Published online Sep 14, 2013. doi: 10.3748/wjg.v19.i34.5615
Revised: July 15, 2013
Accepted: July 18, 2013
Published online: September 14, 2013
Processing time: 110 Days and 17.2 Hours
Colorectal cancer (CRC) is one of the most common malignant diseases worldwide and the prognosis is still poor although much progress has been achieved in recent years. In order to reduce CRC-related deaths, many studies are aimed at identifying novel screening- and prognosis-related biomarkers. MicroRNAs (miRNAs) are a class of 18-27-nucleotide single-stranded RNA molecules that regulate gene expression at the post-transcriptional level. It has been demonstrated that miRNAs regulate a variety of physiological functions, including development, cell differentiation, proliferation, and apoptosis. They play important roles in various physiologic and developmental processes and in the initiation and progression of various human cancers. It has been shown that miRNAs can critically regulate tumor cell gene expression, and evidence suggests that they may function as both oncogenes and tumor suppressor genes. In CRC, miRNAs-21 is one of the most important miRNAs and is rapidly emerging as a novel biomarker in CRC, with good potential as a diagnostic and therapeutic target. In this review, we summarize the latest research findings of the clinicopathological relevance of miRNAs-21 in CRC initiation, development, and progress, highlighting its potential diagnostic, prognostic, and therapeutic application, as well as discussing future prospects.
Core tip: We summarize the latest study findings about microRNA-21 in colorectal cancer through a systematic review of literature. We recommend microRNA-21 as one of the most important microRNAs, which is rapidly emerging as a novel biomarker, with good potential as a diagnostic and therapeutic target.
- Citation: Li T, Leong MH, Harms B, Kennedy G, Chen L. MicroRNA-21 as a potential colon and rectal cancer biomarker. World J Gastroenterol 2013; 19(34): 5615-5621
- URL: https://www.wjgnet.com/1007-9327/full/v19/i34/5615.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i34.5615
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of death in the United States. It was estimated that there were about 142570 new cases diagnosed and 51370 deaths in 2010[1]. Progress in diagnosis and treatment has had a positive effect in improving overall survival, with more patients being diagnosed in the early stage of the disease, but the outcomes of patients diagnosed with advanced stage disease remains quite poor[2]. Long-term survival and better prognosis of patients depend on the stage of the tumor at the time of detection. Fecal occult blood testing and tumor markers (e.g., carcinoembryonic antigen) are used as the primary screening tools, with colonoscopy reserved for patients testing positive. However, they are generally considered to lack the desired convenience, sensitivity and specificity[3]. There are currently no tests or biomarkers that precisely predict the presence of early tumors, recurrence, sensitivity to chemotherapy and long-term survival. It is clear that improvements in early detection of primary and recurrent disease are required.
MicroRNAs (miRNAs) are a family of small, non-coding RNAs (19-22 nucleotides) which post-transcriptionally regulate gene expression. In general, miRNAs are transcribed as a group called the pri-miRNA complex, which is cleaved in the nucleus to form the pre-miRNA which is then translocated to the cytoplasm where they undergo final maturation into a functional miRNA[4]. Once in the cytoplasm, the miRNAs regulate gene expression by binding to the 5’-untranslated region of their target mRNA resulting in degradation of the double-stranded mRNA mediated by the Dicer complex. More than 700 miRNAs have thus far been identified in plants, viruses, animals and humans, and this number continues to increase (http://www.mirbase.org). Studies have shown that about 30% of human genes are regulated by miRNAs[5]. This wide regulation has implications in many important cellular functions including development, differentiation, proliferation, and programmed cell death[6-8]. Given the critical regulatory roles miRNAs serve, it is no surprise that they have been shown to be associated with many cancers[9]. CRC is a complex genetic disease characterized by uncontrolled proliferation, migration, invasion, and failure of apoptotic cell death, due to oncogene activation and tumor suppressor gene defects[10]. Many miRNAs which mediate cell growth and tumor progression have been found to be upregulated in CRC including miR-20, miR-21, miR-17-5p, miR-15b, miR-181b, miR-191 and miR-200c[9,11-14]. While lower levels of mature miRNAs such as miR-34a, miR-126, miR-143, miR-145 and miR-342 are also found, suggesting that they act as tumor suppressor miRNAs[15-18]. This deregulation of various miRNAs has been associated with tumor diagnosis and prognosis indicating that they might be potential biomarker in clinical application[3,19-21]. Multiple studies have identified that miR-21 plays a significant role in cancer biology, diagnostics and prognosis. In this article, we review the literature demonstrating the importance of miR-21 in CRC, summarize the association of miR-21 expression level with CRC diagnosis and prognosis, and discuss the potential therapeutic implications for the future.
