Published online Jun 14, 2013. doi: 10.3748/wjg.v19.i22.3423
Revised: March 21, 2013
Accepted: April 3, 2013
Published online: June 14, 2013
Processing time: 172 Days and 12.6 Hours
AIM: To select characteristic endogenous metabolites in hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) patients and to identify their molecular mechanism and potential clinical value.
METHODS: An ultra performance liquid chromatography and linear trap quadrupole-Orbitrap XL-mass spectrometry platform was used to analyze endogenous metabolites in the homogenate of central tumor tissue, adjacent tissue and distant tissue obtained from 10 HBV-related HCC patients. After pretreatment with Mzmine software, including peak detection, alignment and normalization, the acquired data were treated with Simca-P+software to establish multivariate statistical analysis based on a pattern recognition technique and characteristic metabolites highly correlated with changing trends in metabolic profiling were selected and further identified.
RESULTS: Based on data acquired using Mzmine software, a principal component analysis model (R2X = 66.9%, Q2 = 21.7%) with 6 principal components and an orthogonal partial least squares discriminant analysis model (R2X = 76.5%, R2Y = 93.7%, Q2 = 68.7%) with 2 predicted principal components and 5 orthogonal principal components were established in the three tissue groups. Forty-nine ions were selected, 33 ions passed the 2 related samples nonparametric test (P < 0.05) and 14 of these were further identified as characteristic metabolites that showed significant differences in levels between the central tumor tissue group and distant tumor tissue group, including 9 metabolites (L-phenylalanine, glycerophosphocholine, lysophosphatidylcholines, lysophosphatidylethanolamines and chenodeoxycholic acid glycine conjugate) which had been reported as serum metabolite biomarkers for HCC diagnosis in previous research, and 5 metabolites (beta-sitosterol, quinaldic acid, arachidyl carnitine, tetradecanal, and oleamide) which had not been reported before.
CONCLUSION: Characteristic metabolites and metabolic pathways highly related to HCC pathogenesis and progression are identified through metabolic profiling analysis of HCC tissue homogenates.
Core tip: An ultra performance liquid chromatography-mass spectrometry platform was used in the present study to identify characteristic metabolites in hepatitis B virus-related hepatocellular carcinoma tumor tissues. From an orthogonal partial least squares discriminant analysis model established to determine metabolic profiling in the central tumor tissue group, adjacent tissue group and distant tissue group, 49 ions were selected and 14 of these were identified as characteristic metabolites. The detection of these metabolites in tumor tissue not only confirmed the targeted traceability of previously reported serum biomarkers related to cancer diagnosis, but also provided novel targets for anticancer research.
- Citation: Liu SY, Zhang RL, Kang H, Fan ZJ, Du Z. Human liver tissue metabolic profiling research on hepatitis B virus-related hepatocellular carcinoma. World J Gastroenterol 2013; 19(22): 3423-3432
- URL: https://www.wjgnet.com/1007-9327/full/v19/i22/3423.htm
- DOI: https://dx.doi.org/10.3748/wjg.v19.i22.3423
Hepatocellular carcinoma (HCC) is currently an important research topic in basic and clinical investigations due to its high malignancy, fast development, high mortality, complicated pathogenesis and significant individual differences. There have been a number of previous investigations on the molecular biology and proteomics of HCC[1-3]. However, the occurrence and development of this disease are not simply determined by innate genetic differences. The introduction of metabolomics has provided more information in HCC investigations. Metabolomics is the study of biological systems where changes in metabolites after specific stimulation or interference are determined[4-7]. This type of study focuses on the end products of biological systems, which are a reflection of both the physiological and biochemical status, and are affected by both genotype and environment and are close to biological characteristics. Thus it has the advantage of investigating the objective basis and generative mechanism of individual differences in metabolites.
Current metabolomic investigations in HCC are mainly focused on identifying characteristic metabolites in serum or urine, revealing changes in the metabolic network, and finally selecting potential biomarkers with clinical application and exploring pathologic mechanisms[8,9]. For example, Wu et al[10] used gas chromatography/mass spectrometry (MS) to analyze metabolic profiling in 20 male HCC patients and 20 healthy male volunteers. One hundred and three ions were detected and 66 ions were identified. Following t test analysis, 18 metabolites were significantly different between the central tumor tissue group and the distant tumor tissue group (P < 0.05). The ability to distinguish these 18 ions combined with alpha-fetoprotein was analyzed by the receiver operating characteristic curve (0.9275). This non-invasive method was considered quite promising. Tan et al[11] also used metabolic analytical methods to select serum biomarkers for small HCC diagnosis. Metabolic profiling was performed in a diethylnitrosamine-induced rat HCC model, which is similar to human HCC in the histopathology of liver disease development. The alterations in three metabolites, taurocholic acid, lysophosphoethanolamine 16:0 and lysophosphatidylcholine 22:5, were reported to be correlated with disease progression and considered as potential biomarkers. In addition, serum metabolic profiling was performed in 262 HCC patients, 76 liver cirrhosis patients and 74 HBV patients. The results showed that the sensitivity and specificity of these ions all reached 80%, which was better than alpha-fetoprotein in distinguishing HCC-liver cirrhosis. This research laid the foundation for the clinical application of metabolic biomarkers. At the same time, Soga et al[12] research group reported that gamma-glutamyl dipeptides were used as biomarkers for liver disease diagnosis. The metabolic profiling of 248 serum samples, including patients with 9 types of liver disease, was performed using a capillary electrophoresis-MS platform. Gamma-glutamyl dipeptides were selected and found to be predictive of a decrease in glutathione. Multiple regression analysis showed that the selected biomarkers had the ability to distinguish different liver diseases.
