stem cells and chemoresistance, whereas chemoresistance is lost when SP tumor cells differentiate into non-SP tumor cells[108]. This suggests that the differentiation of hepatic CSC may be a possible therapeutic approach. Recently, Yamashita et al[109] identified an oncostatin M (OSM) receptor in EpCAM+ HCSC. OSM treatment induced hepatocytic differentiation in EpCAM+ HCSC with a reduction of SC-related gene expression and an increase in albumin expression. Furthermore, a combined treatment with OSM and 5-FU eliminated HCSC and non-CSC subpopulations in an efficient manner[109]. A recent study showed that bone morphogenetic protein 4, a critical molecule in hepatogenesis and hepatic stem cell differentiation, can also promote differentiation and inhibit self-renewal in CD133+ HCSC with a high exogenous dose[110].
iPS can be generated from normal tissues by the expression of defined transcription factors, as well as from malignant cells[111]. After transformation with four ectopic reprogramming factors, i.e., OCT4, KLF4, SOX2 and c-MYC, the chronic myeloid leukemia (CML) cell line KBM7 could be reprogrammed into iPS[112]. Moreover, Kumano et al[113] induced iPS in samples isolated from patients with CML sensitive to imatinib. This report was the first example of the reprogramming of human primary cancer cells into iPS. In principle, CSC can also be reprogrammed into iPS using four or less reprogramming factors. Kim et al[39] showed that iPS could be reprogrammed from adult neural SC using only two reprogramming factors. This indicates that the number of reprogramming factors could be reduced using somatic cells that express appropriate levels of complementary factors endogenously. Studies have shown that HCSC exhibit the endogenous expression of SOX2, C-MYC, NANOG and OCT4, and that these endogenous reprogramming factors could facilitate the reprogramming of CSC into iPS, which may reduce the recurrence of HCC.
In this study, we reviewed the expression of transcription factors detected in HCC and summarized the complex mechanisms that contribute to cellular reprogramming in HCC, which then lead to the acquisition and maintenance of self-renewal and stemness features by a population of cancer cells, thereby resulting in the generation of HCSC. There are numerous potential applications of cellular reprogramming in regenerative medicine and cancer therapy. However, we showed that the knowledge obtained through studies of the molecular and cellular mechanisms that underlie reprogramming in HCC will also have deep implications for our understanding and the treatment of HCC, as well as other types of cancer. Furthermore, we also should refine the theory for application since the non-stem cell mediated, mature hepatocyte-derived HCC emerged in mice[114-116].
Recognizing the role of cellular reprogramming in HCC suggests a novel therapeutic concept: reprogramming control therapy. Based on reprogramming in HCC studies, a possible reprogramming control therapy could be developed that targets two opposing: differentiation (or dereprogramming) and reprogramming (or dedifferentiation). The differentiation approach would focus on the differentiation of reprogrammed cells in HCC. Reprogrammed cells exhibit stem cell-like characteristics, including the expression of stemness genes and the activation of specific signaling pathways. Modifications of gene expression and/or signaling pathways could induce the reprogrammed cells to differentiate into mature somatic cells with impaired self-renewal and reversed chemoresistance and/or radioresistance. The reprogramming approach would help to induce the partially reprogrammed cells in HCC to transform in full reprogrammed cells, such as iPS, which can be redifferentiated into various types of mature cells. In vitro experiments and mice model studies have shown that these theoretical therapeutic approaches may have applications in future HCC therapy. Increased knowledge and control of cellular programming could lead to the development of this novel therapeutic concept and its application in clinical HCC therapy, which may be a promising strategy in the future.