Human miR-21 (hsa-miR-21) was cloned from HeLa cell total RNA and is highly conserved among species including human, rat, mouse, fish and frog[22]. It is located on chromosome 17q23-1 overlapping with the TMEM49 gene, a human homologue of rat vacuole membrane protein-1. MiR-21 encodes a single hairpin and is regulated by its own promoter containing binding sites for AP-1 and PU.1 transcription factors[23]. Experimental data has shown that miR-21 functions in many cell types as an anti-apoptotic and pro-survival factor and plays a significant role in cancer biology and prognosis[24-26]. Asangani et al[26] transfected Colo206f cells with miR-21 and found significant suppression of PDCD4 proteins in vitro. Resected normal and tumor tissues of 22 CRC patients demonstrated that miR-21 expression has a direct correlation with tumor invasion and metastasis.
It is clear that the majority of CRCs begin as benign adenomas, and through a series of accumulated genetic events, end up as invasive tumors. However, not all polyps will progress to invasive carcinomas. In fact, it is estimated that up to 20% of benign, subcentimeter adenomas will ultimately regress[27,28]. Therefore, it seems that the key to preventing polyps from progressing to malignant carcinomas is being able to determine which ones have the potential to progress and removing them at the benign stage. Interestingly, increased expression of several miRNAs such as miR-21, miR-31, miR-96, miR-221, miR-191, miR-19a, and miR-135b has been shown to correlate with the presence of adenomas[29,30]. In fact, Yamamichi et al[31] analyzed miR-21 expression patterns in different stages of CRC development using in situ hybrization, and found higher miR-21 expression in precancerous adenomas but not in non tumorigenic polyps. Furthermore, the frequency and extent of miR-21 expression increased during the transition from precancerous colorectal adenoma to advanced carcinoma. This demonstrates that expression of miR21 in benign colon adenomas may represent an early event in the progression to carcinoma.
Current recommendations for CRC screening are found in Table 1[32-37]. Fecal occult blood testing is a widely used test but its low specificity and sensitivity limits its clinical use, particularly for early detection. Newer screening tests are taking advantage of the presence of stem cells from human exfoliated deciduous teeth cells in the stool and are using various molecular tests to examine these cells for genetic events consistent with malignant changes. Expression levels of miRNAs offer attractive new potential biomarkers as they are uniquely stable and may represent some of the earliest changes in adenomas. Ng et al[38] reported high expression levels of miRNAs in colorectal tumors and plasma. Of the panel of 95 miRNAs analyzed by real-time polymerase chain reaction (PCR), five were upregulated in both plasma and tissue. The results were again validated using the plasma of 25 patients with CRC and 20 healthy controls. In these studies, the miRNAs 21, 17-3p, and 92 were elevated in patients with CRC (P < 0.0005). The authors further demonstrated that the plasma levels of these markers were significantly reduced after surgery in 10 patients with CRC (P < 0.05) suggesting that the high levels specifically indicate the presence of a carcinoma. Kristina et al tested the levels of 15 miRNAs in stool and colorectal tissue samples from 15 patients with CRC and five healthy individuals[39]. Although, variability was more pronounced among the stool samples than the tissue samples, the authors concluded that specific miRNA expression profiles could be defined, suggesting that stool is yet another biological material in which miRNAs are preserved and are amenable for early diagnosis of CRC. A stage-independent, sensitive, and specific marker for CRC in plasma or stool would be clinically important, and clearly these promising results support further assessment of miRNAs as potential biomarkers both in adenoma and extracellular fluids.
