Published online Nov 21, 2013. doi: 10.3748/wjg.v19.i43.7735
Revised: August 29, 2013
Accepted: September 16, 2013
Published online: November 21, 2013
Processing time: 161 Days and 15.5 Hours
AIM: To determine the prevalence of SLC25A13 mutations in the Thai population.
METHODS: A total of 1537 subjects representing the Thai population were screened for a novel pathologic allele p.Met1? (c.2T > C) and six previously known common SLC25A13 mutations: [I] (c.851_854delGTAT), [II] (g.IVS11 + 1G > A), [III] (c.1638_1660dup), [IV] (p.S225X), [V] (IVS13 + 1G > A), and [XIX] (g.IVS16ins3kb) using a newly developed TaqMan and established HybProbe assay, respectively. Sanger sequencing was employed for specimens showing an aberrant peak to confirm the targeted mutation as well as the unknown aberrant peaks detected. Frequencies of the mutations identified were compared in each region. Carrier frequency and disease prevalence of citrin deficiency caused by SCL25A13 mutations were estimated.
RESULTS: p.Met1? was identified in the heterozygous state in 85 individuals, giving a carrier frequency of 1/18, which suggests possible selective advantage of this variant. The question of p.Met1? homozygote lethality remains unanswered which may serve as an explanation as to why this homozygote has yet to be identified in patients/controls even with high allele frequency. The p.Met1? mutation has rarely been studied in populations other than Thai and Chinese; therefore, may have been overlooked. Development of the TaqMan assay in the present study would allow a simple, rapid, and cost-effective method for mass screening. Heterozygous mutations: [XIX] and [I] were identified in 17 individuals, giving a carrier rate of 1/90 and a calculated homozygote rate of 1/33000. Two novel variants, g.IVS11 + 17C > G and c.1311C > T, of unknown clinical significance were identified at low frequency.
CONCLUSION: This study highlighted the current underestimation of citrin deficiency and suggests the possible selective advantage of the p.Met1? allele.
Core tip: Citrin deficiency is underestimated in various populations and the high prevalence of some SLC25A13 variants possibly contribute to uncharacterized predisposition/protection of certain disorders.
-
Citation: Wongkittichote P, Sukasem C, Kikuchi A, Aekplakorn W, Jensen LT, Kure S, Wattanasirichaigoon D. Screening of
SLC25A13 mutation in the Thai population. World J Gastroenterol 2013; 19(43): 7735-7742 - URL: https://www.wjgnet.com/1007-9327/full/v19/i43/7735.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i43.7735
Citrin deficiency (CD) is a genetic disorder inherited in an autosomal recessive pattern[1]. It is caused by mutations in the SLC25A13 gene encoding the citrin protein, a mitochondrial aspartate-glutamate carrier, isoform 2 (AGC2) and is expressed mainly in the liver[2-5]. The major function of AGC2 is to export mitochondrial aspartate in exchange for cytosolic glutamate[5] and is involved in several metabolic pathways with major contributions to the urea cycle and malate-aspartate shuttle[1,6,7].
There are three distinct age-dependent clinical phenotypes of CD: the mild phenotype of neonatal intrahepatic cholestasis caused by citrin deficiency [Neonatal Intrahepatic Cholestasis caused by Citrin Deficiency (NICCD), Online Mendelian Inheritance in Man (OMIM): 605814], asymptomatic period or failure to thrive and dyslipidemia in young children, and a fatal phenotype of type II citrullinemia [Adult Onset Type II Citrullinemia (CTLN2), OMIM: 603471] in older children and adults (11-80 years)[1,8,9]. Misdiagnosed or mistreated CTLN2 patients usually have poor outcomes that often result in death due to hyperammonemic encephalopathy[8].
CD is a panethnic disorder, but is relatively more common among East Asian populations. The carrier prevalence among Japanese and Chinese populations has been reported to be approximately 1/65[10-16]. Despite its high prevalence, clinical features and metabolic profiles are diverse among patients, thus making correct diagnosis difficult[1,17,18]. A definitive diagnosis of CD typically requires DNA sequencing analysis of the SLC25A13 gene, which is time consuming and costly. Alternative methods such as polymerase chain reaction restriction fragment length polymorphisms, GeneScan and SNaPshot have been adapted to detect mutations in the SLC25A13 gene[4,13,14,16,18-20]. Recently, Kikuchi et al[21], reported a new method for identifying common mutations using a real-time polymerase chain reaction (PCR)-based technique combined with melting curve analysis using HybProbes. This method appears to be a rapid and economical approach that is also suitable for the use with a high-throughput platform[21].