P- Reviewers: Colnot S, Lin CLS, Pei XT S- Editor: Cui XM L- Editor: A E- Editor: Wu HL
| 1. | Ferlay J SH, Bray F, Forman D, Mathers C, Parkin DM. GLOBOCAN 2008 v2.0, Cancer incidence and mortality worldwide: IARC Cancer Base No. 10 (Internet). Lyon: International Agency for Research on Cancer 2010; Available from: http://globocan.iarc.fr Accessed on 28/8/2013. [Cited in This Article: ] |
| 2. | Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69-90. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23762] [Cited by in F6Publishing: 25182] [Article Influence: 1937.1] [Reference Citation Analysis (3)] |
| 3. | Yamamoto J, Kosuge T, Takayama T, Shimada K, Yamasaki S, Ozaki H, Yamaguchi N, Makuuchi M. Recurrence of hepatocellular carcinoma after surgery. Br J Surg. 1996;83:1219-1222. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 195] [Cited by in F6Publishing: 198] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 4. | Nakai S, Masaki T, Shiratori Y, Ohgi T, Morishita A, Kurokohchi K, Watanabe S, Kuriyama S. Expression of p57(KIP2) in hepatocellular carcinoma: relationship between tumor differentiation and patient survival. Int J Oncol. 2002;20:769-775. [PubMed] [Cited in This Article: ] |
| 5. | Schöniger-Hekele M, Hänel S, Wrba F, Müller C. Hepatocellular carcinoma--survival and clinical characteristics in relation to various histologic molecular markers in Western patients. Liver Int. 2005;25:62-69. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 39] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 6. | EDMONDSON HA, STEINER PE. Primary carcinoma of the liver: a study of 100 cases among 48,900 necropsies. Cancer. 1954;7:462-503. [PubMed] [Cited in This Article: ] |
| 7. | Zavaglia C, De Carlis L, Alberti AB, Minola E, Belli LS, Slim AO, Airoldi A, Giacomoni A, Rondinara G, Tinelli C. Predictors of long-term survival after liver transplantation for hepatocellular carcinoma. Am J Gastroenterol. 2005;100:2708-2716. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 185] [Cited by in F6Publishing: 187] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 8. | Hu TH, Huang CC, Lin PR, Chang HW, Ger LP, Lin YW, Changchien CS, Lee CM, Tai MH. Expression and prognostic role of tumor suppressor gene PTEN/MMAC1/TEP1 in hepatocellular carcinoma. Cancer. 2003;97:1929-1940. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 204] [Cited by in F6Publishing: 207] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 9. | King KL, Li AF, Chau GY, Chi CW, Wu CW, Huang CL, Lui WY. Prognostic significance of heat shock protein-27 expression in hepatocellular carcinoma and its relation to histologic grading and survival. Cancer. 2000;88:2464-2470. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 4] [Reference Citation Analysis (0)] |
| 10. | Endo K, Ueda T, Ueyama J, Ohta T, Terada T. Immunoreactive E-cadherin, alpha-catenin, beta-catenin, and gamma-catenin proteins in hepatocellular carcinoma: relationships with tumor grade, clinicopathologic parameters, and patients’ survival. Hum Pathol. 2000;31:558-565. [PubMed] [Cited in This Article: ] |
| 11. | Villanueva A, Chiang DY, Newell P, Peix J, Thung S, Alsinet C, Tovar V, Roayaie S, Minguez B, Sole M. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972-1983, 1983.e1-e11. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 520] [Cited by in F6Publishing: 562] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 12. | Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499-507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1821] [Cited by in F6Publishing: 1943] [Article Influence: 121.4] [Reference Citation Analysis (0)] |
| 13. | Sarig R, Rivlin N, Brosh R, Bornstein C, Kamer I, Ezra O, Molchadsky A, Goldfinger N, Brenner O, Rotter V. Mutant p53 facilitates somatic cell reprogramming and augments the malignant potential of reprogrammed cells. J Exp Med. 2010;207:2127-2140. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 14. | Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62-73. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 666] [Cited by in F6Publishing: 708] [Article Influence: 50.6] [Reference Citation Analysis (0)] |
| 15. | Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23-28. [PubMed] [Cited in This Article: ] |
| 16. | Park CH, Bergsagel DE, McCulloch EA. Mouse myeloma tumor stem cells: a primary cell culture assay. J Natl Cancer Inst. 1971;46:411-422. [PubMed] [Cited in This Article: ] |
| 17. | Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675-699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 733] [Cited by in F6Publishing: 778] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 18. | Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367:645-648. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3316] [Cited by in F6Publishing: 3222] [Article Influence: 107.4] [Reference Citation Analysis (0)] |