In animal models, serum and urine analysis and sample collection are easy to perform, and HCC serum and urine metabolic profiling can reflect specific signs and reveal pathological mechanisms, however, the targeting ability of these analyses is still limited. The present study used a ultra performance liquid chromatography and linear trap quadrupole (UPLC-LTQ)-Orbitrap XL MS analytical platform to perform metabolic profiling analyses on homogenates of hepatitis B virus (HBV)-related HCC tumors removed by surgery. A metabolic profiling model was established and metabolic pathways highly related to HCC were characterized for further investigations on the genesis and progression of HCC and the potential clinical value of characteristic metabolites.
All solvents were high performance liquid chromatography (HPLC) grade and used without modification. Formic acid and acetonitrile (ACN) were obtained from Merck (KGaA Merck, Germany). Distilled water was produced using a Milli-Q Reagent Water System (Millipore, Billerica, MA, United States). All standard [L-phenylalanine, glycerophosphocholine, chenodeoxycholic acid glycine conjugate and lysophosphatidylcholine (lysoPC) (18:0)] preparations were purchased from Sigma-Aldrich (St. Louis, MO, United States). Ultra performance liquid chromatography was performed on a Thermo Fisher Accela system (Thermo Fisher Scientific, Franklin, MA, United States). MS was performed on a Thermo Fisher (Thermo Fisher Scientific, Franklin, MA, United States) LTQ Orbitrap XL hybrid mass spectrometer. Other equipment included a Multifuge X1R high-speed centrifuge (Thermo Fisher Scientific, United States).
The 10 HBV-related HCC patients included in this study were all from the Department of Hepatobiliary Surgery, Tianjin Third Central Hospital, and included 5 females and 5 males with average age of 54.2 ± 9.2 years. All patients voluntarily joined this study and gave informed consent. Tissue from the central area of the tumor, adjacent tissue (1-2 cm from the tumor) and distant tissue (5 cm from the tumor) were collected. Thirty samples (10 samples in each group) were washed in hypothermic normal saline, dried using neutral filter paper and then stored at -80 °C.
Collection of samples was performed according to the following instructions: (1) all 10 patients had chronic persistent HBV (no other hepatotropic virus infection, no long-term drinking behavior, no autoimmune hepatitis, no schistosomiasis infection and no genetic metabolic liver disease); (2) all 10 patients had solitary tumors < 5 cm in diameter, and Barcelona Clinic Liver Cancer stage A1; (3) tissue pathological immunohistochemistry examination diagnosed HCC using hepatitis B surface antigen (+); the pathological section is shown in Figure 1. Samples were all HCC grade II-III differentiation. Invasive growth was detected using the appropriate specific stain. No cancer cells were detected at the incisional edge; (4) no radiotherapy or chemotherapy was performed before surgery; (5) no evidence of endocrine or metabolic disease; (6) renal function, liver function, blood routine and hydropower solution pH were all in the normal range; (7) no severe infection was detected and parenteral nutrition was used; and (8) patients' dietary requirements were managed by the Nutrition Department of Tianjin Third Central Hospital to a relatively uniform standard, as a result, exogenous dietary influence on metabolic profiling was limited to the lowest level.
Samples were thawed at room temperature and then weighed, 1:3 ultrapure water was added to each sample according to their weight before being homogenized. Tissue homogenate was subjected to ultrasonication at 130 W × 60% for 3 cycles. One cycle included 10 s of ultrasonication and a 10 s interval (ultrasonication was performed in an ice-bath). Homogenate (400 μL) was centrifuged (4 °C, 15000 g, 30 min). Supernatant (100 μL) was mixed with 400 μL methanol to precipitate proteins. The mixture was centrifuged at 4 °C, 15000 g for 30 min. The supernatant was filtered using a 0.22 μm membrane. Quality control (QC) was the equivalent volume mixture in each sample.
Chromatography conditions were as follows: Chromatography was performed on a Thermo Fisher Accela system (Thermo Fisher Scientific, Franklin, MA, United States) which was equipped with a binary solvent delivery manager, and a sample manager. The analytical column was a Thermo Hypersil GOLD (2.1 mm id× 50 mm 1.9 μm) C18 reversed phase column.
Mobile phase: Phase A: 0.1% formic acid (volume ratio), 1 mL of formic acid was added to a 1 L bottle of HPLC-grade water; Phase B: 95% ACN and 0.1% formic acid. And 950 mL ACN, 50 mL HPLC-grade water, and 1 mL formic acid were combined.
Chromatographic separation: Chromatographic separation was performed isocratically within 15 min and the injection volume was 10 μL. The flow rate was set at 200 μL/min. The sample manager and column oven temperature were set at 4 °C and 20 °C, respectively. The chromatographic elution gradient was initialized at 5% Phase B and held for 3 min. In consecutive 10 min periods, Phase B was gradually escalated to 50%, and then a rapid increase in Phase B to 95% was completed within 3 min. After 4 min of maintaining the high volume of organic phase gradient, Phase B was immediately reduced to 5% and this elution gradient was used to balance the analytical column for the final 4 min.