| Method | Sensitivity | Interval | Society |
| Fecal tests | |||
| FOBT | Yearly | USPSTF, ASGE, | |
| USMSTF | |||
| FIT | 65.8%[32,33] | Yearly | |
| Fecal DNA | 50%-60%[34] | Unspecified | USMSTF |
| Serum markers | |||
| CEA | 30%[35] | ||
| CA19-9 | |||
| Imaging tests | |||
| DCBE | 85%-97%[36] | Every 5 years | USMSTF |
| CTC | 55%-94%[37] | Every 5 years | USMSTF |
| Optical tests | |||
| FS | Every 5 years | USPSTF, ASGE, USMSTF | |
| Every 10 years | |||
| FC | USPSTF, ASGE, USMSTF |
The prognosis of patients with CRC is associated with tumor stage and phenotypic characteristics of resected cancer specimens such as tumor grade, positive lymph nodes, and angiolymphatic invasion[40]. Unfortunately, recently identified genomic and proteomic biomarkers, tumor cell mutations, and microsatellite instability cannot be recommended for routine clinical use because of insufficiently available data[41]. However, markers of prognosis are needed to help stratify patients into high risk thereby identifying patients who are likely to benefit from further therapy. Many studies on tumor biomarkers have been undertaken[42]. However, no study has identified a new marker that has been validated in clinical trials. The miRNAs represent particularly attractive markers as they seem to be micromanagers of cellular gene expression and may represent the earliest events responsible for carcinogenesis. In fact, studies have shown that the expression levels of different miRNAs, such as miR-21, miR-320, miR-498, miR-106a and miR-200c, correlate with disease-free and overall survival[43].
miR-21 may be a particularly attractive target as it has been shown to regulate the expression of many genes thought to be important in carcinogenesis. Target validation studies on putative miR-21 targets in breast cancer samples and CRC cells have demonstrated a link between miR-21 expression levels and the p53 tumor suppressor, and also demonstrated that the tumor suppressors PDCD4 and maspin are targets of miR-21[44]. Consistent with the importance in the process of carcinogenesis, Staby et al[45] demonstrated that higher miR-21 expression levels were correlated with advanced cancer stages, worse outcome, poor response to therapy, and shorter disease-free survival. Additionally, they found that miR-21 levels were positively correlated with cancer stage, lymph node involvement, and development of distant metastasis.
The most comprehensive analysis of miRNA expression in CRC performed to date tested two cohorts of 197 colon cancer patients by utilizing microarrays containing 389 miRNAs probes[46]. This analysis revealed 37 miRNAs which were differentially expressed in stage II colonic adenocarcinoma compared with adjacent normal tissue using a test set and two validation cohorts. In one of the cohorts, miR-20a, miR-21, miR-106a, miR-181b, and miR-203 were found to be overexpressed in tumor tissues with high tumor to normal ratios, as well as being associated with poor overall survival. However, the prognostic relevance could be confirmed in the validation set for only one of the candidates, miR-21, and the clinical and biological implications of differential expression of the remaining miRNAs were unclear. Similar conclusions were drawn from a study of 29 tumor samples, in which miR-21 expression was associated with poor survival and therapeutic outcome in stage II and III CRC[46]. Nielsen et al[47] reported the expression of miRNA-21 in 130 colon and 67 rectal stage II cancer specimens using high-affinity locked nucleic acid (LNA) probes. High levels of miR-21 correlated with shorter disease-free survival (hazard ratio: 1.28; 95% confidence interval: 1.06-1.55; P = 0.004) in the stage II colon cancer patient group, whereas no significant correlation with disease-free survival was observed in the stage II rectal cancer group.