Over 30 SLC25A13 mutations have been described[13,14,19] and our group has previously shown that mutation [I] was the most common (8/10) mutated allele identified among NICCD Thai infants with the remaining identified as mutations [III] (c.1638_1660dup) and [XIX] (g.IVS16ins3kb)[16]. In addition, the heterozygous state of a novel p.Met1? (c.2T > C) variant was identified in an infant with idiopathic cholestasis and in 3 out of 100 healthy controls[16]. This variant was also observed in Chinese control individuals with equivalent carrier prevalence[20]. The pathogenic property of p.Met1? has been confirmed in a yeast model of CD and this mutant exhibits loss of citrin function[22]. Here, we conducted a detailed investigation to capture the true prevalence of CD in the Thai population given the small number of Thai patients with CD that have been characterized at the molecular level and with a high prevalence of p.Met1? carriers in an earlier study.
Eligible DNA samples were anonymous specimens previously stored and obtained through the Thai 4th National Health Examination Survey, during August 2008 and March 2009 by the National Health Examination Survey Office, Health System Research Institute. Sample size was calculated based on the estimated carrier prevalence of SLC25A13 mutations, 1/110, which was derived from a previous patient-based study[16]. Using the Power and Sample Size Calculations Program (version 3.0.43), for alpha error of 10% and power of 90%, the target sample size needed to achieve statistical significance was 669. Even in a worst case scenario (carrier rate of 1/200) with the same alpha error and power of testing, the minimum sample size required was 1337. To offset sample loss due to insufficient DNA quantity or the degradation of DNA extract quality, 15%-20% additional specimens were added to the sample size. A computer-based simple randomization was used to randomly select samples from each of the five regions in Thailand including; Bangkok, Central, Northeastern, Northern and Southern (Figure 1). The number of samples from each region was in accordance to the population distribution reported by the National Statistical Office (http://www.nso.go.th/). This ensured an avoidance of ascertainment bias of the study population since there may or may not have been the possibility of ethnic differences of the SLC25A13 variants among subpopulations in Thailand. Moreover, Thais from the Southern region of the country are more ethnically related to Malay descendents and those from the Northeastern region are more ethnically related to descendants of Laos and Cambodia. In total, 1569 specimens were received for the study following the approval of Ramathibodi Hospital institutional review board.
Real-time PCR using hydrolysis probes that were specific to each allele were employed to detect the p.Met1? mutation. Probes and primers were designed according to the SLC25A13 gene sequence (GenBank accession no. NM_014251). Primer sequences were 5’ GTCAGTGGGTCCCGCAGTC 3’ and 5’ GCACCCCATTTTGCTCCG 3’ as a forward and reverse primer, respectively. The probe for detecting the wild-type allele was tagged with 6-FAM at the 5’ end, whereas the mutant allele contained VIC. Sequences of the wild-type and mutant probes were 6-FAM 5’AACCGGGGCGAATCATGGCG 3’ minor groove binder (MGB) and VIC 5’AACCGGGGCGAATCACGGCG 3’ MGB, respectively.
Each real-time PCR reaction contained 0.9 μmol/L of each primer, 0.25 μmol/L of each probe, 20 ng of genomic DNA and 5 μL of TaqMan® Genotyping Master Mix (Applied Biosystem). The thermal profile started with an initial denaturation at 95 °C for 10 min followed by 55 amplification cycles at 30 s, and denaturation at 95 °C. Finally, annealing and extension for 90 s at 62 °C. The ViiA™ 7 System (Applied Biosystem) was used for detection. Positive samples were confirmed using PCR-EagI restriction digest as previously established[16].
Six common mutations accounting for 91%-100% of the mutant alleles previously reported in Japanese, Chinese, and Thai patients were selected for screening in the present study[14,16,21]. These mutations were: [I] (c.851_854delGTAT), [II] (g.IVS11+1G>A), [III] (c.1638_1660dup), [IV] (p.S225X), [V] (IVS13 + 1G > A) and [XIX] (g.IVS16ins3kb). HybProbe assays were performed using probes and primers for SLC25A13 previously validated[21]. Real-time PCR was performed using a LightCycler 480 (Roche Applied Science). A 10-μL real-time PCR reaction contained 0.5 μmol/L of each forward and reverse primer (except 0.1 μmol/L of reverse primer in the probe-primer set B), 0.2 μmol/L of each donor and acceptor probe, 20-40 ng of genomic DNA and 5 μL of LightCycler 480 probe Master (Roche Applied Science) or Premix Ex Taq™ (Perfect Real Time) (Takara Bio Company). All positive samples were confirmed using PCR-HpyCH4IV restriction digest for mutation [I][16], long-range PCR for mutation [XIX][14], and/or direct sequencing (Research Center, Ramathibodi Hospital). Long range PCR conditions were as follows: 94 °C for 5 min; 35 cycles of 98 °C for 20 s, 60 °C for 30 s, 68 °C for 15 min; 72 °C for 20 min and 15 °C for ∞. It should be noted that conventional nomenclature of mutations of the SLC25A13 gene has been widely used; therefore, to ensure that the readers would easily grasp the mutation type concept, conventional nomenclature was used alongside standard nomenclature.