| 19. | Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755-768. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2552] [Cited by in F6Publishing: 2544] [Article Influence: 159.0] [Reference Citation Analysis (0)] |
| 20. | Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ, Tsai TH, Chou SH, Chien CS, Ku HH, Lo JF. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 2008;14:4085-4095. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 475] [Cited by in F6Publishing: 498] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 21. | Chen YC, Hsu HS, Chen YW, Tsai TH, How CK, Wang CY, Hung SC, Chang YL, Tsai ML, Lee YY. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One. 2008;3:e2637. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 340] [Cited by in F6Publishing: 373] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 22. | Nagata T, Shimada Y, Sekine S, Hori R, Matsui K, Okumura T, Sawada S, Fukuoka J, Tsukada K. Prognostic significance of NANOG and KLF4 for breast cancer. Breast Cancer. 2012;Apr 17; Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 97] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 23. | Leis O, Eguiara A, Lopez-Arribillaga E, Alberdi MJ, Hernandez-Garcia S, Elorriaga K, Pandiella A, Rezola R, Martin AG. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene. 2012;31:1354-1365. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 356] [Cited by in F6Publishing: 394] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 24. | Liu CG, Lu Y, Wang BB, Zhang YJ, Zhang RS, Lu Y, Chen B, Xu H, Jin F, Lu P. Clinical implications of stem cell gene Oct-4 expression in breast cancer. Ann Surg. 2011;253:1165-1171. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 25. | Meng HM, Zheng P, Wang XY, Liu C, Sui HM, Wu SJ, Zhou J, Ding YQ, Li JM. Overexpression of nanog predicts tumor progression and poor prognosis in colorectal cancer. Cancer Biol Ther. 2010;Feb 16; Epub ahead of print. [PubMed] [Cited in This Article: ] |
| 26. | Saigusa S, Tanaka K, Toiyama Y, Yokoe T, Okugawa Y, Ioue Y, Miki C, Kusunoki M. Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Ann Surg Oncol. 2009;16:3488-3498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 215] [Cited by in F6Publishing: 238] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 27. | Matsuoka J, Yashiro M, Sakurai K, Kubo N, Tanaka H, Muguruma K, Sawada T, Ohira M, Hirakawa K. Role of the stemness factors sox2, oct3/4, and nanog in gastric carcinoma. J Surg Res. 2012;174:130-135. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 111] [Cited by in F6Publishing: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 28. | Shan J, Shen J, Liu L, Xia F, Xu C, Duan G, Xu Y, Ma Q, Yang Z, Zhang Q. Nanog regulates self-renewal of cancer stem cells through the insulin-like growth factor pathway in human hepatocellular carcinoma. Hepatology. 2012;56:1004-1014. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 219] [Cited by in F6Publishing: 241] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 29. | Huang P, Qiu J, Li B, Hong J, Lu C, Wang L, Wang J, Hu Y, Jia W, Yuan Y. Role of Sox2 and Oct4 in predicting survival of hepatocellular carcinoma patients after hepatectomy. Clin Biochem. 2011;44:582-589. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 57] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 30. | Li Q, Gao Y, Jia Z, Mishra L, Guo K, Li Z, Le X, Wei D, Huang S, Xie K. Dysregulated Krüppel-like factor 4 and vitamin D receptor signaling contribute to progression of hepatocellular carcinoma. Gastroenterology. 2012;143:799-810.e1-e2. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 58] [Cited by in F6Publishing: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 31. | Nirasawa S, Kobayashi D, Tsuji N, Kuribayashi K, Watanabe N. Diagnostic relevance of overexpressed Nanog gene in early lung cancers. Oncol Rep. 2009;22:587-591. [PubMed] [Cited in This Article: ] |
| 32. | Sholl LM, Barletta JA, Yeap BY, Chirieac LR, Hornick JL. Sox2 protein expression is an independent poor prognostic indicator in stage I lung adenocarcinoma. Am J Surg Pathol. 2010;34:1193-1198. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 97] [Cited by in F6Publishing: 115] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 33. | Zhang XY, Dong QG, Huang JS, Huang AM, Shi CL, Jin B, Sha HF, Feng JX, Geng Q, Zhou J. The expression of stem cell-related indicators as a prognostic factor in human lung adenocarcinoma. J Surg Oncol. 2010;102:856-862. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 16] [Cited by in F6Publishing: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 34. | Shimada Y, Okumura T, Sekine S, Moriyama M, Sawada S, Matsui K, Yoshioka I, Hojo S, Yoshida T, Nagata T. Expression analysis of iPS cell - inductive genes in esophageal squamous cell carcinoma by tissue microarray. Anticancer Res. 2012;32:5507-5514. [PubMed] [Cited in This Article: ] |
