Published online Dec 7, 2013. doi: 10.3748/wjg.v19.i45.8163
Revised: August 14, 2013
Accepted: September 16, 2013
Published online: December 7, 2013
Processing time: 186 Days and 11.5 Hours
Colorectal carcinomas (CRCs) are frequently found in industrialized countries and lead to a high incidence of malignancy-related mortality. Defined by histomorphological features, CRCs and their pre-invasive lesions are quite heterogeneous. The underlying molecular mechanisms include genomic instability, genomic mutation of tumor suppressor genes or oncogenes, epigenetic changes, and the microRNA network. The molecular mechanisms are guided by repeated clonal selections. The genotype-to-phenotype relation is assumed to be the great challenge of cancer research and the development of effective targeted therapies. At present a strong genotype-to-phenotype relation is characterized only for a minority of CRCs. Consequently, the molecular characterization of CRCs is essential to interpret histological patterns and to identify prognostic groups as well as patients for targeted therapy.
Core tip: Colorectal carcinomas (CRCs) are frequently found in industrialized countries. At present only a minority of CRCs are characterized by a strong genotype-to-phenotype relation. This is due to several additional factors determining phenotype expression. In conclusion, molecular characterization (genotype) is essential to interpret the histological findings (phenotype) and to identify prognostic groups as well as patients for targeted therapy.
- Citation: Kaemmerer E, Klaus C, Jeon MK, Gassler N. Molecular classification of colorectal carcinomas: The genotype-to-phenotype relation. World J Gastroenterol 2013; 19(45): 8163-8167
- URL: https://www.wjgnet.com/1007-9327/full/v19/i45/8163.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i45.8163
Colorectal carcinoma (CRC) is frequently found in industrialized countries and is a leading cause of cancer-related death[1]. It is suggested by several studies that the genotype-to-phenotype relation remains the great challenge of basic cancer research[2,3]. The important development of DNA sequencing technologies enables clinicians and scientists to perform assessment through to full mutation analysis of CRCs. This approach makes it clear that the number of genetic aberrations in tumor cells is highly variable, including tumors with more than 80 mutations as well as carcinomas harboring fewer than 10 mutations[4,5]. However, in addition to cancer-related mutations, several other genetic and epigenetic mechanisms contribute to CRC heterogeneity. This point of view implies that the genotype-to-phenotype relation is more complex than previously assumed and is not only founded by mutations. The basic molecular events determining and guiding development of histologically defined CRC phenotypes (adenocarcinomas and non-glandular variants classified by the World Health Organization) are not elucidated. At present, several histomorphological tumor variants can be distinguished, but specific molecular characteristics reflecting the histotype only exist in small subgroups. For example, microsatellite instability (MSI) is frequently found in mucinous adenocarcinomas and some signet ring cell carcinomas, but not in all tumors of these classes. In addition, microsatellite stability and cytosine-phosphate-guanine (CpG) island hypermethylation are usually, but not always, found in intestinal cribriform comedo-type adenocarcinomas. Because of the molecular heterogeneity of CRCs, identification of basic principles of carcinogenesis has been studied in hereditary CRCs where distinct molecular events were characterized. Familial adenomatous polyposis and hereditary non-polyposis colon cancer (HNPCC) are the most important syndromes that account for the vast majority of hereditary CRCs.
At present, the combination of multiple genetic alterations and clonal selections modified by lifestyle and environmental factors is recognized as the driving force in colorectal carcinogenesis[6]. The imbalance between mutation development and cell-cycle control further contributes to tumor development. In the review, major classes of molecular events targeting colorectal carcinogenesis are detailed concerning the genotype-to-phenotype relation.
Genomic instability (GIN) is determined by separate molecular phenomena and describes loss of mutation control by the cell-cycle[1]. The important molecular events/pathways of GIN are chromosomal instability (CIN), CpG island methylator phenotype (CIMP), and MSI. In the following, these main pathways are further detailed.
The molecular mechanisms underlying CIN, the most common type of GIN, include chromosome rearrangements, sequence changes, chromosomal number alterations, and chromosomal segregation defects. Loss of 18q with the deletion of genes such as SMAD2, SMAD4, or DCC, which is found in up to 70% of primary CRCs, is a common molecular finding in CIN-related tumors[7]. The carcinomas almost always have a mutation in the APC, while KRAS mutations occur in about 50%. CIN-related molecular lesions are found in dysplastic crypt foci. However, it has not been clear up to now whether CIN is a cause or a consequence of malignant cell growth[1,7]. It is suggested that CIN acts as a molecular founder and promoter of neoplastic growth.