Fisher’s Exact test was used to determine the statistical difference for the frequencies of the mutant alleles identified in different geographic regions. A P value of < 0.05 was considered statistically significant. Analysis of variance to compare sex and age distribution among regions was performed using the SPSS program (version 16.0, SPSS Inc., Chicago, IL, United States).
Complete DNA specimens from 1537 individuals were included in the analysis, 758 males (mean age 47 ± 21 years) and 779 females (mean age 49 ± 21 years), with an overall mean age of 48 ± 21 years. Sex and age distribution were not significantly different in the five different regions, although the mean age of subjects from the Northeastern region was slightly higher than those from the other areas of the country.
The p.Met1? allele was found in 85 individuals giving a 1/18 carrier frequency. This mutation was evenly distributed throughout geographic regions (Table 1). One of these individuals was compound heterozygous with mutation [I]; however, since the specimens in our analysis were anonymous it was impossible to determine the phenotype of this person.
| Region | Mutation [I] (c.851_854delGTAT) | Mutation XIX (g.IVS16ins3kb) | p.Met1? | Total carriers (p.Met1? not included) | Total carriers (p.Met1? included) |
| Bangkok (n = 142) | 2 (1/71)1 | 2 (1/71) | 5 (1/28)1 | 4 (1/36) | 8 (1/18)1 |
| Central (n = 387) | 0 | 3 (1/129) | 17 (1/23) | 3 (1/129) | 20 (1/19) |
| Northeastern (n = 498) | 4 (1/125) | 0 | 27 (1/18) | 4 (1/125) | 31 (1/16) |
| Northern (n = 291) | 1 (1/291) | 3 (1/97) | 21 (1/14) | 4 (1/74) | 25 (1/12) |
| Southern (n = 219) | 1 (1/219) | 1 (1/219) | 15 (1/15) | 2 (1/110) | 17 (1/13) |
| Total (n = 1537) | 8 (1/192) | 9 (1/171) | 85 (1/18) | 17 (1/90) | 101 (1/15) |
| P value2 | 0.18 | 0.067 | 0.366 | NA | 0.413 |
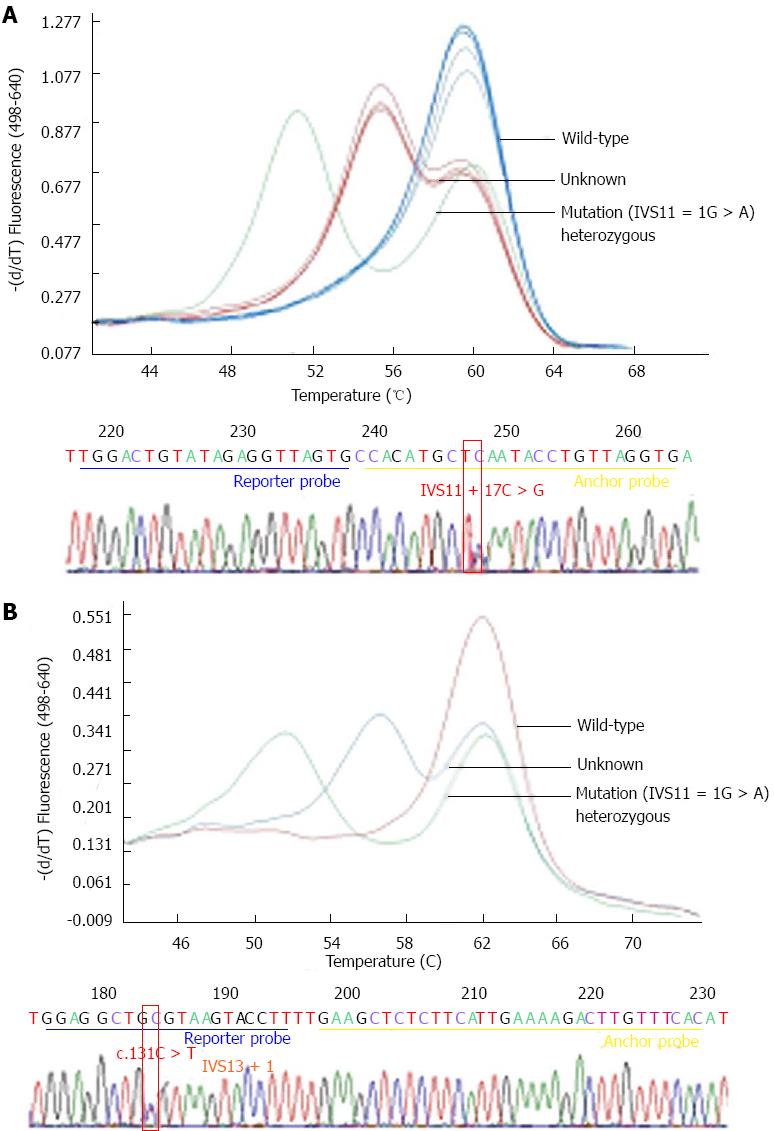
A total of 17 individuals were heterozygotes for mutant alleles. Excluding the p.Met1? variant, 8 mutation [I] and 9 mutation [XIX] (Table 1) were observed, giving a 1/90 carrier rate. Frequencies of mutations [I] and [XIX] identified in each region were not statistically different (P = 0.180 and 0.067, respectively). The four other mutations were not found. Melting point analysis revealed four specimens with heterozygous peaks that were distinct from mutation [II] (IVS11 + 1G > A) (Figure 2A). Sequencing of these specimens revealed a novel single nucleotide polymorphism (SNP), IVS11 + 17C > G. This SNP was located in the anchor-probe binding sequence. Prediction by NNSplice[23] (http://www.fruitfly.org/seq_tools/splice.html) indicated no change in the splice-site score.
Screening of mutation [V] revealed one specimen with an abnormal melting peak (Figure 2B) where direct sequencing revealed an SNP, c.1311C > T, at the binding site of the reporter probe. This novel variant was located at the last base of a codon and at the last base of exon 13, likely resulting in a synonymous change, c.C437C. The calculated donor splice score remained unchanged (0.97) suggesting that this variant is likely to be a rare polymorphism.