| 35. | Wang Q, He W, Lu C, Wang Z, Wang J, Giercksky KE, Nesland JM, Suo Z. Oct3/4 and Sox2 are significantly associated with an unfavorable clinical outcome in human esophageal squamous cell carcinoma. Anticancer Res. 2009;29:1233-1241. [PubMed] [Cited in This Article: ] |
| 36. | Hamano R, Miyata H, Yamasaki M, Sugimura K, Tanaka K, Kurokawa Y, Nakajima K, Takiguchi S, Fujiwara Y, Mori M. High expression of Lin28 is associated with tumour aggressiveness and poor prognosis of patients in oesophagus cancer. Br J Cancer. 2012;106:1415-1423. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 77] [Cited by in F6Publishing: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 37. | Cheng L, Thomas A, Roth LM, Zheng W, Michael H, Karim FW. OCT4: a novel biomarker for dysgerminoma of the ovary. Am J Surg Pathol. 2004;28:1341-1346. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 123] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 38. | Lu L, Katsaros D, Shaverdashvili K, Qian B, Wu Y, de la Longrais IA, Preti M, Menato G, Yu H. Pluripotent factor lin-28 and its homologue lin-28b in epithelial ovarian cancer and their associations with disease outcomes and expression of let-7a and IGF-II. Eur J Cancer. 2009;45:2212-2218. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 74] [Cited by in F6Publishing: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 39. | Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Araúzo-Bravo MJ, Ruau D, Han DW, Zenke M. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646-650. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 723] [Cited by in F6Publishing: 766] [Article Influence: 47.9] [Reference Citation Analysis (0)] |
| 40. | Riggi N, Suvà ML, De Vito C, Provero P, Stehle JC, Baumer K, Cironi L, Janiszewska M, Petricevic T, Suvà D. EWS-FLI-1 modulates miRNA145 and SOX2 expression to initiate mesenchymal stem cell reprogramming toward Ewing sarcoma cancer stem cells. Genes Dev. 2010;24:916-932. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 209] [Cited by in F6Publishing: 213] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 41. | Chiba T, Zheng YW, Kita K, Yokosuka O, Saisho H, Onodera M, Miyoshi H, Nakano M, Zen Y, Nakanuma Y. Enhanced self-renewal capability in hepatic stem/progenitor cells drives cancer initiation. Gastroenterology. 2007;133:937-950. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 180] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 42. | Nagata S, Hirano K, Kanemori M, Sun LT, Tada T. Self-renewal and pluripotency acquired through somatic reprogramming to human cancer stem cells. PLoS One. 2012;7:e48699. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 43. | Liu Y, Clem B, Zuba-Surma EK, El-Naggar S, Telang S, Jenson AB, Wang Y, Shao H, Ratajczak MZ, Chesney J. Mouse fibroblasts lacking RB1 function form spheres and undergo reprogramming to a cancer stem cell phenotype. Cell Stem Cell. 2009;4:336-347. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 44. | Chiou SH, Wang ML, Chou YT, Chen CJ, Hong CF, Hsieh WJ, Chang HT, Chen YS, Lin TW, Hsu HS. Coexpression of Oct4 and Nanog enhances malignancy in lung adenocarcinoma by inducing cancer stem cell-like properties and epithelial-mesenchymal transdifferentiation. Cancer Res. 2010;70:10433-10444. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 448] [Cited by in F6Publishing: 469] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 45. | Kumar SM, Liu S, Lu H, Zhang H, Zhang PJ, Gimotty PA, Guerra M, Guo W, Xu X. Acquired cancer stem cell phenotypes through Oct4-mediated dedifferentiation. Oncogene. 2012;31:4898-4911. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 225] [Cited by in F6Publishing: 247] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 46. | Su YJ, Lai HM, Chang YW, Chen GY, Lee JL. Direct reprogramming of stem cell properties in colon cancer cells by CD44. EMBO J. 2011;30:3186-3199. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 141] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 47. | Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1254] [Cited by in F6Publishing: 1212] [Article Influence: 75.8] [Reference Citation Analysis (0)] |
| 48. | Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313-317. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3332] [Cited by in F6Publishing: 3003] [Article Influence: 176.6] [Reference Citation Analysis (0)] |
| 49. | Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663-676. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17989] [Cited by in F6Publishing: 17055] [Article Influence: 947.5] [Reference Citation Analysis (0)] |