CIN-related CRCs demonstrate no characteristic histomorphological pattern. They differ in tumor grading, occurrence of necrosis, and accumulation of extracellular mucin. The putative molecular founder event/mutation for the intestinal phenotype of CRCs has not been characterized up to now.
MSI is found in up to 15% of so-called sporadic CRCs and in almost all HNPCC (Lynch syndrome) associated CRCs due to either somatic inactivation of both alleles or an inherited germline mutation to one allele with additional somatic inactivation of the other[8]. A mismatch repair function usually corrects deletion/insertion errors during DNA replication. In MSI, sequence corrections resulting in alleles of varying length are not performed. The differences in length are diagnostic in PCR-based strategies using consensus primer panels. In standardized panels for MSI testing, two mononucleotides (BAT25 and BAT26) and three dinucleotide microsatellites (D5S346, D2S123, D17S250) were used[9]. MSI CRCs are not usually associated with mutations in KRAS or TP53. However, genes containing simple repeats such as EGFR, BAX, and TGFbetaRII are often mutated in these tumors. The BRAF status is another variable in MSI CRCs and a prognostic factor. Disease-free survival and overall survival are significantly improved in patients with MSI and non-mutated BRAF[10]. MSI CRCs do not have chromosomal abnormalities.
On microscopic examination, MSI CRCs are often poorly differentiated, containing mucinous components, have intratumoral lymphocytes, displaying Crohn’s like inflammatory response near to the tumor edge, and are plump infiltrative. These morphological features are variably expressed and sometimes absent in MSI CRCs.
The CIMP was originally grouped together with MSI tumors. The islands are CpG rich regions within the genome and especially found in promoter sequences. In carcinogenesis, methylation of CpG islands (so-called type C methylation) leads to transcriptional silencing of genes involved in tumor suppression, apoptosis, DNA repair, and cell-cycle control[11]. Genes that are frequently affected by this non-covalent epigenetic modification are p16, MGMT, and hMLH1. The age-related methylation of genes is designated as type A methylation. Based on molecular data, different subgroups of CIMP CRCs are defined. CIMP1 includes carcinomas with frequent MSI and BRAF mutation, whereas CIMP2 refers to microsatellite stable carcinomas with a high frequency of KRAS mutations. Microsatellite stable carcinomas with frequent TP53 mutations are commonly CIMP negative[12].
Clinically, CIMP CRCs are commonly found in a proximal location and often have methylation of the hMLH1 mismatch repair gene. However, over 50% of the CIMP CRCs are microsatellite stable. In general, CIMP CRCs have a poor prognosis and are associated with mutations in KRAS and/or BRAF. The histological phenotype of CIMP CRCs is not well characterized or defined. In these carcinomas a poor degree of histomorphological differentiation is frequently found reflecting some aspects of MSI. However, despite methylation of the hMLH1 mismatch repair gene, histomorphological MSI-related histological features are not fully expressed in CIMP CRCs.
It has been recognized that about 20% of CRCs arise from a distinct pathway including special molecular and histomorphological features[1]. The so-called serrated pathway is associated with a sequence of genetic and epigenetic alterations. Activating mutations of the BRAF gene, coding a pro-proliferative, anti-apoptotic serine-threonine kinase, are an early event. The anti-apoptotic BRAF function and probably additional failures in the apoptotic pathway of enterocytes are assumed to be crucial in the establishment of serration, where an accumulation of cells is found. The BRAF associated proliferative burst is probably followed by up-regulation of p16INK4a, acting as a tumor suppressor, and increased secretion of an insulin-like, growth-factor-binding protein 7 (IGFBP7). Silencing of either p16INK4a or IGFBP7 CIMP sensible cells via methylation is proposed as essential in the progression to sessile serrated adenoma/polyp (SSA/P)[13].
The phenotypes of serrated polyps vary considerably and the entity mixed polyp reflects the considerable overlap among these lesions[14]. SSA/P constitutes about 20% of all serrated polyps and is morphologically defined by the elongation of serrated crypts and distortion of the proliferative zone[15]. Several crypts display a dilated, L-shaped, inverted T-shaped or anchor-shaped morphology with excess serration at the crypt base. Progression of SSA/P is associated with the occurrence of cytological dysplasia and development of invasive adenocarcinoma. SSA/P and related adenocarcinomas are preferentially found in the right hemicolon.