We have demonstrated a carrier rate of at least 1 in 90, and a homozygote/compound homozygote rate of 1 in 33000 for the known and previously identified SLC25A13 mutations, excluding the p.Met1? variant among the general Thai population. This number is different, although similar, to the estimated carrier prevalence of 1/110 and homozygotes/compound homozygote rate of 1/48000 that was obtained from the only other available patient-based study[16]. Our findings are also consistent with the prevalence of SLC25A13 homozygotes/compound heterozygotes of 1/17000 in the Japanese population and the disease frequency of 1/17000-34000 for NICCD, and 1/100000-230000 for CTLN2[8,24,25].
No studies of the p.Met1? variant had been carried out prior to the recent description in affected and unaffected populations of China and Thailand[8,14-16,20]. This mutant allele has a high carrier frequency of 1/18 and a predicted homozygote rate of 1/1300. Based on our analysis, approximately 50000 individuals homozygous for p.Met1? are predicted to be present in the Thai population. Proven deleterious effects of the p.Met1? variant in a yeast model[22] and identification of a compound heterozygote between (paternally inherited) p.Met1? and the (maternally inherited) SLC25A13 pathologic allele (r.16_212dup) in a Chinese NICCD patient[20] supports the pathogenicity of the p.Met1? allele. When taking the p.Met1? allele into account with the other six previously described common mutations identified in the present study, a very high carrier rate, 1 in 15, and homozygote rate, 1 in 900, is predicted for SLC25A13 mutations.
Patient- and population-based analyses of the prevalence of the p.Met1? variant in other ethnic backgrounds would help reveal its clinical relevance in CD and other disorder (s), if any. Based on the yeast model, the p.Met1? variant is expected to cause citrin protein production loss. It is also possible that in the p.Met1? variant, an alternative translation initiation site may produce a truncated citrin protein lacking 34 amino acid residues. However, the loss of the 34 N-terminal citrin residues causes the mislocalization and impaired function of the aberrant protein[22], indicating that even if this truncated protein was produced, it would not contribute to normal citrin activity. Currently, we have not identified a p.Met1? homozygotic individual and it is possible that p.Met1? may lead to embryonic lethality, similar to homozygotes of the Southeast Asian Ovalocytosis mutant allele in the anion-exchanger 1[26,27]. Several questions regarding the p.Met1? mutation still remain unanswered. Despite its deleterious nature in the yeast model, high prevalence in Thai and Chinese populations from tropical areas, resistance to infectious diseases, and unexplainable unidentified homozygotes in patients even with a high allele frequency, its clinical pathology in humans, ethnic and regional prevalence, selective advantage against infectious diseases, and its lethality warrant further study.