MS was performed on a Thermo Fisher (Thermo Fisher Scientific, Franklin, MA, United States) LTQ Orbitrap XL hybrid mass spectrometer[13], operating in the positive ion mode with an ion source voltage of 4.5 kV, a capillary voltage of 30 V, cone voltage of 150 V, desolvation temperature of 275 °C, sheath gas flow of 30 arb and assistant gas flow of 5 arb (99.999% nitrogen). Data were collected over 15 min in centroid mode over the mass range 50-1000 m/z. The MS resolution was at 100000 full width half maximum (FMHW) and the calibration standards were provided by Thermo Fisher Scientific (caffeine, Ultramark 1621 and L-methionyl-arginyol-phenylalanylalanine acetate H2O). MS/MS analysis was carried out with collision-induced dissociation (CID) collision energy 35 (normalization collision energy) and the collision gas was 99.999% helium.
There were 14 QC samples throughout the test (equal volume mixture of each analyzed sample). Before the samples were tested, 8 QC samples were analyzed continuously and the remaining QC samples were inserted into the sequence after every 5 samples were analyzed.
The sequence of samples was randomly generated by the excel function before and after sample analysis (including QC), and cross-contamination was avoided by inserting a blank between adjacent samples. The whole experiment lasted 780 min.
All samples were collected in accordance with the ethical guidelines and written consent protocols mandated by the Tianjin Third Central Hospital. The Institutional Review Board approved the collection of serum for comprehensive metabolite characterization. All patients and all control individuals were approached using approved ethical guidelines and those who agreed to participate in this study, were required to sign consent forms. Patients could refuse entry, discontinue participation, or withdraw from the study at any time without prejudice for further treatment or management. All participants provided written consent.
MZmine 2.0 was used for peak detection, alignment and normalization. The filter conditions were: chromatography peak intensity signal/noise > 30, retention time tolerance = ± 0.1 min, and m/z tolerance = ± 0.01.
One of the aims of this study was to establish a model to predict the tissue metabolic profile in HCC. With the help of Simca-P+12.0.1.0 (Umetrics, Sweden), software based on chemometrics methods, the OPLS-DA supervised model was established. The detailed process was as follows: The variables were firstly traded by Pareto scaling. The principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) model of all the samples were established and checked by cross validation[14,15]. The preliminary selection of characteristic metabolites was accomplished using the corresponding variable importance (VIP) value, confidence interval and coefficient plot generated by the OPLS-DA model. The selected metabolites were then preliminarily confirmed by the S and SUS diagram. Finally, the variances were evaluated by SPSS 16.0 software (SPSS, United States), using the 2 related samples nonparametric test.
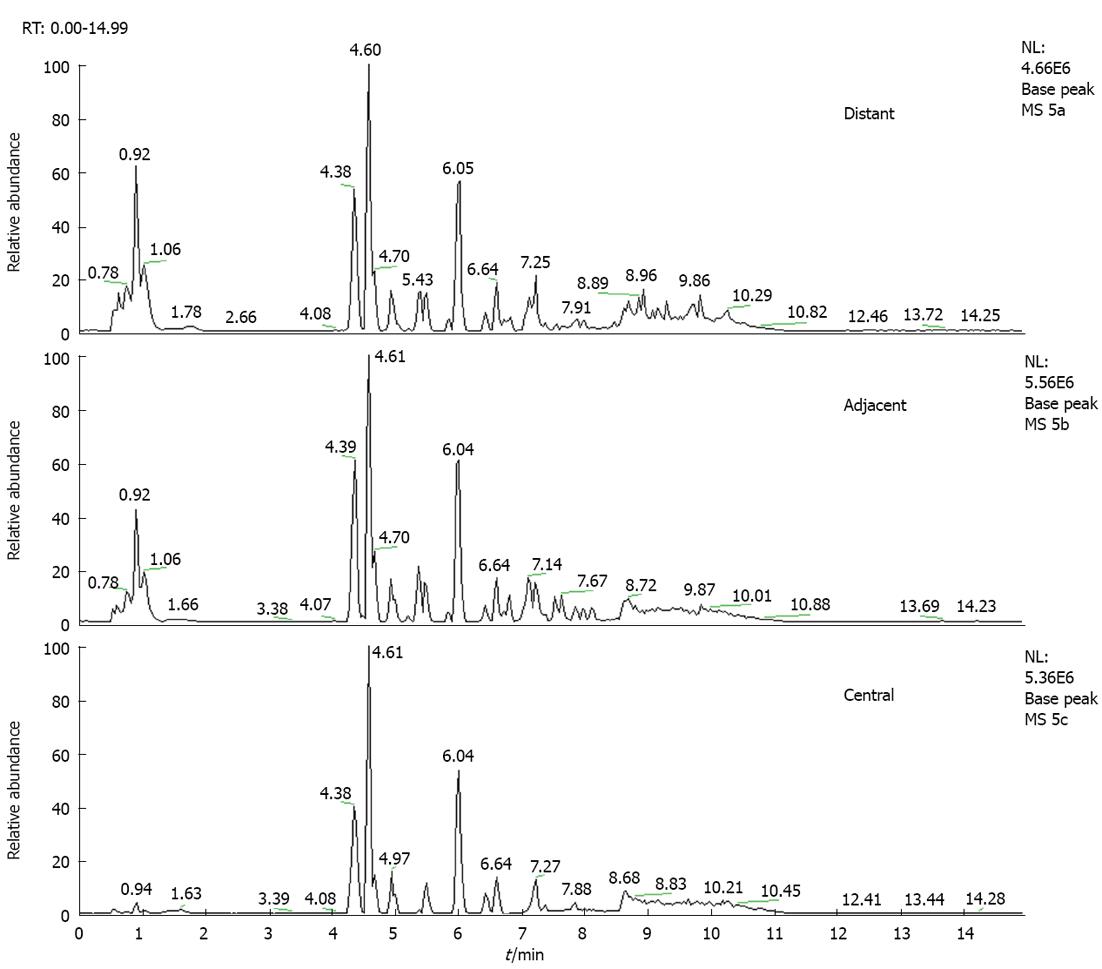
The total ion chromatogram of the central tumor tissue group, adjacent tissue group and distant tissue group acquired by the UPLC-MS platform are shown in Figure 2. After pretreatment and standardization using MZmine 2.0, there were 211 integral peaks following extraction ion chromatography detected in QC samples and 215 peaks in test samples.