miR-21 and response to chemotherapy: The current treatment for CRC involves a multidisciplinary approach including surgery supplemented with chemotherapy and radiation therapy in certain instances. In general, patients with node-positive disease benefit from chemotherapy. However, there may be a subgroup of patients with node-positive disease who are at low risk of recurrence, as nearly 40% of patients randomized to a no-treatment arm in chemotherapy trials did not develop a recurrence[48]. In addition, it is clear that some patients with node-negative disease who have advanced T-stage tumors are at high risk of developing recurrences[49]. A test that would allow for the accurate stratification of patients with stage II and III disease into low and high risk would be very clinically useful. Recently, the role of miRNAs in predicting the response to 5-fluorouracil (5-FU)-based chemotherapy in CRC treatment has been explored. A significant focus has been placed on the value of miR-21 expression levels and their abilities to predict both response to and need for chemotherapy[50].
Rossi et al[51] utilized two subclones from the human CRC cell lines HT29 and HCT116 to investigate the effect of 5-FU on miRNA expression and also to determine patterns of expression that correlated with response to therapy. Quantitative real-time PCR revealed that 5-FU upregulated 19 and downregulated three miRNAs. While some changes in miRNA expression were consistent with the antitumor effects of the drug, others were not, such as upregulation of miR-21 and the polycistronic miR-17-92 cluster (which include miR-19a and miR-20). In fact, a number of miRNAs that are already overexpressed in neoplastic tissues, including miR-21, have been shown to be upregulated in colon cancer cell lines treated with 5-FU[51]. Tomimaru et al[52] found that hepatocellular carcinoma cells transfected with pre-miR-21 were significantly resistant to 5-FU, while the 5-FU sensitivity of transfected anti-miR-21 was weakened by transfection with siRNAs of the target molecules, PTEN and PDCD4. This finding may be a cell-specific defense mechanism to survive 5-FU treatment. Svoboda et al[53] found significant changes in miRNA expression in 35 patients with rectal carcinoma undergoing preoperative capecitabine chemoradiation therapy. Tumor biopsies were taken before starting therapy and after 2 wk of therapy. The extent of the tumor response to the therapy was investigated microscopically by an experienced pathologist according to Mandard’s tumor regression criteria. In addition, the levels of miRNAs were evaluated using real-time PCR. The authors found dramatic changes in the expression levels of many miRNAs including miR-21, miR-10a, miR-145, miR-212, miR-339, miR-361. However, only two miRNAs, miR-125b and miR-137, were found to be significantly increased after 2 wk of therapy and these miRNA expression levels had a positive correlation with a poorer tumor regression response[53]. These types of studies highlight the potential for using miRNA expression profiles to predict the response to chemotherapy. However, verification of the targets in adequately designed clinical panels is the important next step that has yet to be taken.
miR-21 maybe a potential therapeutic target in colorectal cancer treatment: miRNAs are important regulators of gene expression and may present potentially interesting therapeutic targets in cancer. The synthesis, maturation and activity of miRNAs can be manipulated with various oligonucleotides that encode the sequences complementary to mature miRNAs. By influencing particular miRNAs, a cascade of pathways could be modified to inhibit tumor growth.
Wong et al[54] reported the application of 20-O-methyl- and/or DNA/LNA-mixed oligonucleotides to specifically inhibit miR-21 in cultured glioblastoma and breast cancer cells suppressed cell growth in vitro in association with increased caspase-mediated apoptosis[55]. Suppression of miR-21 also significantly reduced invasion and lung metastasis in MDA-MB-231 metastatic breast cancer[56]. Although there are at present no clinical reports describing therapy targeting miR-21 in CRC treatment, most seem to have optimistic views on the future utility of miR-21 as therapeutic targets but further studies are clearly needed[57-59]. Expression of microRNA-21 and the clinical relevance in CRC are summarized in Table 2.