There are two noteworthy limitations of the current study: subject age and the investigation of other mutations. Given that 48 million people in Thailand are represented in our study, based on the inclusion age of 15 or older, over 1455 individuals may possibly have CD considering the confirmed disease causing mutations (excluding p.Met1?). The carrier rate derived from the present study may be underestimated due to the age of the population and mutations screened. Subjects involved in the study were living adults with an average age of 48 years. Some homozygotes/compound heterozygotes might not have survived prior to the start of the survey due to severe phenotypes, and thus would not have been included in our analysis. However, the age of the subjects may not have such a significant effect on the results of this study since the severe phenotype is relatively rare. Another limitation of the present study was that only six mutations were screened, thus the contribution of other mutations was not included in our analysis. The other two less common mutations among the Chinese and Japanese population: [VI] (c.1799_1800insA) and [VIII] (c.1801G > T)[21] were not selected for screening due to budget limitations.
CD is relatively common in East Asian populations[13,14]. Data from this study suggest that it is also common in Thais. While our analysis did not reveal any significant changes in mutation distribution in the five regions of Thailand, it is possible that an increase in sample size from each region may be necessary to reveal any other variations. CD has also been reported in Vietnamese and Malaysians[14,15,28]. Additional population studies in Southeast Asian populations will shed more light on the geographical distribution of this disease.
When considering each individual confirmed disease causing mutations, [XIX] and [I] were the two leading mutations identified in the general Thai population with equivalent frequency, 9/1537 and 8/1537, respectively (Table 2), whereas patient-based studies indicate a higher frequency of mutation [I] (8/10 mutated alleles)[16]. The discrepancy between the frequencies of mutant alleles identified from population-based studies and those obtained from patient-based analyses is also evident in other studies[13,21].
| Mutation | Number of carriers (allele frequency) | |||||
| Japanese[21] | Japanese[13,14] | Korean[13] | Chinese[13] | Thai (present study) | ||
| I | 851del4 | 4 (0.48) | 4 (0.15) | 11 (0.22) | 45 (0.54) | 8 (0.26) |
| II | IVS11 + 1G > A | 3 (0.36) | 9 (0.33) | 8 (0.16) | 0 (0.00) | 0 (0.00) |
| III | 1638ins23 | 0 (0.00) | 1 (0.04) | 1 (0.02) | 3 (0.04) | 0 (0.00) |
| IV | S225X | 0 (0.00) | 5 (0.18) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| V | IVS13 + 1G > A | 2 (0.24) | 1 (0.04) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| VII | R605X | 0 (0.00) | 0 (0.00) | 2 (0.04) | 0 (0.00) | NA |
| VIII | E601X | 1 (0.12) | 0 (0.00) | 0 (0.00) | 0 (0.00) | 0 (0.00) |
| X | IVS6 + 5G > A | 0 (0.00) | 0 (0.00) | 0 (0.00) | 15 (0.18) | NA |
| XI | R184X | 0 (0.00) | 0 (0.00) | 0 (0.00) | 1 (0.01) | NA |
| XIX | IVS16ins3kb | 0 (0.00) | 1 (0.04) | NA | NA | 9 (0.29) |
| p.Met1? | NA | NA | NA | NA | 85 (2.77) | |
| n | 420 | 1372 | 2455 | 4169 | 1537 | |
| p.Met1? not included | ||||||
| Number of carriers | 10 | 20 | 22 | 64 | 17 | |
| Carrier rate | 1/42 | 1/65 | 1/112 | 1/65 | 1/90 | |
| Homozygote rate | 1/7100 | 1/17000 | 1/50000 | 1/17000 | 1/33000 | |
| p.Met1? included | ||||||
| Number of carriers | NA | 101 | ||||
| Carrier rate | 1/15 | |||||
| Homozygote rate | 1/900 | |||||
Of the known disease causing mutations, [I] and [XIX] are most common in Thai and Chinese populations, whereas mutations [II] and [I] variants are most common among Japanese populations and patients (Table 2)[13,14,21]. In further exploration of this difference in ethnic mutation, when comparing between Chinese and Japanese, there is a possibility that the Thai ethnic background is more closely related to that of the Chinese. This may be linked to the ancient migration of certain Chinese ethnic groups to Thailand[29].
