INTRODUCTION
Crohn’s disease (CD) and ulcerative colitis, the main clinical phenotypes of (idiopathic, relapsing-remitting) inflammatory bowel disease (IBD) are systemic disorders affecting the gastrointestinal-tract with frequent extraintestinal manifestations and associated autoimmune conditions[1]. IBD is considered as a polygenic immune disorder with complex multifactor etiology. Generally, IBD is arising in susceptible individuals in whom upon environmental triggers a sustained disturbed, deleterious mucosal immune reaction is provoked towards commensal microbiota[2]. In chronic inflammatory conditions, when organs with large epithelial surfaces are affected, like in IBD the epithelial barrier function is critical for the disease onset. Since the epithelium is densely inhabited by a resident microbial flora the role of native immunity is particularly appreciated in recognizing and distinguishing commensal enteric bacteria from the invading ones, and thus, in maintaining tolerance and homeostasis[2]. Subsequently, the chronic unrestrained inflammatory response that occurs in IBD is mainly driven by a disintegrated host immune regulatory network. In IBD development the host genetic susceptibility represents an important etiologic factor. In CD the genetic component is strongly indicated by familial aggregation, and further, by an approx. Twenty-six-fold greater population-based sibling risk, and an approximately 30%-35% of concordance rate in monozygotic twins[3,4]. The introduction of genome-wide association studies (GWAS) has yielded an expansion in studying the genetic basis of IBD. Nowadays more than 70 loci are associated with CD[5]. Further, in CD pathogenesis GWAS highlighted on certain earlier not really suspected biological pathways, such as autophagy. In polygenic diseases functional variants of single genes could be identified. Indeed, many of the recently identified genetic risk loci in CD are related to various cell types and pathways, suggesting the involvement of fairly different aspects of host immune responses in the IBD phenotype. Missing heritability in CD cannot be simply explained by genetic alterations[2]. Moreover, the fact of the worldwide considerable increase in disease incidence and prevalance emphasizes the importance of additional, environmental and epigenetic contributions[6,7]. The interplay of genes regulating immune functions is strongly affected by the environment, especially gut resident microbiota. On the basis of genetic alterations in CD impaired sensing and handling of intracellular bacteria by the innate immunity, that is closely interrelated with the autophagic and unfolded protein pathways seem to be the most relevant pathophysiologic features[8].
AUTOPHAGY MACHINERY
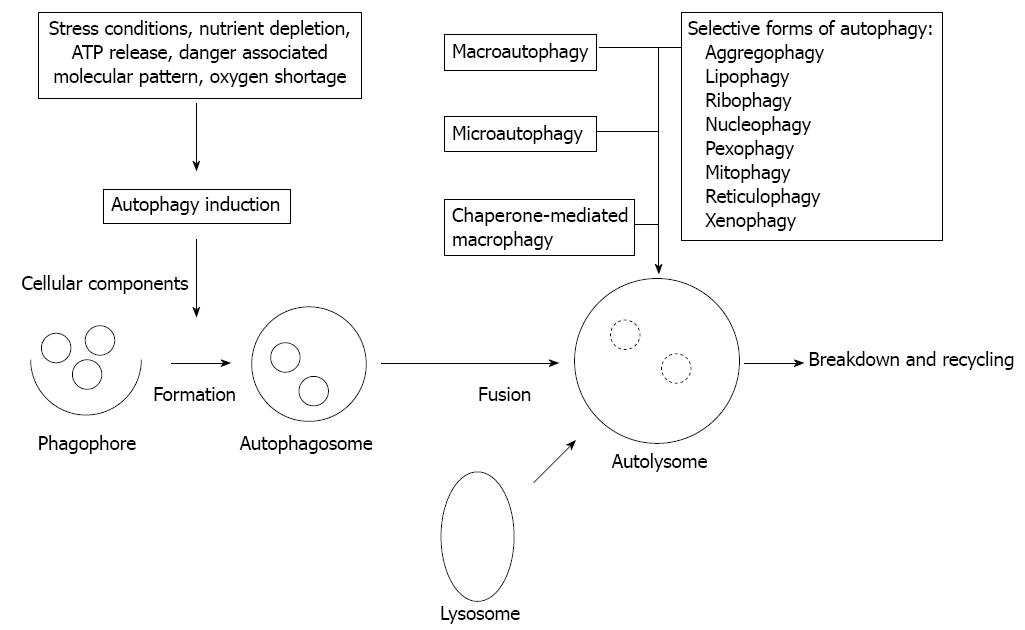
Besides the proteasomal degradation pathway autophagy represents an additional, evolutionarily highly conserved multi-step process of cellular self-digestion due to sequestration of excessive, damaged, or aged proteins and intracellular organelles in double-membranous vesicles of autophagosomes, terminally self-digested in lysosomes[9]. Autophagy is deeply implicated in the regulation of numerous physiologic functions including cell development and differentiation, survival and senescence, and it also affects fundamentally the inflammatory process, and the innate and adaptive arms of immune responses[10]. On a basal level intact autophagy serves constantly and constitutively as a critical adaptive and surveillance mechanism in maintaining cellular homeostasis[11]. Nevertheless, autophagy is inducible in response to different cellular metabolic stress conditions, such as nutrient and growth factor deprivation in order to preserve cell viability. Further, autophagy is upregulated in cases of protein aggregation and accumulation of misfolded proteins, i.e., when the structural remodeling is mandatory. In respect of innate immunity, however, autophagy plays an essential role during infections by degrading intracellular pathogens[10,12]. Different types of autophagy according to the route of delivery to lysosomes and the