| Ref. | Regulation | Biological material tested | Detection method | Clinical relevance | Comment |
| Tumor development | |||||
| Fassan et al[60] | Up | 300 polypoid lesions of the colon mucosa | RT-PCRISH | Significant miR-21 upregulation in preneoplastic/neoplastic samples | High miR-21 expression is consistent with PDCD4 downregulation |
| Yantiss et al[61] | Up | 24 patients < 40 years45 patients ≥ 40 years | RT-PCR | Significantly higher expression | |
| Tumor diagnosis | |||||
| Link et al[62] | Up | Stool samples | RT-PCR | Higher expression in patients with adenomas and CRCs | May be an excellent candidate of a noninvasive screening test for colorectal neoplasms |
| Tumor prognosis | |||||
| Chang et al[63] | Up | 48 colorectal tumors, 61 normal tissues , 7 polyps | RT-PCR | Disease recurrence | miR-21 post-transcriptionallymodulates PDCD4 via mRNA degradation |
| Nielsen et al[47] | Up | 130 stage II colon and 67 stage II rectal cancer specimens | ISH | Shorter disease-free survival in colon cancer, but not in rectal cancer | |
| Kulda et al[64] | Up | 46 paired tissue samples30 tissue samples with live metastasis | RT-PCR | Disease-free interval | |
| Schetter et al[46] | Up | 196 paired tissues | RT-PCR | Association with cancer-specific mortality, including stage II patients alone | miR-21 expression are independent predictors of colon cancer prognosis and may provide a clinically useful tool to identify high-risk patients |
| Schetter et al[46] | Up | US cohort: 84 patients; Hong Kong cohort: 113 patients | MicroRNA microarray, RT-PCR | Poor survival and poor therapeutic outcome | |
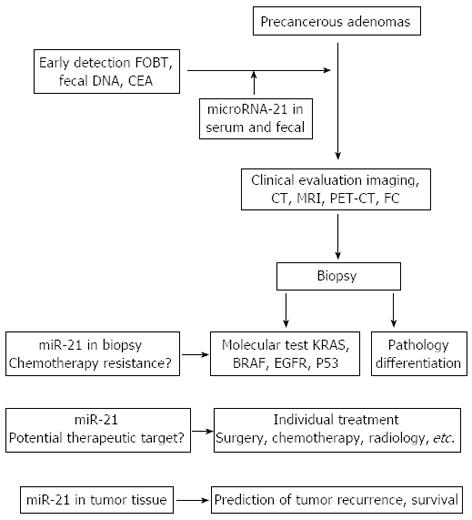
The role of miRNAs in CRC presents potentially exciting new opportunities for future investigations to determine the use of miRNAs as potential biomarkers for prognosis at the time of diagnosis as well as to determine their ability to predict the response to chemotherapy. In addition, the potential for miRNAs to serve as targets for new chemotherapeutic treatments has yet to be realized. Among the many miRNAs that have been associated with clinical outcomes, miR-21 has been consistently shown to be dysregulated in CRC. Many of the early studies relating miR-21 to CRC have been performed in vitro on established cell lines. In addition, studies using human tumor tissue have been performed in a retrospective fashion, which limits the conclusions because of inherent study bias. If these micromanagers of cell processes are to be useful tools in the diagnosis or treatment of colon cancer, we will have to study them in well-designed prospective trials. It remains to be discovered if these types of molecular expression profiles will be used in clinical practice (Figure 1).