The discovery of g.IVS11 + 17C > G and c.1311C > T variants which are located on the anchor/reporter probe binding sites raised the possibility that an SNP located in the same area may affect the dissociation of probes from the target DNA and interfere with detection of the target SNP. Therefore, direct sequencing should be performed on positive subjects in order to confirm the presence of the target SNP. Moreover, the presence of double SNPs in cis could possibly obscure detection of the target SNP; however, this possibility remains to be demonstrated. Bioinformatic analysis of the g.IVS11 + 17C > G and c.1311C > T variants showed no change in splice score or amino acid sequence, suggesting a benign nature of these SNPs. However, the possibility of either being a pathogenic allele cannot be completely excluded due to its surprisingly low prevalence, 4 and 1 in 1537, respectively.
Overall, this study revealed that CD is not uncommon in the Thai population and there is a high frequency of the p.Met1? allele. Once the optimization for TaqMan/HybProbe analysis for each mutation is complete, it will serve as a rapid, efficient, robust, convenient, and cost-saving method for large scale analysis that will enable general population and newborn screening across the country. This has already been demonstrated by the successful establishment of the TaqMan assay for the p.Met1? mutation. Procedures utilized in the present study should prove valuable in examining the distribution and frequencies of SLC25A13 mutations including p.Met1? among Southeast Asian populations.
Further investigations are required to establish the clinical relevance of p.Met1? both in patients and controls. Demonstration of the molecular pathogenic mechanism of p.Met1? in human/mammalian models, although it is predicted to be a loss of function mutation[22], will also aid in further understanding. The unusually high frequency of the p.Met1? mutation suggests it may have a role in the predisposition and/or protection of disorder (s), perhaps similar to the selection of red cell polymorphisms in areas endemic for malarial infection[30-32]. Our further work will examine the possible connection between the p.Met1? mutation and protection against infectious tropical diseases.
The authors thank Professors Takeyori Saheki and Keiko Kobayashi for their wholehearted support, Dr. Chusak Okascharoen for statistical advice, and Ms. Porawee Kodcharin for sample preparation assistance.
Citrin deficiency (CD) leads to three distinct phenotypes; two of which are non-fatal. These phenotypes include: Neonatal intrahepatic cholestasis caused by citrin deficiency, failure-to-thrive and dyslipidemia. The third, a fatal phenotype of type II citrullinemia, has marked elevation of ammonia level mimicking a primary disorder of the urea cycle.
Treatment for CD is distinct from other urea cycle disorders. Epidemiological data are needed to predict disease prevalence and aid in creating awareness for diagnosis among physicians. In this study, the authors screened 1537 subjects from the general population for a novel pathologic allele, p.Met1?, and six common mutations using newly developed Taqman and established HybProbe assays. They demonstrated a carrier frequency of 1/18 for p.Met1? allele, and the carrier rate of 1/90 for mutations: [XIX] and [I], and calculated homozygote rate of 1/33000 for the two latter mutations.
The question of p.Met1? homozygote lethality remains unanswered which may serve as an explanation as to why this homozygote has yet to be identified in patients/controls even with high allele frequency. The p.Met1? mutation has been rarely studied in populations other than the Thai and Chinese and therefore, may have been overlooked. The high carrier rate of p.Met1? suggests the possible selective advantage of this particular allele.
The unusually high frequency of the p.Met1? mutation suggests its possible role in the predisposition and/or protection of disorder(s), which is perhaps similar to that of the selection of red cell polymorphisms in endemic areas for malarial infection. The established TaqMan assay would allow for a simple, rapid, and cost-effective method for p.Met1? mass screening.
CD is caused by a mutation in the SLC25A13 gene encoding for an aspartate-glutamate carrier isoform 2 (AGC2) and is expressed mainly in the liver. The major function of AGC2 is to export mitochondrial aspartate in exchange for cytosolic glutamate and it is involved in several metabolic pathways with major contributions to the urea cycle and malate-aspartate shuttle.
The article covers a topic area of increasing interest. It contains novel information and data affirms on available literature. 1537 subjects from general population were screened for a novel pathologic allele p.Met1? and six common mutations using newly developed Taqman and established HybProbe assays. The study highlighted the current underestimation of citrin deficiency and suggested the possible selective advantage of the p.Met1? allele.