main physiological functions have been characterized, such as macro- and micro-autophagy, and chaperon-mediated autophagy[11]. Upon specific targeted degradation of cytosolic aggregated proteins, lipids, and organelles (ribosomes, nucleosomes, peroxisomes, mitochondria, endoplasmic reticulum), selective forms of autophagy (aggregophagy, lipophagy, ribophagy, nucleophagy, pexophagy, mitophagy, reticulophagy) can further be classified[9,11]. In addition, elimination of intracellularly infective pathogens represents another selective form of autophagy, namely xenophagy (Figure 1). Xenophagy can be considered as a substantial element of the innate immune system. Generally (macro)autophagy refers to cytoplasmatic bulk, non-selective degradation of subcellular constituents. Within this complex catabolic pathway tightly regulated by a limited number of autophagy genes (ATGs) various morphologic stages of the assembly process are distinguishable[12]. It is initiated with the phagophore (isolation membrane) formation around different molecules or particles to be sequestred, and is followed by elongation and maturation into the autophagosome, leading finally to fusion with lysosomes[9,12]. Subsequently, the phagophore is controlled by Beclin1 (ATG6) and ATG14 genes, and both the inhibitory class1 canonical phosphatidylinositol 3’-kinase/AKT (PI3K/AKT) mammalian target of rapamycin (mTOR) and the promoting c-Jun N-terminal kinases (JNK1) pathways[13,14]. The intricate formation of autophagosome is regulated mainly by the ATG5-ATG12 complex, then stabilized by ATG16L1, and further processed by microtubule-associated protein light chain (LC3/ATG8) under the strict control of ubiquitin-like conjugation systems (ATG10, ATG7, ATG3). The engulfment of random or selective cargo, closure of the autophagosome, and fusion with the lysosomal compartment is orchestrated by LC3, and the Beclin1-UV-irradiation resistance-associated gene (UVRAG) complex[13,14]. Defects in basal autophagy may yield accumulation of cytotoxic materials, damaged DNA, and thus, genomic instability, while alterations of induced autophagy especially lead to reduced cell survival[10,12]. By compromising cellular fitness defective autophagy has been ultimately related to several chronic inflammatory disease conditions, such as inflammatory bowel disease, like CD and cancer, neurodegeneration, and infectious disorders[10,11,15]. Generally autophagy deficiency is closely related to accelerated tumorigenesis. In autophagy-incompetent cells upon induced oxidative stress cell-autonomous mechanisms are exhibited in forms of accumulated DNA damage and chromatin instability[16]. However, inflammatory events as a non-cell-autonomous mechanism along with defective apoptosis could independently contribute to malignant transformation and cancer progression, partly by favoring cell necrosis[17]. Similar situation has been found in human IBD with high risk of malignancy, and in experimental cases of Atg5-/- or Atg7-/- mice displaying abnormalities resembling human IBD[18]. Autophagy and stress-responsive cellular degradation pathways of intrinsic and extrinsic apoptosis can fundamentally alter, activate or inhibit each other via an extensive molecular crosstalk, and in fact, cell destiny is determined by their actual functional status and interplay[19]. Their crosstalk is primarily regulated by the current status of the ATG6/Beclin-1 complex, a Bcl-2/Bcl-xL interacting element, since Bcl2 is a potent autophagy inhibitor. Dissociation of this complex can be achieved by toll-like receptor (TLR) adaptors (MyD88, TRIF), or activation of mitogen activated phosphokinase (MAPK)-JNK cascade, as well as by translocation of the damage-associated molecular pattern (DAMP) protein high-mobility-group B (HMGB)-1[13,19]. There is also diverse interaction between autophagy and the nuclear factor-κB (NF-κB) signaling pathways through positive and negative feedback regulatory loops[14]. The tumor suppressor p53 gene exerts a typical dual role in autophagy regulation, depending primarily on its subcellular, nuclear or cytoplasmic distribution[13,14].