P- Reviewer Zorcolo L S- Editor Wen LL L- Editor Cant MR E- Editor Zhang DN
| 1. | Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277-300. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10002] [Cited by in RCA: 10453] [Article Influence: 696.9] [Reference Citation Analysis (0)] |
| 2. | Malafosse R, Penna C, Sa Cunha A, Nordlinger B. Surgical management of hepatic metastases from colorectal malignancies. Ann Oncol. 2001;12:887-894. [PubMed] |
| 3. | Aslam MI, Taylor K, Pringle JH, Jameson JS. MicroRNAs are novel biomarkers of colorectal cancer. Br J Surg. 2009;96:702-710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 90] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 4. | Agostini M, Pucciarelli S, Calore F, Bedin C, Enzo M, Nitti D. miRNAs in colon and rectal cancer: A consensus for their true clinical value. Clin Chim Acta. 2010;411:1181-1186. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 6. | Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580-587. [PubMed] |
| 7. | Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 8. | Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1888] [Cited by in RCA: 2001] [Article Influence: 125.1] [Reference Citation Analysis (0)] |
| 9. | Lujambio A, Calin GA, Villanueva A, Ropero S, Sánchez-Céspedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556-13561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 795] [Cited by in RCA: 822] [Article Influence: 48.4] [Reference Citation Analysis (0)] |
| 10. | Hagan JP, Croce CM. MicroRNAs in carcinogenesis. Cytogenet Genome Res. 2007;118:252-259. [PubMed] |
| 11. | Peacock O, Lee AC, Larvin M, Tufarelli C, Lund JN. MicroRNAs: relevant tools for a colorectal surgeon? World J Surg. 2012;36:1881-1892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 12. | Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257-2261. [PubMed] |
| 13. | Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E. The colorectal microRNAome. Proc Natl Acad Sci USA. 2006;103:3687-3692. [PubMed] |
| 14. | Xi Y, Formentini A, Chien M, Weir DB, Russo JJ, Ju J, Kornmann M, Ju J. Prognostic Values of microRNAs in Colorectal Cancer. Biomark Insights. 2006;2:113-121. [PubMed] |
| 15. | Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472-15477. [PubMed] |
| 16. | Guo C, Sah JF, Beard L, Willson JK, Markowitz SD, Guda K. The noncoding RNA, miR-126, suppresses the growth of neoplastic cells by targeting phosphatidylinositol 3-kinase signaling and is frequently lost in colon cancers. Genes Chromosomes Cancer. 2008;47:939-946. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 313] [Cited by in RCA: 329] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 17. | Michael MZ, O’ Connor SM, van Holst Pellekaan NG, Young GP, James RJ. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882-891. [PubMed] |
| 18. | Grady WM, Parkin RK, Mitchell PS, Lee JH, Kim YH, Tsuchiya KD, Washington MK, Paraskeva C, Willson JK, Kaz AM. Epigenetic silencing of the intronic microRNA hsa-miR-342 and its host gene EVL in colorectal cancer. Oncogene. 2008;27:3880-3888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 228] [Cited by in RCA: 229] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 19. | Okayama H, Schetter AJ, Harris CC. MicroRNAs and inflammation in the pathogenesis and progression of colon cancer. Dig Dis. 2012;30 Suppl 2:9-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 20. | Faber C, Kirchner T, Hlubek F. The impact of microRNAs on colorectal cancer. Virchows Arch. 2009;454:359-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 21. | Tang JT, Fang JY. MicroRNA regulatory network in human colorectal cancer. Mini Rev Med Chem. 2009;9:921-926. [PubMed] |
| 22. | Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853-858. [PubMed] |
| 23. | Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957-1966. [PubMed] |
| 24. | Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029-6033. [PubMed] |
| 25. | Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, Calin GA, Volinia S, Liu CG, Scarpa A. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677-4684. [PubMed] |
| 26. | Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128-2136. [PubMed] |