P- Reviewers: Guo ZS, Han T, Palmirotta R, Syam AF S- Editor: Gou SX L- Editor: Webster JR E- Editor: Wu HL
| 1. | Saheki T, Kobayashi K. Mitochondrial aspartate glutamate carrier (citrin) deficiency as the cause of adult-onset type II citrullinemia (CTLN2) and idiopathic neonatal hepatitis (NICCD). J Hum Genet. 2002;47:333-341. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 219] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 2. | Del Arco A, Agudo M, Satrústegui J. Characterization of a second member of the subfamily of calcium-binding mitochondrial carriers expressed in human non-excitable tissues. Biochem J. 2000;345 Pt 3:725-732. [PubMed] |
| 3. | Iijima M, Jalil A, Begum L, Yasuda T, Yamaguchi N, Xian Li M, Kawada N, Endou H, Kobayashi K, Saheki T. Pathogenesis of adult-onset type II citrullinemia caused by deficiency of citrin, a mitochondrial solute carrier protein: tissue and subcellular localization of citrin. Adv Enzyme Regul. 2001;41:325-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 46] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Kobayashi K, Sinasac DS, Iijima M, Boright AP, Begum L, Lee JR, Yasuda T, Ikeda S, Hirano R, Terazono H. The gene mutated in adult-onset type II citrullinaemia encodes a putative mitochondrial carrier protein. Nat Genet. 1999;22:159-163. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 293] [Cited by in RCA: 312] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 5. | Palmieri L, Pardo B, Lasorsa FM, del Arco A, Kobayashi K, Iijima M, Runswick MJ, Walker JE, Saheki T, Satrústegui J. Citrin and aralar1 are Ca(2+)-stimulated aspartate/glutamate transporters in mitochondria. EMBO J. 2001;20:5060-5069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 365] [Cited by in RCA: 391] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 6. | Begum L, Jalil MA, Kobayashi K, Iijima M, Li MX, Yasuda T, Horiuchi M, del Arco A, Satrústegui J, Saheki T. Expression of three mitochondrial solute carriers, citrin, aralar1 and ornithine transporter, in relation to urea cycle in mice. Biochim Biophys Acta. 2002;1574:283-292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 48] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 7. | Saheki T, Kobayashi K, Iijima M, Moriyama M, Yazaki M, Takei Y, Ikeda S. Metabolic derangements in deficiency of citrin, a liver-type mitochondrial aspartate-glutamate carrier. Hepatol Res. 2005;33:181-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 52] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 8. | Kobayashi K, Saheki T, Song Y. Citrin Deficiency Sep 16 [Updated 2012 Jan 5]. GeneReviews™ [Internet]. Seattle: University of Washington, Seattle 1993; Available from: http://www.ncbi.nlm.nih.gov/books/NBK1181/. |
| 9. | Ohura T, Kobayashi K, Tazawa Y, Nishi I, Abukawa D, Sakamoto O, Iinuma K, Saheki T. Neonatal presentation of adult-onset type II citrullinemia. Hum Genet. 2001;108:87-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 97] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 10. | Dimmock D, Kobayashi K, Iijima M, Tabata A, Wong LJ, Saheki T, Lee B, Scaglia F. Citrin deficiency: a novel cause of failure to thrive that responds to a high-protein, low-carbohydrate diet. Pediatrics. 2007;119:e773-e777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 11. | Dimmock D, Maranda B, Dionisi-Vici C, Wang J, Kleppe S, Fiermonte G, Bai R, Hainline B, Hamosh A, O’Brien WE. Citrin deficiency, a perplexing global disorder. Mol Genet Metab. 2009;96:44-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 76] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 12. | Fiermonte G, Parisi G, Martinelli D, De Leonardis F, Torre G, Pierri CL, Saccari A, Lasorsa FM, Vozza A, Palmieri F. A new Caucasian case of neonatal intrahepatic cholestasis caused by citrin deficiency (NICCD): a clinical, molecular, and functional study. Mol Genet Metab. 2011;104:501-506. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 13. | Lu YB, Kobayashi K, Ushikai M, Tabata A, Iijima M, Li MX, Lei L, Kawabe K, Taura S, Yang Y. Frequency and distribution in East Asia of 12 mutations identified in the SLC25A13 gene of Japanese patients with citrin deficiency. J Hum Genet. 2005;50:338-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 88] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 14. | Tabata A, Sheng JS, Ushikai M, Song YZ, Gao HZ, Lu YB, Okumura F, Iijima M, Mutoh K, Kishida S. Identification of 13 novel mutations including a retrotransposal insertion in SLC25A13 gene and frequency of 30 mutations found in patients with citrin deficiency. J Hum Genet. 2008;53:534-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 98] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 15. | Thong MK, Boey CC, Sheng JS, Ushikai M, Kobayashi K. Neonatal intrahepatic cholestasis caused by citrin deficiency in two Malaysian siblings: outcome at one year of life. Singapore Med J. 2010;51:e12-e14. [PubMed] |