| 50. | Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318-324. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2048] [Cited by in F6Publishing: 1876] [Article Influence: 110.4] [Reference Citation Analysis (0)] |
| 51. | Yuan F, Zhou W, Zou C, Zhang Z, Hu H, Dai Z, Zhang Y. Expression of Oct4 in HCC and modulation to wnt/β-catenin and TGF-β signal pathways. Mol Cell Biochem. 2010;343:155-162. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 52. | Lin ZS, Chu HC, Yen YC, Lewis BC, Chen YW. Krüppel-like factor 4, a tumor suppressor in hepatocellular carcinoma cells reverts epithelial mesenchymal transition by suppressing slug expression. PLoS One. 2012;7:e43593. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 53. | Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai P, Chu PW, Lam CT, Poon RT, Fan ST. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153-166. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 883] [Cited by in F6Publishing: 897] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 54. | Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, Zheng BJ, Guan XY. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542-2556. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 899] [Cited by in F6Publishing: 896] [Article Influence: 52.7] [Reference Citation Analysis (0)] |
| 55. | Sceusi EL, Loose DS, Wray CJ. Clinical implications of DNA methylation in hepatocellular carcinoma. HPB (Oxford). 2011;13:369-376. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 56. | Zhai JM, Yin XY, Hou X, Hao XY, Cai JP, Liang LJ, Zhang LJ. Analysis of the genome-wide DNA methylation profile of side population cells in hepatocellular carcinoma. Dig Dis Sci. 2013;58:1934-1947. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 11] [Cited by in F6Publishing: 13] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 57. | De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20:609-617. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 152] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 58. | Wang XQ, Ongkeko WM, Chen L, Yang ZF, Lu P, Chen KK, Lopez JP, Poon RT, Fan ST. Octamer 4 (Oct4) mediates chemotherapeutic drug resistance in liver cancer cells through a potential Oct4-AKT-ATP-binding cassette G2 pathway. Hepatology. 2010;52:528-539. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 175] [Cited by in F6Publishing: 185] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 59. | Wang J, Lu F, Ren Q, Sun H, Xu Z, Lan R, Liu Y, Ward D, Quan J, Ye T. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011;71:7238-7249. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 173] [Cited by in F6Publishing: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 60. | Zhang C, Xu Y, Zhao J, Fan L, Jiang G, Li R, Ling Y, Wu M, Wei L. Elevated expression of the stem cell marker CD133 associated with Line-1 demethylation in hepatocellular carcinoma. Ann Surg Oncol. 2011;18:2373-2380. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 18] [Cited by in F6Publishing: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 61. | You H, Ding W, Rountree CB. Epigenetic regulation of cancer stem cell marker CD133 by transforming growth factor-beta. Hepatology. 2010;51:1635-1644. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 168] [Cited by in F6Publishing: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 62. | Teng IW, Hou PC, Lee KD, Chu PY, Yeh KT, Jin VX, Tseng MJ, Tsai SJ, Chang YS, Wu CS. Targeted methylation of two tumor suppressor genes is sufficient to transform mesenchymal stem cells into cancer stem/initiating cells. Cancer Res. 2011;71:4653-4663. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 73] [Cited by in F6Publishing: 80] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 63. | Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;294:797-799. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 229] [Cited by in F6Publishing: 192] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 64. | Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488-498. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 740] [Cited by in F6Publishing: 631] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 65. | Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459-461. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 552] [Cited by in F6Publishing: 592] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 66. | Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376-388. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 900] [Cited by in F6Publishing: 869] [Article Influence: 66.8] [Reference Citation Analysis (0)] |