Figure 1 The autophagic process and types of autophagy.
NOD-LIKE RECEPTORS AND CROHN'S DISEASE
NOD-like receptors (NLRs) are pattern recognition receptors (PRRs) and belong to the family of innate immune receptors sensing pathogen-associated molecular patterns (PAMPs). Nucleotide-binding oligomerization domain-containing protein 2 (NOD2) is constitutively expressed intracellularly in macrophages and dendritic cells, and to lesser extent in intestinal epithelial cells and T cells. The centrally located motifs of NLRs are referred to NOD domains that are interacted with the caspase activation and recruitment domain ones. NOD2 recognizes N-acetyl--muramyl-peptide (MDP), a bacterial peptidoglycan component, and upon activation the induced receptorial conformation changes result a multiprotein, the inflammasome (NLRP3). Ligation of NOD2 triggers recruiment of the adaptor protein receptor interacting protein 2 (RIP2) causing a TRAF6-mediated ubiquitination of inhibitor of κB-kinase gamma (IKKγ; NEMO), and hence results in activation of downstream signaling pathways implicating NF-κB, MAPKs and proinflammatory caspases[20,21]. The Crohn’s disease-associated NOD2 genetic variants are located in the leucine-rich repeat (LRR) region of NOD2, i.e., in the ligand-binding domain of this intracellular PRR[22]. The altered amino acid sequence is related either to insertion resulting in a frame-shift mutation, or to non-synonymous SNPs resulting in amino acid exchanges. The more commonly observed genetic variants (of missense or nonsense mutations) in CD are the SNP8 (R702W), SNP12 (G908R), and SNP13 (L1007fsC), respectively, however a number of rare NOD2 variants have also been discovered, being localized again almost exclusively to the LRR region[22,23]. Upon MDP ligation the Crohn’s disease-associated “loss-of-function” NOD2 variants abrogate RIP2 binding, and so fail to activate NF-κB[24,25]. Further, NOD2 is involved in the modulation of TLR signaling, as well. Thus, in case of Crohn’s disease-related gene polymorphisms the TLR2-induced NF-κB activation is also decreased[26,27]. However, yet it is difficult to correctly interpret the real functional consequences of given mutations since they may activate additional, compensatory mechanisms resulting in a definite inflammatory phenotype. On the other hand NOD2 has a pivotal role in direct antibacterial defense by the induced release of defensins. NOD2-/- mice and patients with the CD NOD2 variants display diminished expression of antimicrobial α-defensins in Paneth cells, that contributes to impaired antibacterial capacity and decreased epithelial barrier function[28,29]. In contrast to hypomorphic functions the frame-shift gene mutation variant encodes a “gain-of-function” by actively suppressing interleukin-10 (IL-10) transcription[30].
AUTOPHAGY AND CROHN'S DISEASE
The autophagy machinery in IBD represents a recently developed pathway fundamentally contributing to the pathogenesis[10]. Functional polymorphisms of the autophagy genes ATG16L1 (T300A) and immunity-related GTPase family M protein (IRGM; C313T) have been found as definite risk factors for CD[31-34]. The ATG16L1 protein is widely expressed in intestinal epithelial cells, and also in macrophages and lymphocytes. The ubiquitous ATG16L1 seems to be fundamental in selective autophagy, i.e., in xenophagy, nonetheless its defect has only been described within the gut[35].