| 16. | Treepongkaruna S, Jitraruch S, Kodcharin P, Charoenpipop D, Suwannarat P, Pienvichit P, Kobayashi K, Wattanasirichaigoon D. Neonatal intrahepatic cholestasis caused by citrin deficiency: prevalence and SLC25A13 mutations among Thai infants. BMC Gastroenterol. 2012;12:141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Ohura T, Kobayashi K, Tazawa Y, Abukawa D, Sakamoto O, Tsuchiya S, Saheki T. Clinical pictures of 75 patients with neonatal intrahepatic cholestasis caused by citrin deficiency (NICCD). J Inherit Metab Dis. 2007;30:139-144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 105] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 18. | Yamaguchi N, Kobayashi K, Yasuda T, Nishi I, Iijima M, Nakagawa M, Osame M, Kondo I, Saheki T. Screening of SLC25A13 mutations in early and late onset patients with citrin deficiency and in the Japanese population: Identification of two novel mutations and establishment of multiple DNA diagnosis methods for nine mutations. Hum Mutat. 2002;19:122-130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 85] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Yasuda T, Yamaguchi N, Kobayashi K, Nishi I, Horinouchi H, Jalil MA, Li MX, Ushikai M, Iijima M, Kondo I. Identification of two novel mutations in the SLC25A13 gene and detection of seven mutations in 102 patients with adult-onset type II citrullinemia. Hum Genet. 2000;107:537-545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 20. | Zhang ZH, Lin WX, Deng M, Zhao XJ, Song YZ. Molecular analysis of SLC25A13 gene in human peripheral blood lymphocytes: Marked transcript diversity, and the feasibility of cDNA cloning as a diagnostic tool for citrin deficiency. Gene. 2012;511:227-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 21. | Kikuchi A, Arai-Ichinoi N, Sakamoto O, Matsubara Y, Saheki T, Kobayashi K, Ohura T, Kure S. Simple and rapid genetic testing for citrin deficiency by screening 11 prevalent mutations in SLC25A13. Mol Genet Metab. 2012;105:553-558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 41] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 22. | Wongkittichote P, Tungpradabkul S, Wattanasirichaigoon D, Jensen LT. Prediction of the functional effect of novel SLC25A13 variants using a S. cerevisiae model of AGC2 deficiency. J Inherit Metab Dis. 2013;36:821-830. [PubMed] |
| 23. | Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1248] [Cited by in RCA: 1354] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 24. | Kobayashi K, Shaheen N, Kumashiro R, Tanikawa K, O’Brien WE, Beaudet AL, Saheki T. A search for the primary abnormality in adult-onset type II citrullinemia. Am J Hum Genet. 1993;53:1024-1030. [PubMed] |
| 25. | Shigematsu Y, Hirano S, Hata I, Tanaka Y, Sudo M, Sakura N, Tajima T, Yamaguchi S. Newborn mass screening and selective screening using electrospray tandem mass spectrometry in Japan. J Chromatogr B Analyt Technol Biomed Life Sci. 2002;776:39-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 104] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Liu SC, Jarolim P, Rubin HL, Palek J, Amato D, Hassan K, Zaik M, Sapak P. The homozygous state for the band 3 protein mutation in Southeast Asian Ovalocytosis may be lethal. Blood. 1994;84:3590-3591. [PubMed] |
| 27. | Wrong O, Bruce LJ, Unwin RJ, Toye AM, Tanner MJ. Band 3 mutations, distal renal tubular acidosis, and Southeast Asian ovalocytosis. Kidney Int. 2002;62:10-19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 64] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 28. | Chew HB, Ngu LH, Zabedah MY, Keng WT, Balasubramaniam S, Hanifah MJ, Kobayashi K. Neonatal intrahepatic cholestasis associated with citrin deficiency (NICCD): a case series of 11 Malaysian patients. J Inherit Metab Dis. 2010;33 Suppl 3:S489-S495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 29. | Baker CJ, Pasuk P. A history of Thailand. 2th edition; New York: Cambridge University Press 2009; 1-334. |
| 30. | Nagel RL. Innate resistance to malaria: the intraerythrocytic cycle. Blood Cells. 1990;16:321-339; discussion 340-349. [PubMed] |
| 31. | Williams TN. Human red blood cell polymorphisms and malaria. Curr Opin Microbiol. 2006;9:388-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 114] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 32. | Williams TN. Red blood cell defects and malaria. Mol Biochem Parasitol. 2006;149:121-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 68] [Article Influence: 3.6] [Reference Citation Analysis (0)] |