| 67. | Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633-638. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 565] [Cited by in F6Publishing: 535] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 68. | Card DA, Hebbar PB, Li L, Trotter KW, Komatsu Y, Mishina Y, Archer TK. Oct4/Sox2-regulated miR-302 targets cyclin D1 in human embryonic stem cells. Mol Cell Biol. 2008;28:6426-6438. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 383] [Cited by in F6Publishing: 389] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 69. | Khalili M, Sadeghizadeh M, Ghorbanian K, Malekzadeh R, Vasei M, Mowla SJ. Down-regulation of miR-302b, an ESC-specific microRNA, in Gastric Adenocarcinoma. Cell J. 2012;13:251-258. [PubMed] [Cited in This Article: ] |
| 70. | Li R, Qian N, Tao K, You N, Wang X, Dou K. MicroRNAs involved in neoplastic transformation of liver cancer stem cells. J Exp Clin Cancer Res. 2010;29:169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 51] [Cited by in F6Publishing: 54] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 71. | Jia Y, Liu H, Zhuang Q, Xu S, Yang Z, Li J, Lou J, Zhang W. Tumorigenicity of cancer stem-like cells derived from hepatocarcinoma is regulated by microRNA-145. Oncol Rep. 2012;27:1865-1872. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 72. | Yan H, Dong X, Zhong X, Ye J, Zhou Y, Yang X, Shen J, Zhang J. Inhibitions of epithelial to mesenchymal transition and cancer stem cells-like properties are involved in miR-148a-mediated anti-metastasis of hepatocellular carcinoma. Mol Carcinog. 2013;Jul 17; Epub ahead of print. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 73. | Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472-480. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 397] [Cited by in F6Publishing: 436] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 74. | Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237-241. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 574] [Cited by in F6Publishing: 552] [Article Influence: 36.8] [Reference Citation Analysis (0)] |
| 75. | van Malenstein H, Gevaert O, Libbrecht L, Daemen A, Allemeersch J, Nevens F, Van Cutsem E, Cassiman D, De Moor B, Verslype C. A seven-gene set associated with chronic hypoxia of prognostic importance in hepatocellular carcinoma. Clin Cancer Res. 2010;16:4278-4288. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 48] [Cited by in F6Publishing: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 76. | Botchkina IL, Rowehl RA, Rivadeneira DE, Karpeh MS, Crawford H, Dufour A, Ju J, Wang Y, Leyfman Y, Botchkina GI. Phenotypic subpopulations of metastatic colon cancer stem cells: genomic analysis. Cancer Genomics Proteomics. 2009;6:19-29. [PubMed] [Cited in This Article: ] |
| 77. | Casetti L, Martin-Lannerée S, Najjar I, Plo I, Augé S, Roy L, Chomel JC, Lauret E, Turhan AG, Dusanter-Fourt I. Differential contributions of STAT5A and STAT5B to stress protection and tyrosine kinase inhibitor resistance of chronic myeloid leukemia stem/progenitor cells. Cancer Res. 2013;73:2052-2058. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 78. | Rountree CB, Senadheera S, Mato JM, Crooks GM, Lu SC. Expansion of liver cancer stem cells during aging in methionine adenosyltransferase 1A-deficient mice. Hepatology. 2008;47:1288-1297. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 79. | Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CM, Hubaud A, Stadler B, Choi M, Bar M. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640-4652. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 353] [Cited by in F6Publishing: 392] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 80. | Haraguchi N, Ishii H, Mimori K, Tanaka F, Ohkuma M, Kim HM, Akita H, Takiuchi D, Hatano H, Nagano H. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest. 2010;120:3326-3339. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 449] [Cited by in F6Publishing: 462] [Article Influence: 33.0] [Reference Citation Analysis (0)] |
| 81. | Tang Y, Kitisin K, Jogunoori W, Li C, Deng CX, Mueller SC, Ressom HW, Rashid A, He AR, Mendelson JS. Progenitor/stem cells give rise to liver cancer due to aberrant TGF-beta and IL-6 signaling. Proc Natl Acad Sci USA. 2008;105:2445-2450. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 234] [Cited by in F6Publishing: 271] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 82. | Meng F, Glaser SS, Francis H, DeMorrow S, Han Y, Passarini JD, Stokes A, Cleary JP, Liu X, Venter J. Functional analysis of microRNAs in human hepatocellular cancer stem cells. J Cell Mol Med. 2012;16:160-173. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 86] [Cited by in F6Publishing: 101] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 83. | Arzumanyan A, Friedman T, Ng IO, Clayton MM, Lian Z, Feitelson MA. Does the hepatitis B antigen HBx promote the appearance of liver cancer stem cells? Cancer Res. 2011;71:3701-3708. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 80] [Cited by in F6Publishing: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 84. | Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell. 2009;4:301-312. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 282] [Cited by in F6Publishing: 265] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 85. | Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012-1024. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 891] [Cited by in F6Publishing: 905] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 86. | Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55-63. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1567] [Cited by in F6Publishing: 1537] [Article Influence: 73.2] [Reference Citation Analysis (0)] |
| 87. | Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746-755. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 401] [Cited by in F6Publishing: 402] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 88. | Lluis F, Pedone E, Pepe S, Cosma MP. Periodic activation of Wnt/beta-catenin signaling enhances somatic cell reprogramming mediated by cell fusion. Cell Stem Cell. 2008;3:493-507. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 117] [Cited by in F6Publishing: 100] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 89. | Yamashita T, Budhu A, Forgues M, Wang XW. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007;67:10831-10839. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 322] [Cited by in F6Publishing: 356] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 90. | Yang W, Yan HX, Chen L, Liu Q, He YQ, Yu LX, Zhang SH, Huang DD, Tang L, Kong XN. Wnt/beta-catenin signaling contributes to activation of normal and tumorigenic liver progenitor cells. Cancer Res. 2008;68:4287-4295. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 286] [Cited by in F6Publishing: 318] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 91. | Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell. 2009;5:491-503. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 632] [Cited by in F6Publishing: 594] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 92. | Chen CL, Tsukamoto H, Liu JC, Kashiwabara C, Feldman D, Sher L, Dooley S, French SW, Mishra L, Petrovic L. Reciprocal regulation by TLR4 and TGF-β in tumor-initiating stem-like cells. J Clin Invest. 2013;123:2832-2849. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 107] [Cited by in F6Publishing: 118] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 93. | Burdon T, Stracey C, Chambers I, Nichols J, Smith A. Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev Biol. 1999;210:30-43. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 435] [Cited by in F6Publishing: 411] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 94. | Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 624] [Cited by in F6Publishing: 602] [Article Influence: 40.1] [Reference Citation Analysis (0)] |
| 95. | Tang KH, Ma S, Lee TK, Chan YP, Kwan PS, Tong CM, Ng IO, Man K, To KF, Lai PB. CD133(+) liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology. 2012;55:807-820. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 178] [Cited by in F6Publishing: 173] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 96. | Piao LS, Hur W, Kim TK, Hong SW, Kim SW, Choi JE, Sung PS, Song MJ, Lee BC, Hwang D. CD133+ liver cancer stem cells modulate radioresistance in human hepatocellular carcinoma. Cancer Lett. 2012;315:129-137. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 141] [Cited by in F6Publishing: 148] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 97. | Ding W, Mouzaki M, You H, Laird JC, Mato J, Lu SC, Rountree CB. CD133+ liver cancer stem cells from methionine adenosyl transferase 1A-deficient mice demonstrate resistance to transforming growth factor (TGF)-beta-induced apoptosis. Hepatology. 2009;49:1277-1286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 71] [Cited by in F6Publishing: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 98. | Chiba T, Miyagi S, Saraya A, Aoki R, Seki A, Morita Y, Yonemitsu Y, Yokosuka O, Taniguchi H, Nakauchi H. The polycomb gene product BMI1 contributes to the maintenance of tumor-initiating side population cells in hepatocellular carcinoma. Cancer Res. 2008;68:7742-7749. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 155] [Cited by in F6Publishing: 172] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 99. | Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4043] [Cited by in F6Publishing: 3969] [Article Influence: 180.4] [Reference Citation Analysis (0)] |
| 100. | Zheng YW, Tsuchida T, Taniguchi H. A novel concept of identifying precancerous cells to enhance anti-cancer therapies. J Hepatobiliary Pancreat Sci. 2012;19:621-625. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 101. | Fragola G, Germain PL, Laise P, Cuomo A, Blasimme A, Gross F, Signaroldi E, Bucci G, Sommer C, Pruneri G. Cell reprogramming requires silencing of a core subset of polycomb targets. PLoS Genet. 2013;9:e1003292. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 102. | Chiba T, Suzuki E, Negishi M, Saraya A, Miyagi S, Konuma T, Tanaka S, Tada M, Kanai F, Imazeki F. 3-Deazaneplanocin A is a promising therapeutic agent for the eradication of tumor-initiating hepatocellular carcinoma cells. Int J Cancer. 2012;130:2557-2567. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 81] [Cited by in F6Publishing: 86] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 103. | Choi YJ, Lin CP, Ho JJ, He X, Okada N, Bu P, Zhong Y, Kim SY, Bennett MJ, Chen C. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol. 2011;13:1353-1360. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 299] [Cited by in F6Publishing: 316] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 104. | Zhang J, Luo N, Luo Y, Peng Z, Zhang T, Li S. microRNA-150 inhibits human CD133-positive liver cancer stem cells through negative regulation of the transcription factor c-Myb. Int J Oncol. 2012;40:747-756. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 105. | Ma S, Tang KH, Chan YP, Lee TK, Kwan PS, Castilho A, Ng I, Man K, Wong N, To KF. miR-130b Promotes CD133(+) liver tumor-initiating cell growth and self-renewal via tumor protein 53-induced nuclear protein 1. Cell Stem Cell. 2010;7:694-707. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 296] [Cited by in F6Publishing: 313] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 106. | Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469-1483. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 151] [Cited by in F6Publishing: 159] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 107. | Ma S, Lee TK, Zheng BJ, Chan KW, Guan XY. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749-1758. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 567] [Cited by in F6Publishing: 610] [Article Influence: 35.9] [Reference Citation Analysis (0)] |
| 108. | Chow EK, Fan LL, Chen X, Bishop JM. Oncogene-specific formation of chemoresistant murine hepatic cancer stem cells. Hepatology. 2012;56:1331-1341. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 109. | Yamashita T, Honda M, Nio K, Nakamoto Y, Yamashita T, Takamura H, Tani T, Zen Y, Kaneko S. Oncostatin m renders epithelial cell adhesion molecule-positive liver cancer stem cells sensitive to 5-Fluorouracil by inducing hepatocytic differentiation. Cancer Res. 2010;70:4687-4697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 65] [Cited by in F6Publishing: 76] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 110. | Zhang L, Sun H, Zhao F, Lu P, Ge C, Li H, Hou H, Yan M, Chen T, Jiang G. BMP4 administration induces differentiation of CD133+ hepatic cancer stem cells, blocking their contributions to hepatocellular carcinoma. Cancer Res. 2012;72:4276-4285. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 68] [Cited by in F6Publishing: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 111. | Ramos-Mejia V, Fraga MF, Menendez P. iPSCs from cancer cells: challenges and opportunities. Trends Mol Med. 2012;18:245-247. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 57] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 112. | Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, Wernig M, Jaenisch R, Brummelkamp TR. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039-4042. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 174] [Cited by in F6Publishing: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 113. | Kumano K, Arai S, Hosoi M, Taoka K, Takayama N, Otsu M, Nagae G, Ueda K, Nakazaki K, Kamikubo Y. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234-6242. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 115] [Cited by in F6Publishing: 116] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 114. | Ali N, Allam H, May R, Sureban SM, Bronze MS, Bader T, Umar S, Anant S, Houchen CW. Hepatitis C virus-induced cancer stem cell-like signatures in cell culture and murine tumor xenografts. J Virol. 2011;85:12292-12303. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 115. | Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol. 2011;12:79-89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 486] [Cited by in F6Publishing: 465] [Article Influence: 35.8] [Reference Citation Analysis (0)] |
| 116. | Alison MR, Islam S, Lim S. Stem cells in liver regeneration, fibrosis and cancer: the good, the bad and the ugly. J Pathol. 2009;217:282-298. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 164] [Cited by in F6Publishing: 178] [Article Influence: 11.9] [Reference Citation Analysis (0)] |