In CD patients homozygous for the risk ATG16L1 allele the “loss-of-function” deficiency due to failures of autophagosome formation results in impaired engulfment and degradation of cytoplasmic content (microbes), defective presentation of bacterial antigens to CD4+ T cells, and further, in alterations of Paneth cell granule formation causing a disrupted granule exocytosis[18,36-38]. Additionally, ATG16L1-deficient Paneth cells in CD display a “gain-of-function” defect by increasing expression of inflammatory cytokines[18,38]. Moreover, upon stimulation with NOD2 ligands or with lipopolysaccharides (LPS) through TLR4, macrophages and myeloid cells with the ATG16L1 risk variant generate high levels of reactive oxygen species (ROS), and respond with inflammasome overactivation leading to enhanced IL-1β and IL-18 production via Myd88 and TRIF-dependent activation of caspase-1[36-38]. Generally, aberrant activation of PRR signaling pathways may result critically severe inflammation. IRGM is the only human gene representative for innate immunity-related GTPases, necessary for γ-interferon-mediated resistance to intracellular pathogens[39]. During initiation of autophagy IRGM expression is essentially required for the proper clearance of bacteria. The risk polymorphism of IRGM due to the impaired protein expression can lead to functional abnormalities in xenophagy[32,33]. Since IRGM is possibly regulated in a cell specific manner the CD risk allele may cause cell specific phenotypes.
NOD2 and autophagy
Functionally NOD2 is closely associated with autophagy, and yet interacts mechanically (i.e., immunoprecipitated) with ATG16L1, therefore autophagy seems to be a key factor in CD[37,40,41]. Autophagy is mainly activated due to sensors of the innate immunity, i.e., by PRR signaling upon recognition of PAMPs (MDP, LPS, ss/ds RNA, methylated DNA/CpG), but it could also be induced by DAMPs (like ATP, ROS, and misfolded proteins), pathogen receptors (like CD46), IKK, JNK and HMGB proteins[10,12,19,38]. Sensory PRR-molecules include TLRs, NLRs and RIG-I-like receptors (RLRs). Induction of NOD2 in dendritic and epithelial cells by bacterial ligands and leaving bacteria results in ATG16L1-dependent formation of autophagic vacuoles. However, the NOD2 variants of CD lack this activity, and further MDP-induced autophagy is also absent in cells with the ATG16L1 risk variant, suggesting that both NOD2 and ATG16L1 co-localized on plasma membrane are required for an optimal innate immune signaling[40-42]. In addition, a NOD2-dependent failure in autophagy-induction and consequently a diminished bacterial killing was found for Salmonella typhimurium, Shigella flexneri, and enteroadherent invasive Escherichia coli (E. coli)(AIEC)[40,41,43]. The normal NOD2, but not the CD-associated variants recruits ATG16L1 to the plasma membrane preferentially at the bacterial entry side, so physiologically NOD2 is critical for engulfing invading pathogens by autophagosomes[41,42]. Furthermore, in dendritic cells NOD2-dependent autophagy is also essential for the appropriate antigen processing and presentation and a subsequent induction of CD4+ T-cells[41]. Dendritic cells from CD patients with either NOD2 or ATG16L1 variants display a failure to translocate bacteria to lysosomes and relocate MHC II to cell surface, as well[40]. When the disease-associated ATG16L1 and NOD2 alleles are present in combination, a synergistic genetic epistasis, i.e., an increase in CD susceptibility was observed, underscoring the importance of a signaling crosstalk regarding the inflammasome and autophagy[44].