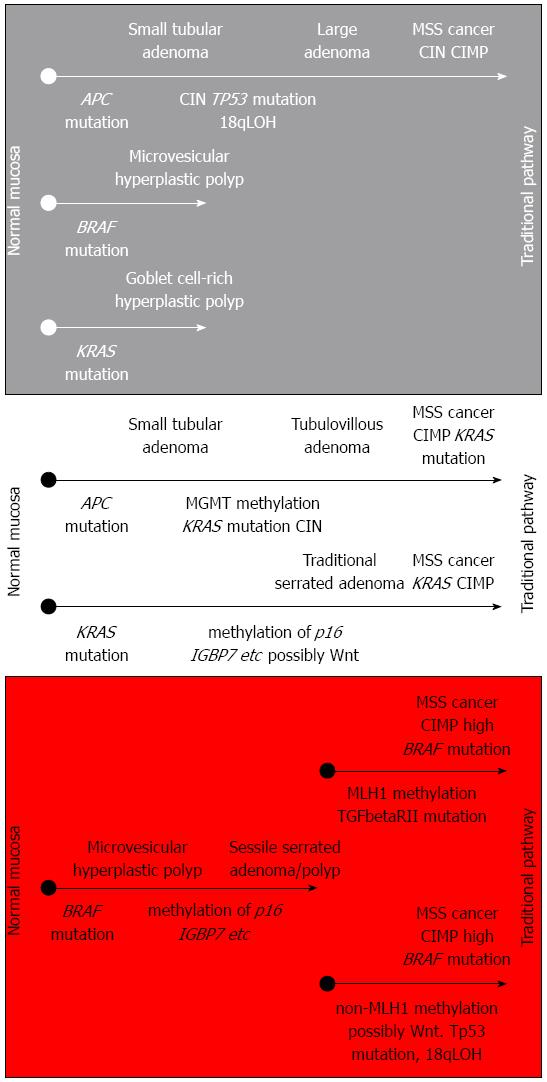
Traditional serrated adenomas (TSA) are morphological variants of serrated adenomas and show considerable differences from SSA/P concerning mutation (KRAS mutation in about 25%), localization (left-sided), and methylation status (increased methylation, but not methylation of MLH1)[16]. In terms of histomorphology, TSAs display an overall complex, often filiform configuration with tall columnar cells, eosinophilic cytoplasm, a centrally placed, elongated, hyperchromatic nucleus, and the formation of so-called ectopic crypts (the relationship of crypts with the adjacent muscularis mucosae is not preserved). The filiform serrated adenoma may represent a TSA subset. Conventional adenomas with serrated architecture are defined as an additional subgroup in the alternative serrated neoplastic pathway[1]. These morphologically defined entities are reflected by molecular findings and the model of an alternate pathway in colorectal carcinogenesis[13]. Important pathways in colorectal carcinogenesis are summarized in Figure 1.
Given the malignant potential of serrated polyps, two important serrated pathways of colorectal carcinogenesis were characterized: (1) sessile serrated pathway; and (2) traditional serrated pathway. The resulting serrated adenocarcinoma has architectural similarity to a SSA/P that may be accompanied by additional morphological features including trabecular and mucinous areas. However, these CRCs can have MSI-L or MSI-H, BRAF- or KRAS-mutations, and CIMP[13,14]. Given the molecular heterogeneity of serrated adenocarcinomas, a strong genotype-to-phenotype relation is not well established at present.
In addition to the molecular mechanisms and pathways in colorectal carcinogenesis detailed above, several other molecular events have been characterized[1]. These molecular lesions include mutational inactivation of tumor suppressor genes such as APC, TP53, and TGF-beta, activation of oncogene pathways driven by the RAS-RAF-MAPK or the PI3K-Akt signaling, injury of the miRNA network (common changes in CRCs are: up-regulation of miR-31, miR-183, and miR-17-5; down-regulation of miR-143, and miR-145), and epigenetic changes such as histone modification[1,17-19]. Experimental data describing and characterizing the molecular network behind colorectal carcinogenesis are continuously growing and should give more insight into the genotype-to-phenotype relation.