| 27. | O’Brien MJ, Winawer SJ, Zauber AG, Gottlieb LS, Sternberg SS, Diaz B, Dickersin GR, Ewing S, Geller S, Kasimian D. The National Polyp Study. Patient and polyp characteristics associated with high-grade dysplasia in colorectal adenomas. Gastroenterology. 1990;98:371-379. [PubMed] |
| 28. | Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044-2058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 945] [Cited by in RCA: 857] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 29. | Nana-Sinkam SP, Fabbri M, Croce CM. MicroRNAs in cancer: personalizing diagnosis and therapy. Ann N Y Acad Sci. 2010;1210:25-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 30. | Hogan NM, Joyce MR, Kerin MJ. MicroRNA expression in colorectal cancer. Cancer Biomark. 2012;11:239-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 31. | Yamamichi N, Shimomura R, Inada K, Sakurai K, Haraguchi T, Ozaki Y, Fujita S, Mizutani T, Furukawa C, Fujishiro M. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin Cancer Res. 2009;15:4009-4016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 154] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 32. | Geiger TM, Ricciardi R. Screening options and recommendations for colorectal cancer. Clin Colon Rectal Surg. 2009;22:209-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 33. | Morikawa T, Kato J, Yamaji Y, Wada R, Mitsushima T, Shiratori Y. A comparison of the immunochemical fecal occult blood test and total colonoscopy in the asymptomatic population. Gastroenterology. 2005;129:422-428. [PubMed] |
| 34. | Imperiale TF, Ransohoff DF, Itzkowitz SH, Turnbull BA, Ross ME. Fecal DNA versus fecal occult blood for colorectal-cancer screening in an average-risk population. N Engl J Med. 2004;351:2704-2714. [PubMed] |
| 35. | Kim HJ, Yu MH, Kim H, Byun J, Lee C. Noninvasive molecular biomarkers for the detection of colorectal cancer. BMB Rep. 2008;41:685-692. [PubMed] |
| 36. | Rex DK, Rahmani EY, Haseman JH, Lemmel GT, Kaster S, Buckley JS. Relative sensitivity of colonoscopy and barium enema for detection of colorectal cancer in clinical practice. Gastroenterology. 1997;112:17-23. [PubMed] |
| 37. | Cotton PB, Durkalski VL, Pineau BC, Palesch YY, Mauldin PD, Hoffman B, Vining DJ, Small WC, Affronti J, Rex D. Computed tomographic colonography (virtual colonoscopy): a multicenter comparison with standard colonoscopy for detection of colorectal neoplasia. JAMA. 2004;291:1713-1719. [PubMed] |
| 38. | Ng EK, Chong WW, Jin H, Lam EK, Shin VY, Yu J, Poon TC, Ng SS, Sung JJ. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58:1375-1381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 855] [Cited by in RCA: 901] [Article Influence: 56.3] [Reference Citation Analysis (0)] |
| 39. | Schee K, Fodstad Ø, Flatmark K. MicroRNAs as biomarkers in colorectal cancer. Am J Pathol. 2010;177:1592-1599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 40. | Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, Starling N. Colorectal cancer. Lancet. 2010;375:1030-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1208] [Cited by in RCA: 1178] [Article Influence: 78.5] [Reference Citation Analysis (0)] |
| 41. | Locker GY, Hamilton S, Harris J, Jessup JM, Kemeny N, Macdonald JS, Somerfield MR, Hayes DF, Bast RC. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J Clin Oncol. 2006;24:5313-5327. [PubMed] |
| 42. | Bedeir A, Krasinskas AM. Molecular diagnostics of colorectal cancer. Arch Pathol Lab Med. 2011;135:578-587. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 43. | Nugent M, Miller N, Kerin MJ. MicroRNAs in colorectal cancer: function, dysregulation and potential as novel biomarkers. Eur J Surg Oncol. 2011;37:649-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 44. | Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 802] [Cited by in RCA: 862] [Article Influence: 50.7] [Reference Citation Analysis (0)] |
| 45. | Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, Nenutil R, Vyzula R. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72:397-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 543] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 46. | Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425-436. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1037] [Cited by in RCA: 1196] [Article Influence: 70.4] [Reference Citation Analysis (0)] |