Endoplasmic reticulum stress and autophagy
The unfolded protein response (UPR) induced by endoplasmatic reticulum (ER) stress represents another pathway in IBD pathophysiology[45]. Genetically ER stress is associated with both forms of IBD and occurs upon excessive accumulation of misfolded or unfolded proteins in the ER, leading to UPR especially in cells with high secretory capacity, like goblet cells and Paneth cells[35,46]. UPR is regulated by different pathways (and related transcription factors) with the preference of the inositol-requiring enzyme 1/X-box binding protein 1 (IRE1/XBP1) axis[47]. Via this axis there is a conserved link between innate immunity (TLR and NOD signaling) and the UPR[47]. GWAS-based candidate gene studies revealed the role of XBP1 SNPs in IBD-related ER-stress[48,49]. Decreased or absent XBP1 function in the intestinal epithelial cell (IEC) compartment through IRE1 hyperactivation results in uncontrolled ER-stress, i.e., a proinflammatory overactivation, and further in dysfunction and premature apoptotic depletion of Paneth cells, with the consequent impaired handling of the microbiota[49]. Under ER stress autophagy is induced via JNK (dowstream of IRE1), which is overactivated by the hypomorphic XBP1[50,51]. However, even defective autophagy per se is able to provoke ER stress, especially when the ATG7 protein involved in regulation of autophagosome formation is also depressed[52]. Regarding PI3K there is an antagonistic action, since in UPR it is responsible for the activation of XBP1, but in the contrary autophagy is suppressed by the canonical AKT-TOR pathway[53,54]. In IBD, IECs presumably are affected both by impaired UPR signaling and aberrant autophagy, but their exact interplay needs to be further clarified.
GUT MICROBIOTA AND CROHN'S DISEASE
The intestinal microbiota, which normally colonize mucosal surfaces in symbiotic mutualism with the host is unique and quite stable over time[55]. The basic challenge for the intestinal immune recognition is the requirement of a simultaneous delicate balance between tolerance and responsiveness towards microbes[56]. Several data suggest the existence of immune tolerance to antigens of the individual own bacterial flora, whereas its breakdown definitely contributes to IBD pathogenesis[37,57]. In CD there is a profound and complex host defect in sensing and responding intestinal (lumenal and mucosal) microbiome. Accordingly, reprogramming in the microbial composition, i.e., a significant decreased load of commensal, protective resident bacteria (like Bifidobacteria, Lactobacilli and Firmicutes) along with the impaired immunity against the putative pathogenic (harmful) ones (such as Bacteriodetes, and Proteobacteria, including E. coli) provoke a deleterious inflammatory condition, corresponding to CD[58]. The exact nature of the distinct mucosal flora (dysbiosis), however has not yet been fully elicited. Specific strains of E. coli (termed AIEC) in CD affect especially the epithelial layer with the ability to adhere, invade and replicate in IECs, and further, a subpopulation even resides and survives within macrophages, and thereby induces increased production of tumor necrosis factor-α[43,59]. ATG16L1 and IRGM-deficient autophagosomes promote the AIEC survival as well[43]. Moreover, in the presence of CD-associated NOD2 variants or hypomorphic XBP1 dendritic cells exhibit diminished intracellular bacterial killing[41]. It is hypothesized, that AIEC possesses the capacity to circumvent innate immune responses leading to activation of NF-κB[60]. Thus, regarding the host interactions with microbes genetic risk factors of CD functionally render pathways of the innate immunity to converge to a deeply impaired autophagic process.