CRC is a heterogeneous disease and a leading cause of cancer-related mortality. At present, a strong genotype-to-phenotype relation, which is assumed to be the great challenge of cancer research and the development of effective targeted therapies, is only defined in a small number of CRC variants. Nevertheless, the molecular understanding of key events and modifying pathways in colorectal carcinogenesis has been essentially improved through CRC classification and therapeutic regimes in molecular terms. However, the scientific progress in the molecular understanding of CRCs calls the paradigm of a strong genotype-to-phenotype relation into question. Factors that govern the expression of pathogenic mutations include genomic aberration in a heterozygote background, the network of products from mutant and wild type genes, and environmental factors. In summary, the molecular characterization of CRCs is essential to interpret histological patterns and to identify prognostic groups as well as patients for targeted therapy.
We would like to thank Akens P for her help in typing and proofreading the manuscript.
P- Reviewers: Bujanda L, El-Tawil AM, Sgourakis G S- Editor: Gou SX L- Editor: A E- Editor: Ma S
| 1. | Kanthan R, Senger JL, Kanthan SC. Molecular events in primary and metastatic colorectal carcinoma: a review. Patholog Res Int. 2012;2012:597497. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 2. | Heinen CD. Genotype to phenotype: analyzing the effects of inherited mutations in colorectal cancer families. Mutat Res. 2010;693:32-45. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108-1113. [PubMed] |
| 4. | Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268-274. [PubMed] |
| 5. | Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66-72. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1123] [Cited by in RCA: 964] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 6. | Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397-411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 430] [Cited by in RCA: 451] [Article Influence: 32.2] [Reference Citation Analysis (0)] |
| 7. | Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 2010;138:2059-2072. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 8. | Moreira L, Balaguer F, Lindor N, de la Chapelle A, Hampel H, Aaltonen LA, Hopper JL, Le Marchand L, Gallinger S, Newcomb PA. Identification of Lynch syndrome among patients with colorectal cancer. JAMA. 2012;308:1555-1565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 360] [Cited by in RCA: 395] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 9. | Zhang X, Li J. Era of universal testing of microsatellite instability in colorectal cancer. World J Gastrointest Oncol. 2013;5:12-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 45] [Cited by in RCA: 56] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 10. | French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, Shepherd L, Windschitl HE, Thibodeau SN. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008;14:3408-3415. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 188] [Cited by in RCA: 192] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 11. | Goel A, Nagasaka T, Arnold CN, Inoue T, Hamilton C, Niedzwiecki D, Compton C, Mayer RJ, Goldberg R, Bertagnolli MM. The CpG island methylator phenotype and chromosomal instability are inversely correlated in sporadic colorectal cancer. Gastroenterology. 2007;132:127-138. [PubMed] |
| 12. | Suehiro Y, Wong CW, Chirieac LR, Kondo Y, Shen L, Webb CR, Chan YW, Chan AS, Chan TL, Wu TT. Epigenetic-genetic interactions in the APC/WNT, RAS/RAF, and P53 pathways in colorectal carcinoma. Clin Cancer Res. 2008;14:2560-2569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 83] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 13. | Leggett B, Whitehall V. Role of the serrated pathway in colorectal cancer pathogenesis. Gastroenterology. 2010;138:2088-2100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 677] [Cited by in RCA: 722] [Article Influence: 48.1] [Reference Citation Analysis (1)] |
| 14. | Noffsinger AE. Serrated polyps and colorectal cancer: new pathway to malignancy. Annu Rev Pathol. 2009;4:343-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 161] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 15. | Torlakovic EE, Gomez JD, Driman DK, Parfitt JR, Wang C, Benerjee T, Snover DC. Sessile serrated adenoma (SSA) vs. traditional serrated adenoma (TSA). Am J Surg Pathol. 2008;32:21-29. [PubMed] |
| 16. | East JE, Saunders BP, Jass JR. Sporadic and syndromic hyperplastic polyps and serrated adenomas of the colon: classification, molecular genetics, natural history, and clinical management. Gastroenterol Clin North Am. 2008;37:25-46, v. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 169] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 17. | Goel A, Boland CR. Recent insights into the pathogenesis of colorectal cancer. Curr Opin Gastroenterol. 2010;26:47-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 58] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 18. | Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143:1442-1460.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 199] [Article Influence: 15.3] [Reference Citation Analysis (0)] |