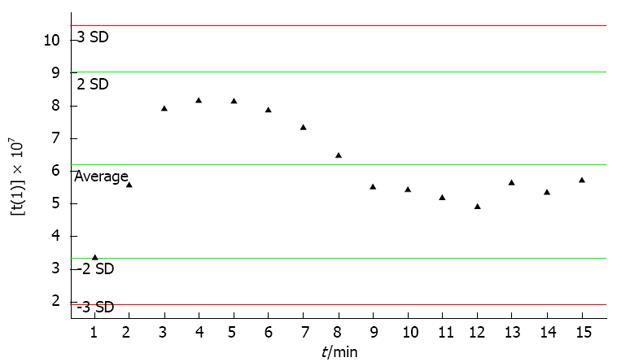
The stability of the UPLC-MS system was adequately assessed by the analysis of QC samples during the entire experimental period[16]. Through PCA of 14 data sets of QC samples, a PCA model with 2 principal components was established (Figure 3). Figure 3 shows the score plot of QC sample sequence versus first principal component (the most influential factor which varied with time). From the QC principal component score plot we found that the UPLS-MS system was stable after the first 8 continuous QC injections. In the test sample sequence, a QC sample was inserted after every 5 test samples to evaluate the stability of the system during the entire analytical process. The results showed that the detection system was stable throughout the experiment after the first 8 QC samples were injected (no outliers exceeding ± 2 SD were detected in the QC samples). According to a related article[17], the QC standard was set as follows: (1) ion peaks were defined as reliable peaks when their intensity was in the range of ± 30% average ion intensity; (2) a QC sample was qualified if its 70% ion peaks were reliable; and (3) experimental data were accepted only when 60% QC was qualified. In the present experiment, 11 of 14 QC samples (reliable ion peaks distributed among 70.5%-86.3%) inserted into the test sample sequence qualified and the qualified ratio was 78.5%, which meant that the analytical results were trusted.
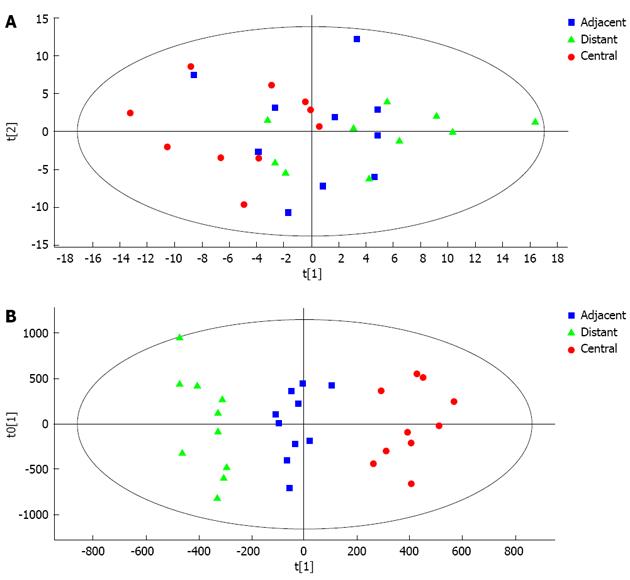
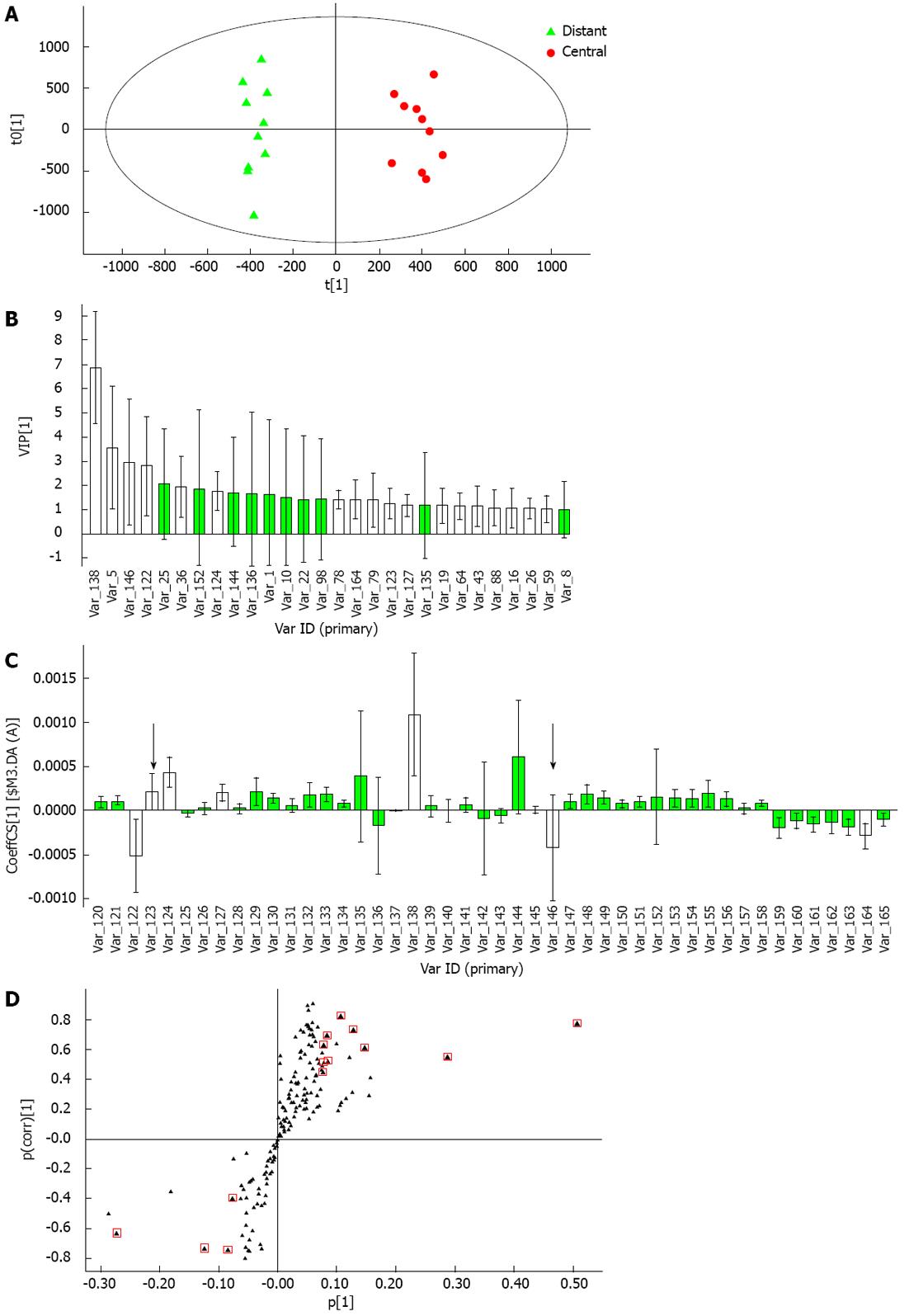
A PCA model with 6 principal components was established (R2X = 66.9%, Q2 = 21.7%) and the score plot of its first two principal components is shown in Figure 4A. It can be seen that the central tumor tissue group and distant tissue group showed a clustering tendency in the direction of first predictive principal component (X axis). The adjacent tissue group was located between the central tumor tissue and distant tissue groups. Although the clustering tendency was not significant, its distribution could assess the development of HCC. An OPLS-DA model was established using all 30 samples. The model had 2 predictive principal components and 5 orthogonal principal components (R2X = 76.5%, R2Y = 93.7%, Q2 = 68.7%). As shown in Figure 4B, the score plot of the first predictive principal component and first orthogonal principal component showed significant clustering tendency. In the X axis direction, the changing tendency of metabolic profiling in the three groups reflected the development of HCC, at the same time differences between the three groups in metabolic profiling were significant. As a result, it was concluded that the first principal component could reflect the development of HCC.
Through metabolic profiling analysis of the homogenates of HCC tumor tissue, endogenous metabolites highly related to HCC were detected. The main focus of the present study was the metabolites which had an obvious impact on clustering tendency of the central tumor tissue group and distant tissue group. First, the OPLS-DA model was established for the central tumor tissue group and distant tissue group (Figure 5A), which had 1 predictive principal component and 3 orthogonal principal components (R2Y = 97.6%, Q2 = 83.8%). The characteristic metabolites with the ability to distinguish disease were selected in the established OPLS-DA model after the following two processes: (1) Among ions with VIP > 1, excluded ions which included zero in the confidence interval in the VIP diagram (Figure 5B) and excluded ions which included zero in the confidence interval in the coefficient plot (Figure 5C, excluded ions were marked by black arrows)[18]; and (2) From the S-plot (Figure 5B), ions with a high degree of variation (high horizontal ordinate value) and reliability (high vertical ordinate value) were selected. After these two steps, 14 ions were selected. To limit the cover-up effect, a “blank’’ OPLS-DA model was established which excluded the 14 selected ions. The OPLS-DA model had 1 predictive principal component and 3 orthogonal principal components (R2Y = 97.1%, Q2 = 74.2%). Because the ability of the “blank’’ model to distinguish disease was still high, other characteristic metabolites were selected following the two steps previously mentioned, and 35 ions were selected. Among the total 49 ions selected, 33 ions passed the 2 related samples nonparametric test (P < 0.05).
Some characteristic metabolites were identified by MS graphs when compared with the chromatographic peaks and mass spectrographic peaks of the standard (including MS1 and MS2). Identification of other selected ions was performed as follows: Firstly, because of the high resolution of the Orbitrap XL mass spectrometer (resolution set as 100000 FMHW), characteristic metabolites were preliminarily identified by checking accurate m/z on the Human Metabolome DataBase (http://hmdb.ca/). The matching metabolites were retained for further identification according to the rules that m/z deviation was below 0.01, with equal charge number and with suitable ionization mode. Secondly, after MS/MS scanning, MS2 graphs of the characteristic ions were obtained and then compared with theoretical fragments of previous preliminary results according to the rules that MS2 m/z deviation was below 0.2, matching the top three peaks and at least 80% of secondary MS graphs (secondary MS fragments were generated by the ion trap CID model). The theoretical fragments were derived using Mass Frontier 6.0. The final identified results and statistical differences among the three groups are shown in Table 1 (only 14 identified metabolites are listed).
| m/z | Retention time (min) | Metabolite | Adduct2 | Content3 | ||
| Normal/adjacent | Adjacent/tumor | Normal/tumor | ||||
| 453.345 | 4.39632 | Sitosterol-β | M + K [1+] | Upa | Upa | |
| 166.086 | 1.05638 | L-phenylalanine1 | M + H [1+] | Upa | Upb | |
| 520.341 | 6.84401 | LysoPC [18:2 (9Z, 12Z)] | M + H [1+] | Upa | Upb | |
| 258.111 | 0.77819 | Glycerophosphocholine1 | M + H [1+] | Upb | Upb | |
| 476.276 | 7.10121 | LysoPE [18:3 (9Z, 12Z, 15Z)/0:0] | M + H [1+] | Upa | Upb | |
| 450.322 | 5.88844 | Chenodeoxycholic acid glycine conjugate1 | M + H [1+] | Upb | Upb | |
| 568.342 | 6.78248 | LysoPC [22:6 (4Z, 7Z, 10Z, 13Z, 16Z, 19Z)] | M + H [1+] | Upa | Upb | |
| 212.02 | 5.54796 | Quinaldic acid | M + K [1+] | Upb | Upa | |
| 456.406 | 8.78787 | Arachidyl carnitine | M + H [1+] | Downa | Downb | |
| 504.308 | 7.89672 | LysoPE (18:0/0:0) | M + Na [1+] | Upa | Upa | |
| 546.357 | 7.08546 | LysoPC (18:0)1 | M + Na [1+] | Upa | Upb | |
| 566.323 | 6.82403 | LysoPC [20:4 (5Z, 8Z, 11Z, 14Z)] | M + Na [1+] | Upb | Upb | |
| 230.249 | 6.09076 | Tetradecanal | M + NH4 [1+] | Downb | Downb | |
| 282.28 | 9.02044 | Oleamide | M + H [1+] | Downb | Downb | |
HCC, the fifth commonest cancer and the third most common cause of cancer-related death, accounts for 6% of all cancers worldwide[19,20]. Fifty percent of diagnosed HCC cases occur in China[21-23]. Because it takes a long time for HCC to occur in liver cirrhosis patients, early diagnosis of HCC is of great importance. Lots of effort has gone in discovering early biomarkers in human fluid, especially in serum, to monitor the occurrence and development of HCC[11,24,25]. Unfortunately, although the biomarkers discovered in serum have great potential value in clinical and subclinical areas, their targeting ability and specificity are still limited. The present study determined serum metabolic profiling of HCC and provided new research targets for molecular biology.
Tissue metabolic profiling, especially human tissue metabolic profiling has several advantages in metabolomics investigations[26,27]. For example, compared with serum metabolic profiling, tissue metabolic profiling reflects the metabolic changes in certain target lesions in tissues or organs instead of changes in the whole metabolic system. Thus, it has higher targeting ability, specificity and is affected by fewer exogenous influencing factors, such as psychological changes in patients. Compared with animal tissue metabolic profiling, where the model is generally induced by a single factor, human tissue metabolic profiling reflects the natural physiological changes of lesions and is trusted in investigations on the occurrence and development of certain diseases.
Following tissue metabolite profiling of HBV-related HCC, 215 ions were detected and after multivariate statistical analysis of their relative levels (integral peak area of extraction ion), 14 ions were selected as characteristic metabolites and then identified. Of these characteristic metabolites, 9 had been suggested as serum metabolite biomarkers for HCC diagnosis in previous reports, including L-phenylalanine, glycerophosphocholine, lysoPCs, lysophosphatidylethanolamines (lysoPEs) and chenodeoxycholic acid glycine conjugate[11,28-30]. The present results enhanced the targeting ability of these characteristic metabolites, as alterations in these metabolites occurred in HCC cells.
The remaining 5 identified characteristic metabolites have seldom been reported, including Beta-Sitosterol, Quinaldic acid, Arachidyl carnitine, Tetradecanal, and Oleamide. This was possibly due to alterations in these metabolites which were too small to be observed in serum or were masked by alterations in other metabolites. Although these metabolites did not show great value in clinical application as potential biomarkers, they still provided new targets for anticancer research.
Previous research has shown that the levels of 11 metabolites including beta-sitosterol, L-phenylalanine, lysoPCs, glycerophosphocholine, lysoPEs, chenodeoxycholic acid glycine conjugate and quinaldic acid were significantly high in the distant tissue group compared with the central tumor tissue group. In contrast, the levels of Arachidyl carnitine, tetradecanal and oleamide were significantly lower in the distant tissue group compared with the central tumor tissue group. A comparison of the levels of these metabolites in the adjacent tissue group showed that beta-sitosterol, quinaldic acid and oleamide were significantly lower in the central tumor tissue group, but relatively close to those in the central tumor tissue group. From these changes in metabolite levels, we can speculate that these endogenous metabolites correlate with the progression of adjacent tissue to central tumor tissue and contain important information on the occurrence and development of HCC. Further attention should be focused on these metabolites which will hopefully lead to the discovery of novel potential biomarkers for cancer diagnosis and disease course monitoring and provide new targets for tumor therapy.
In summary, the establishment of a metabolic profiling model and the analysis of the characteristic metabolites provided new perspectives for further research into the pathophysiology of HCC. Following investigations on homogenates of tumor tissue obtained during radical surgery of HCC, detection and identification of these previously reported potential biomarkers in human tissue enhanced their targeted traceability and a comparison of the changes in these metabolites in the central tumor tissue group, adjacent tissue group and distant tissue group revealed metabolic information highly related to HCC development. More attention should be focused on these characteristic metabolites which will provide more potential biomarkers for HCC diagnosis and development. In addition, further research into the pathways related to these metabolites would provide new clinical targets for HCC therapy.
We would like to express my gratitude to Jie Gao and Yi-Jun Wang who helped with the sample collection and to Li Zhang who helped with the data collection. We also acknowledge the Liver Surgery Department of Tianjin Third Central Hospital for sample submission.
Hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) is the most common type of liver cancer in Asian populations with an extremely high incidence and poor survival rate. Unfortunately, there are no satisfactory definitive positive and negative markers for HCC diagnosis in clinical application at the present time.
Recently, metabolomics research provided a series of novel biomarkers in serum or urine with clinical value in the diagnosis and prognosis of HCC. However, the application of these biomarkers is still limited due to the lack of target traceability in the diseased organ. The detection of these previously reported characteristic metabolites in tumor tissue in the present research provided supporting proof for these novel biomarkers.
The targeting ability of animal models, serum and urine analysis are limited although experiments and sample collection are easier to perform. The present research used homogenates of central, adjacent and distant HBV-related HCC tumor tissue removed by surgery to identify these metabolites. Thus, this method had higher targeting ability, specificity and was less affected by exogenous influencing factors. In addition, the analysis detected alterations in these metabolites that were barely observed in serum or urine.
By detecting characteristic metabolites in tumor tissue, this research confirmed the traceability of 9 previously reported biomarkers detected in serum or urine provided supporting evidence for their potential clinical applications. The introduction of homogenates also suggested new avenues of research in metabolic analysis.
Principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA) analysis are data analysis methods based on projection technology. PCA can visually represent sample correlations (results distribution) such as clustering and migration. However, this method is easily affected by noise signals such as instrumental bias and operation error. In this situation, OPLS-DA analysis is often introduced in data analysis to focus experimental data in a particular direction according to a priori knowledge.
The manuscript selects 14 characteristic metabolites in liver tumor tissue that not only supports the clinical application of previous reported serum or urine biomarkers but also provides novel research targets for anticancer investigations.
P- Reviewer Grizzi F S- Editor Zhai HH L- Editor A E- Editor Zhang DN
| 1. | Masuzaki R, Karp SJ, Omata M. New serum markers of hepatocellular carcinoma. Semin Oncol. 2012;39:434-439. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 2. | Zhong DN, Ning QY, Wu JZ, Zang N, Wu JL, Hu DF, Luo SY, Huang AC, Li LL, Li GJ. Comparative proteomic profiles indicating genetic factors may involve in hepatocellular carcinoma familial aggregation. Cancer Sci. 2012;103:1833-1838. [PubMed] |
| 3. | Fu WM, Zhang JF, Wang H, Tan HS, Wang WM, Chen SC, Zhu X, Chan TM, Tse CM, Leung KS. Apoptosis induced by 1,3,6,7-tetrahydroxyxanthone in Hepatocellular carcinoma and proteomic analysis. Apoptosis. 2012;17:842-851. [PubMed] |
| 4. | Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153-161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1516] [Cited by in RCA: 1373] [Article Influence: 59.7] [Reference Citation Analysis (1)] |
| 5. | Verma M, Khoury MJ, Ioannidis JP. Opportunities and challenges for selected emerging technologies in cancer epidemiology: mitochondrial, epigenomic, metabolomic, and telomerase profiling. Cancer Epidemiol Biomarkers Prev. 2013;22:189-200. [PubMed] |
| 6. | Monteiro MS, Carvalho M, Bastos ML, Guedes de Pinho P. Metabolomics analysis for biomarker discovery: advances and challenges. Curr Med Chem. 2013;20:257-271. [PubMed] |
| 7. | Davis VW, Schiller DE, Eurich D, Sawyer MB. Urinary metabolomic signature of esophageal cancer and Barrett’s esophagus. World J Surg Oncol. 2012;10:271. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 61] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 8. | Xiao JF, Varghese RS, Zhou B, Nezami Ranjbar MR, Zhao Y, Tsai TH, Di Poto C, Wang J, Goerlitz D, Luo Y. LC-MS based serum metabolomics for identification of hepatocellular carcinoma biomarkers in Egyptian cohort. J Proteome Res. 2012;11:5914-5923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 102] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 9. | Wang X, Zhang A, Sun H. Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma. Hepatology. 2013;57:2072-2077. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 174] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 10. | Wu H, Xue R, Dong L, Liu T, Deng C, Zeng H, Shen X. Metabolomic profiling of human urine in hepatocellular carcinoma patients using gas chromatography/mass spectrometry. Anal Chim Acta. 2009;648:98-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 136] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 11. | Tan Y, Yin P, Tang L, Xing W, Huang Q, Cao D, Zhao X, Wang W, Lu X, Xu Z. Metabolomics study of stepwise hepatocarcinogenesis from the model rats to patients: potential biomarkers effective for small hepatocellular carcinoma diagnosis. Mol Cell Proteomics. 2012;11:M111.010694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 118] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 12. | Soga T, Sugimoto M, Honma M, Mori M, Igarashi K, Kashikura K, Ikeda S, Hirayama A, Yamamoto T, Yoshida H. Serum metabolomics reveals γ-glutamyl dipeptides as biomarkers for discrimination among different forms of liver disease. J Hepatol. 2011;55:896-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 192] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 13. | Makarov A, Scigelova M. Coupling liquid chromatography to Orbitrap mass spectrometry. J Chromatogr A. 2010;1217:3938-3945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 142] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 14. | Trygg J, Holmes E, Lundstedt T. Chemometrics in metabonomics. J Proteome Res. 2007;6:469-479. [PubMed] |
| 15. | Eriksson L, Johansson E, Kettaneh-Wold N, Wold S. Multi-and megavariate data analysis: principles and applications. Umetrics: Umea 2001; . |
| 16. | Gika HG, Theodoridis GA, Wingate JE, Wilson ID. Within-day reproducibility of an HPLC-MS-based method for metabonomic analysis: application to human urine. J Proteome Res. 2007;6:3291-3303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 405] [Cited by in RCA: 417] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 17. | Dunn WB, Broadhurst D, Brown M, Baker PN, Redman CW, Kenny LC, Kell DB. Metabolic profiling of serum using Ultra Performance Liquid Chromatography and the LTQ-Orbitrap mass spectrometry system. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;871:288-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 126] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 18. | Yin P, Wan D, Zhao C, Chen J, Zhao X, Wang W, Lu X, Yang S, Gu J, Xu G. A metabonomic study of hepatitis B-induced liver cirrhosis and hepatocellular carcinoma by using RP-LC and HILIC coupled with mass spectrometry. Mol Biosyst. 2009;5:868-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 173] [Cited by in RCA: 176] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 19. | Patel M, Shariff MI, Ladep NG, Thillainayagam AV, Thomas HC, Khan SA, Taylor-Robinson SD. Hepatocellular carcinoma: diagnostics and screening. J Eval Clin Pract. 2012;18:335-342. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Patterson AD, Maurhofer O, Beyoglu D, Lanz C, Krausz KW, Pabst T, Gonzalez FJ, Dufour JF, Idle JR. Aberrant lipid metabolism in hepatocellular carcinoma revealed by plasma metabolomics and lipid profiling. Cancer Res. 2011;71:6590-6600. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 218] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 21. | Llovet JM, Beaugrand M. Hepatocellular carcinoma: present status and future prospects. J Hepatol. 2003;38 Suppl 1:S136-S149. [PubMed] |
| 22. | Pei Y, Zhang T, Renault V, Zhang X. An overview of hepatocellular carcinoma study by omics-based methods. Acta Biochim Biophys Sin (Shanghai). 2009;41:1-15. [PubMed] |
| 23. | Khwaja A, Silverman D, Sloan F, Wang Y. Are mature smokers misinformed? J Health Econ. 2009;28:385-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 7] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Ressom HW, Xiao JF, Tuli L, Varghese RS, Zhou B, Tsai TH, Ranjbar MR, Zhao Y, Wang J, Di Poto C. Utilization of metabolomics to identify serum biomarkers for hepatocellular carcinoma in patients with liver cirrhosis. Anal Chim Acta. 2012;743:90-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 25. | Zhou L, Wang Q, Yin P, Xing W, Wu Z, Chen S, Lu X, Zhang Y, Lin X, Xu G. Serum metabolomics reveals the deregulation of fatty acids metabolism in hepatocellular carcinoma and chronic liver diseases. Anal Bioanal Chem. 2012;403:203-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 127] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 26. | York B, Sagen JV, Tsimelzon A, Louet JF, Chopra AR, Reineke EL, Zhou S, Stevens RD, Wenner BR, Ilkayeva O. Research resource: tissue- and pathway-specific metabolomic profiles of the steroid receptor coactivator (SRC) family. Mol Endocrinol. 2013;27:366-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 27. | Budczies J, Denkert C, Müller BM, Brockmöller SF, Klauschen F, Györffy B, Dietel M, Richter-Ehrenstein C, Marten U, Salek RM. Remodeling of central metabolism in invasive breast cancer compared to normal breast tissue - a GC-TOFMS based metabolomics study. BMC Genomics. 2012;13:334. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 111] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 28. | Yang Y, Li C, Nie X, Feng X, Chen W, Yue Y, Tang H, Deng F. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res. 2007;6:2605-2614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 196] [Cited by in RCA: 187] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 29. | Chen F, Xue J, Zhou L, Wu S, Chen Z. Identification of serum biomarkers of hepatocarcinoma through liquid chromatography/mass spectrometry-based metabonomic method. Anal Bioanal Chem. 2011;401:1899-1904. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 30. | Chen T, Xie G, Wang X, Fan J, Qiu Y, Zheng X, Qi X, Cao Y, Su M, Wang X. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics. 2011;10:M110.004945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 246] [Article Influence: 17.6] [Reference Citation Analysis (0)] |