CONCLUSION
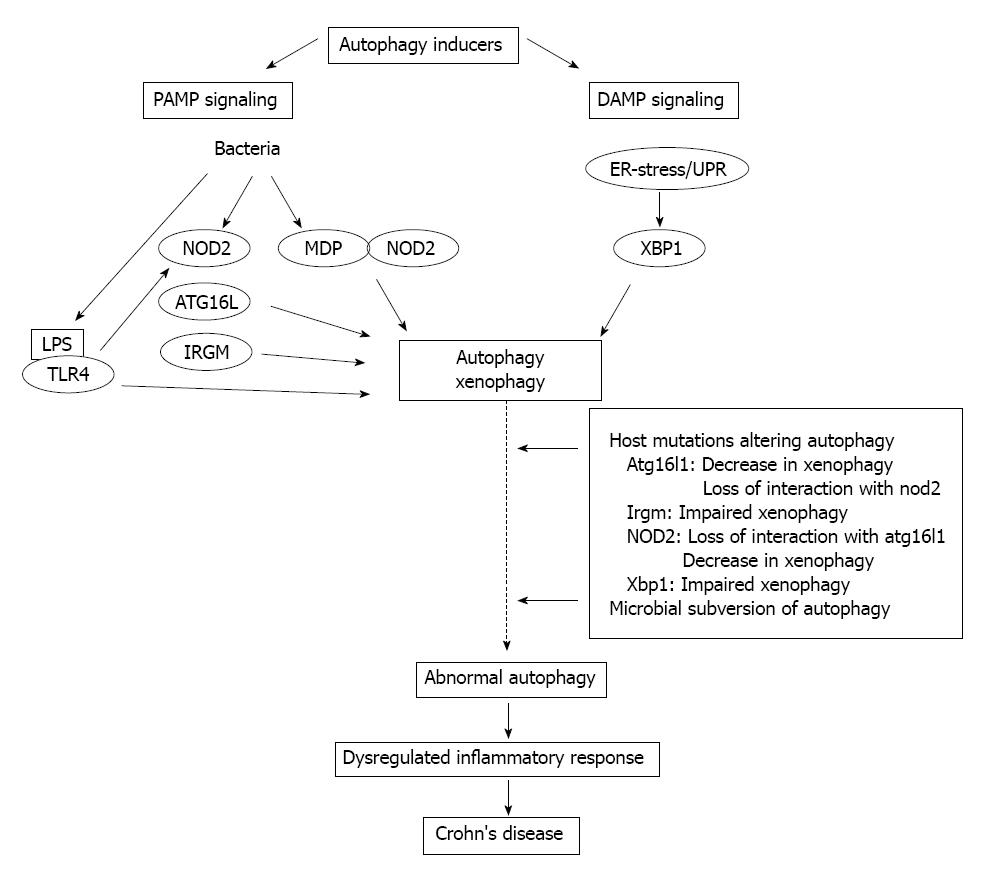
Overall, there is no doubt that autophagy can be considered as an apparently difficult regulatory network, being in close connection with several signal transduction pathways and cellular programs. Principle elements of immunological autophagy include the direct cell-autonomous pathogen elimination, the regulation of PRRs, and inflammasome activation, and the cytoplasmic antigen processing for MHC presentation to T cells. Recently significant advances have been achieved in understanding the importance of autophagy in CD, which previously had not been implicated in IBD pathology. In CD functional consequences of the underlying autophagy-related gene defects (ATG16L1, IRGM, NOD2, XBP1), in particular the inappropriate stimulation of antimicrobial and inflammasome pathways eventually result in uncontrolled inflammation (Figure 2). Therefore, autophagy in CD is predicted as a key regulator mechanism with the capacity to integrate several aspects of disease pathogenesis. Theoretically the complex autophagy signaling in CD offers a promising novel therapeutic target, since due to its induction potentially not only the load of cytoinvasive bacteria, and the perturbed immune responses, but the resulting inflammatory process, as well may simultaneously be reduced. Thus, autophagy boosting would represent an efficient biologic manipulation, and could provide an alternative therapeutic option. Several candidate pathways, e.g., inhibition of mTOR, decrease of ER-stress, lowering of inositol triphosphate (IT3), etc., could be considered. On the other hand, however, much cautiousness is required regarding its pleiotropic physiological repertoire, since pharmacologic autophagy modulation can initiate additional biologic effects not expected in CD. Further detailed functional analyses of the Crohn’s disease-associated genetic polymorphisms are needed to explore and define more precisely the subcellular and molecular basis of the crosstalk between autophagy and the innate immune axis, hopefully allowing the introduction of selective new therapeutic approaches into daily practice.
Figure 2 Schematic illustration of the crosstalk between autophagy and innate immunity in Crohn’s disease.
PAMP: Pathogen-associated molecular patterns; DAMP: Damage-associated molecular pattern; ER: Endoplasmic reticulum; UPR: Unfolded protein response; NOD: Nucleotide-binding oligomerization domain-containing protein; MDP: N-acetyl-muramyl-peptide; LPS: Lipopolysaccharide; TLR: Toll-like receptor; XBP1: X-box binding protein 1; IRGM: Immunity-related GTPase family M protein.
P- Reviewer Tommasini A S- Editor Gou SX L- Editor A E- Editor Zhang DN