| 47. | Nielsen BS, Jørgensen S, Fog JU, Søkilde R, Christensen IJ, Hansen U, Brünner N, Baker A, Møller S, Nielsen HJ. High levels of microRNA-21 in the stroma of colorectal cancers predict short disease-free survival in stage II colon cancer patients. Clin Exp Metastasis. 2011;28:27-38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 203] [Cited by in RCA: 226] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 48. | Wolpin BM, Mayer RJ. Systemic treatment of colorectal cancer. Gastroenterology. 2008;134:1296-1310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 385] [Cited by in RCA: 369] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 49. | Simunovic M, Smith AJ, Heald RJ. Rectal cancer surgery and regional lymph nodes. J Surg Oncol. 2009;99:256-259. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 50. | Urso ED, Nascimbeni R, Pucciarelli S, Agostini M, Casella C, Moneghini D, Di Lorenzo D, Maretto I, Sullivan M, Mammi I. Factors affecting the treatment of multiple colorectal adenomas. Surg Endosc. 2013;27:207-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 51. | Rossi L, Bonmassar E, Faraoni I. Modification of miR gene expression pattern in human colon cancer cells following exposure to 5-fluorouracil in vitro. Pharmacol Res. 2007;56:248-253. [PubMed] |
| 52. | Tomimaru Y, Eguchi H, Nagano H, Wada H, Tomokuni A, Kobayashi S, Marubashi S, Takeda Y, Tanemura M, Umeshita K. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-α/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer. 2010;103:1617-1626. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 150] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 53. | Svoboda M, Izakovicova Holla L, Sefr R, Vrtkova I, Kocakova I, Tichy B, Dvorak J. Micro-RNAs miR125b and miR137 are frequently upregulated in response to capecitabine chemoradiotherapy of rectal cancer. Int J Oncol. 2008;33:541-547. [PubMed] |
| 54. | Wong ST, Zhang XQ, Zhuang JT, Chan HL, Li CH, Leung GK. MicroRNA-21 inhibition enhances in vitro chemosensitivity of temozolomide-resistant glioblastoma cells. Anticancer Res. 2012;32:2835-2841. [PubMed] |
| 55. | Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799-2803. [PubMed] |
| 56. | Zhu H, Luo H, Li Y, Zhou Y, Jiang Y, Chai J, Xiao X, You Y, Zuo X. MicroRNA-21 in Scleroderma Fibrosis and its Function in TGF-β- Regulated Fibrosis-Related Genes Expression. J Clin Immunol. 2013;33:1100-1109. [PubMed] |
| 57. | Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369-5380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 663] [Cited by in RCA: 695] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 58. | Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685-689. [PubMed] |
| 59. | Wang V, Wu W. MicroRNA-based therapeutics for cancer. BioDrugs. 2009;23:15-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 128] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 60. | Fassan M, Croce CM, Rugge M. miRNAs in precancerous lesions of the gastrointestinal tract. World J Gastroenterol. 2011;17:5231-5239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 22] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 61. | Yantiss RK, Goodarzi M, Zhou XK, Rennert H, Pirog EC, Banner BF, Chen YT. Clinical, pathologic, and molecular features of early-onset colorectal carcinoma. Am J Surg Pathol. 2009;33:572-582. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 135] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 62. | Link A, Balaguer F, Shen Y, Nagasaka T, Lozano JJ, Boland CR, Goel A. Fecal MicroRNAs as novel biomarkers for colon cancer screening. Cancer Epidemiol Biomarkers Prev. 2010;19:1766-1774. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 240] [Cited by in RCA: 251] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 63. | Chang KH, Miller N, Kheirelseid EA, Lemetre C, Ball GR, Smith MJ, Regan M, McAnena OJ, Kerin MJ. MicroRNA signature analysis in colorectal cancer: identification of expression profiles in stage II tumors associated with aggressive disease. Int J Colorectal Dis. 2011;26:1415-1422. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 86] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 64. | Kulda V, Pesta M, Topolcan O, Liska V, Treska V, Sutnar A, Rupert K, Ludvikova M, Babuska V, Holubec L. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet. 2010;200:154-160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |