Published online Mar 7, 2012. doi: 10.3748/wjg.v18.i9.965
Revised: November 10, 2011
Accepted: December 16, 2011
Published online: March 7, 2012
AIM: To investigate the genetic characteristics and pathogenicity of hepatitis E virus (HEV) and assess the potential risk factors for sporadic hepatitis E.
METHODS: Sixty-two serum samples from the patients with acute hepatitis E were collected, including 23 cases coinfected with hepatitis B virus. Anti-HEV detection and partial HEV RNA amplification were performed by enzyme immunoassays and reverse transcription-nested polymerase chain reaction (RT-nPCR) method, respectively, and PCR products were sequenced. The isolated human HEV sequences were analyzed phylogenetically.
RESULTS: The positive rate of serum HEV RNA were 21.0% (13/62), including 5 cases of liver failure. All the 13 isolates shared a 82.1%-98.0% nucleotide homology with each other and had identities of 74.7%-81.0%, 75.3%-78.6%, 75.3%-80.0% and 82.1%-96.1% with the corresponding regions of HEV genotypes 1-4, respectively. The human HEV strain GS-NJ-12 shared a 100% nucleotide identity with the swine HEV strain swIM6-43 isolated from Inner Mongolia, China.
CONCLUSION: Swine may be a principal risk factor for occurrence of sporadic hepatitis E in eastern China, and genotype 4 HEV can induce acute liver failure.
- Citation: Geng JB, Wang MR, Wang L, Wang J, Yang ZG, Cheng Y, Qiao F, Wang M. Genetic characteristics and pathogenicity of human hepatitis E virus in Nanjing, China. World J Gastroenterol 2012; 18(9): 965-970
- URL: https://www.wjgnet.com/1007-9327/full/v18/i9/965.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i9.965
Hepatitis E virus (HEV) is a single-strand, positive-sense RNA, non-enveloped virus which is classified into the Hepeviridae family with a single serotype and at least 4 known main genotypes of mammalian HEV, one avian HEV and a new HEV genotype have been isolated from rabbits recently[1]. Genotype 1 and 2 of mammalian strains are predominant in humans and associated with large waterborne epidemics in endemic regions[2]. However, genotype 3 and 4, which were suggested to be zoonotically transmitted between animals and humans, are mainly responsible for sporadic cases of hepatitis E clinically manifested as icterus, malaise, anorexia, fever, hepatomegaly and pruritus. Additionally, increasing reports suggest that different HEV genotypes are associated with different disease severity. HEV genotype 1 and 2, which have similar epidemiological and sporadic features, can result in acute hepatitis, acute liver failure, and acute-on-chronic liver failure. However, HEV genotype 3 and 4, which were generally considered to cause acute, self-limiting illness followed by a complete recovery, seem to be less virulent in humans than genotype 1 and 2[3], and do not cause severe liver diseases[4]. In mainland China, HEV genotype 4 has become the dominant genotype instead of genotype 1 since 2004[5].
Since the first swine HEV strain was isolated in 1997, many strains of HEV have been identified from human and other mammalian reservoirs (swine, wild boar, deer, mongooses, rabbits and rats), and swine was considered to be the principal reservoir of HEV[6-10]. Accumulating data indicates that hepatitis E is a zoonotic disease. Transmissions through the consumption of contaminated food products such as pork have provided further direct evidence. Thus, zoonotic transmission of hepatitis E raises an important public health concern over food safety and zoonotic risk[11].
In China, seroepidemiological studies in patients with viral hepatitis have shown a high superinfection rate (32.4%) with two or more types of hepatitis virus; and HEV superinfection in patients with chronic hepatitis B (CHB) accounts for 17.6%[12]. HEV could result in severe disease and a poor outcome in patients with pre-existing liver diseases[13,14]. However, there were few reports on the association between genetic characteristics and pathogenicity of HEV infection. In addition, whether genotype 3 and 4 HEV could induce liver failure in normal population and patients with chronic liver disease (CLD) is still unclear. This study was designed to investigate the genotype of HEV prevalent in eastern China, the pathogenicity of HEV in patients with or without CLD and the phylogenetic relationship between human and swine HEV.
A total of 62 serum samples were collected from the hospitalized patients with hepatitis E during the period from November 2008 to December 2010. The diagnostic criteria of hepatitis E are as follows: the elevation of alanine aminotransferase (ALT) level (> 2ULN); the positive result for anti-HEV IgM or at least 4-fold increase of IgG levels during hospitalization. Patients coinfected with hepatitis B virus had positive serum HBsAg and HBV DNA. All patients were negative for anti-human immunodeficiency virus, anti-hepatitis A virus, anti-hepatitis c virus antibodies and autoantibodies. As some patients did not seek medical care in the early stages of their illness, the presence of HEV-IgG was used to diagnose acute hepatitis E in this study[14-16]. The clinical data of patients with acute liver failure or acute-on-chronic liver failure were recorded.
All the serum samples were detected for anti-HEV IgM, anti-HEV IgG, anti-HAV IgM, HBsAg, anti-HCV IgG using commercial enzyme immunoassay (EIA) kits (Beijing Wantai Biological Pharmacy Enterprise Co., Beijing, China). All assay procedures were carried out according to the manufacturer’s instructions. All anti-HEV antibody positive specimens were confirmed by Wantai EIA kit one more time.
All serum samples were tested for presence of HEV RNA by reverse transcription-nested polymerase chain reaction (RT-nPCR). RNA was extracted from 200 μL of serum according to the instructions of TRIzol reagent (Invitrogen). The viral RNA was reverse-transcribed to cDNA for 1h at 42 °C with M-MuLV reverse transcriptase (Promega) and specific external anti-sense primers in a 10 μL reaction volume. Nested PCRs for open reading frame (ORF) 2 and ORF1 were performed to detect HEV sequences using two sets of consensus oligonucleotide primers. The primer sequences and amplification parameters were as described previously[10]. The final PCR product was analyzed by 15 g/L agarose gel electrophoresis.
The target second-round PCR products were purified and double-ends sequenced by ABI model 3730 sequencer. Nucleotide sequences were analyzed with the MEGA 4.0, ALIGNX and Bioedit v7.0.9 software. Phylogenetic trees were constructed by the neighbor-joining method and the interior branch test with the aid of MEGA 4.0 software package. One thousand resemblings of the data were used to calculate percentages of the branches obtained.
Designations and accession numbers of full-length reference sequences representing different genotypes for analysis of HEV ORF1 and ORF2 were retrieved from GenBank as follows: Genotype 1: Abb-2B (AF185822); Bur86 (D10330), Sar-55 (M80581), Uigh179 (D11093), FHF (X98292), Morocco (AY230202), T3-Chad (AY204877); Genotype 2: M1 (M74506); Genotype 3: US2 (AF060669), Osh-205 (AF455784), JBOAR-1Hyo04 (AB189070), swArkell (AY115488), HE-JA10 (AB089824), JKN-Sap (AB074918), JMY-HAW (AB074920); Genotype 4: 4a: ChH-S-1 (EF077063), swGX32 (EU366959), JKO-ChiSai98C (AB197673); 4b: swGX40 (EU676172), swDQ (DQ279091); 4c: swJ13-1 (AB097811), HE-JA1 (AB097812), HE-JK4 (AB099347), JSN-SAP-FH02 (AB200239); 4d: T1 (AJ272108), swCH25 (AY594199); 4g: ccc220 (AB108537).
In addition, the reference sequences used for analysis of partial ORF2 regions included subtype 4e: IND-SW1 (AF324501), IND-SW2 (AF324502), IND-SW3 (AF324503); 4d: swIM6-26 (AB550622), swIM6-43 (AB550624); subtype 4f: HE-JA2 (AB082558).
The clinical data of patients with liver failure is summarized in Table 1. Among 62 cases, 10 developed liver failure (4 with acute liver failure and 6 with acute-on-chronic failure), 5 with CLD died of acute-on-chronic liver failure, and one 21-year-old female patient died of acute liver failure.
| Patient No. | Sex | Age (yrs) | TBIL (μmol/L) | DBIL (μmol/L) | ALT (U/L) | AST (U/L) | PA (g/L) | ALB (g/L) | CHE (U/L) | PT (s) | PTA (%) |
| 1II III | Male | 23 | 507.4 | 442.9 | 183 | 390 | 30 | 26.1 | 1.6 | 19 | 34.7 |
| 2III III | Male | 61 | 590.9 | 481.2 | 779 | 787 | 12 | 25 | 1.2 | 14.5 | 46.4 |
| 3I | Male | 69 | 179.2 | 136.7 | 5056 | 4754 | 45 | 34.6 | 3.3 | 16.9 | 34.2 |
| 4III III | Female | 21 | 636.4 | 437.5 | 355 | 611 | 12 | 31.8 | 1.1 | 65.7 | 4.3 |
| 5III III | Female | 44 | 993.5 | 775.4 | 166 | 451 | 29 | 25 | 0.9 | 20.1 | 31.8 |
| 6III | Male | 45 | 711.4 | 603.4 | 409 | 209 | 30 | 28.4 | 1.4 | 16.6 | 43 |
| 7III III | Male | 57 | 639.9 | 494.2 | 2100 | 2820 | 9 | 33.9 | 1.7 | 28.2 | 18.9 |
| 8I | Female | 58 | 443.1 | 302.5 | 689 | 1493 | 15 | 27.8 | 3.1 | 21.4 | 23.8 |
| 9III III | Male | 36 | 759.8 | 541.9 | 319 | 278 | 27 | 29.7 | 1.7 | 25.3 | 18.4 |
| 10IIII | Male | 79 | 506.8 | 428.7 | 329 | 548 | 4 | 30 | 1.4 | 15.7 | 38.9 |
Out of 62 serum samples, 33 were positive for anti-HEV IgM but negative for anti-HEV IgG, while 23 were positive only for anti-HEV IgG, 6 were positive for both IgG and IgM. The overall positivity rate for HEV RNA was 21.0% (13/62).
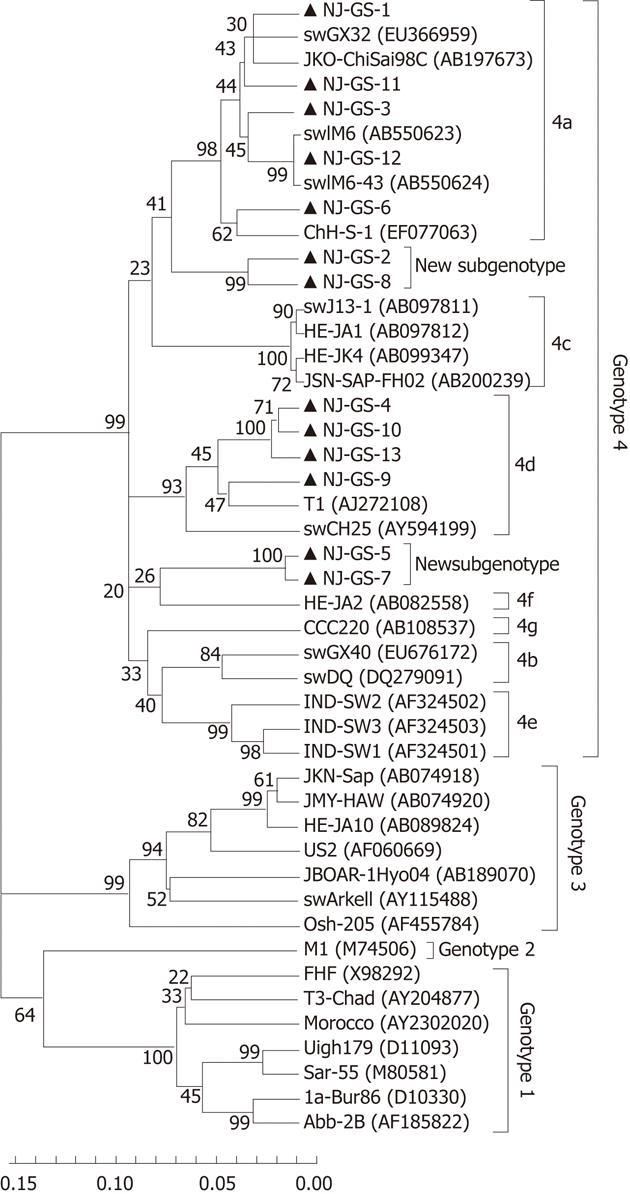
HEV cDNA was amplified from 13 serum samples using primer S4, and the 365nt PCR products of partial ORF2 sequences were determined. The 13 isolated strains were designated as GS-NJ-1 to GS-NJ-13 (GenBank accession numbers JF309208 - JF309220). These 13 isolates shared 82.1%-98.0% nucleotide homology, and had identities of 74.7%-81.0%, 75.3%-78.6%, 75.3%-80.0% and 82.1%-96.1% with the corresponding regions of HEV genotypes 1-4, respectively. Phylogenetic analysis based on partial ORF2 (286 bp) showed that the 13 sequences could be clearly grouped into four main clades (Figure 1), one of which consisted of 4 HEV isolates sharing an 88.4%-97.2% identity with HEV subtype 4d. The second clade included 5 isolates sharing a 90.6%-96.1% identity with HEV subtype 4a. The third clades were formed by GS-NJ-5 and GS-NJ-7 sharing a 98.0% identity with each other and 82.1%-87.3% nucleotide homology with subtypes 4a-4g. The last clade contained 2 isolates sharing a 94.2% identity with each other and 82.5%-89.0% identity with subtypes 4a-4g. Phylogenetic tree showed that both the last two branches were formed individually, separating from the established subgenotypes 4a-4g, which indicated that they may belong to novel subgenotypes. The phylogenetic analysis based on partial ORF1 sequences confirmed the genotyping results (Figure 2).
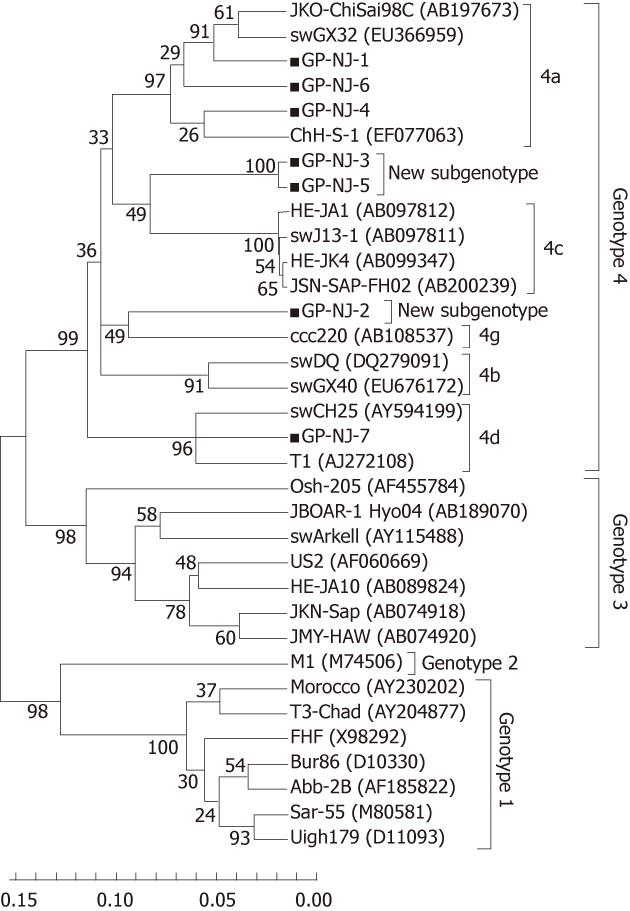
To exclude the possibility of contamination and confirm the results of phylogenetic analysis based on partial ORF2 sequences, 13 positive samples were amplified using ORF1 primers (P primer) and 11 were positive. Sequence analysis of partial HEV ORF1 fragments showed that there were 7 groups, designated as GP-NJ-1 to GP-NJ-7 with GenBank accession numbers JF309201-JF309207. The 7 strains shared an 82.0%-99.1% sequence identity with each other, and had identities of 73.2%-78.1%, 75.3%-79.0%, 74.0%-82.7% and 80.6%-95.0% with the corresponding regions of HEV genotypes 1-4, respectively. Phylogenetic analysis (Figure 2) confirmed the subtyping result based on partial sequences of ORF2.
In China, a substantial proportion of people had serological evidence of prior HEV exposure but no disease[17]. Moreover, according to the national investigation of 2002, the positive rate of HBsAg was 9.09% in persons over 3 years of age and most of the HBsAg carriers were asymptomatic and under recognized. During the long period of CHB, there was a chance for patients to be sporadically superinfected by HEV. Studies had demonstrated that patients with CLD coinfected with HEV had more severe liver diseases with higher rates of compensated cirrhosis, hepatic failure, and complications[13,14]. In present study, 10 of 62 patients suffered from hepatic failure and 6 patients died (1 died of acute liver failure and 5 died of acute-on-chronic liver failure) (Table 1). Therefore, all susceptible people, especially the patients with CLD should take appropriate strategies to decrease the incidence of HEV infection, such as consumption of boiled water and well-cooked food and hand washing with soap.
As many studies reported, HEV was transmitted primarily by fecal-oral route, usually via the consumption of contaminated water or food. However, recent investigations have not consistently found well-defined water sources of HEV, suggesting other possible modes of transmission[18,19]. Transmissions through blood transfusions, mother-to-fetus, and person-to-person were also reported[20,21], but these routes of transmission were not thought to be frequent. In this study, a substantial proportion of patients had not consumed any raw meat in the recent past, and no clear source of infection was found, raising the suspicion of transmission through contaminated water or food. It is worth mentioning that in eastern China, where there are many rivers, water are usually contaminated by domestic pig feces and used to fertilize crops without special treatment. As HEV was isolated frequently from swine feces in most parts of the world, people who drink unboiled water or inadequately cooked food may be infected by HEV. Therefore, clinicians should be vigilant and should consider hepatitis E in the differential diagnosis of unexplained jaundice and clinical indications, such as the lack of evidence for other causes of abnormal liver failure, should increase the threshold for testing for HEV infection.
Phylogenetically, sequence analysis of partial ORF2 and ORF1 showed the highest identity with genotype 4, and phylogenetic tree conformed that all HEV strains belonged to genotype 4 and could further subdivided into 4 subgenotypes (Figures 1 and 2). These results indicate that patients in eastern China are currently infected with divergent genotype 4 HEV strains that may be indigenous; genotype 4 has emerged as the predominant genotype in this region with at least 4 subgenotypes. Additionally, further phylogenetic analysis showed a very close relationship and a high nucleotide identity between human and swine HEVs by blasting the partial human HEV sequences in Genbank (Table 2). The GS-NJ-12 strain shared a 100% nucleotide identity with two swine HEVs (swIM6-43 and swIM6-41) isolated from Inner Mongolia, China. Moreover, all swine HEVs showing the highest nucleotide identities comparable with human HEVs were isolated from different areas of mainland China, indicating that sporadic hepatitis E was acquired domestically, and swine may be a principal risk factor for occurrence of sporadic hepatitis E.
| Human HEV | Swine HEV | Identity (%) | Coverage (%) |
| GP-NJ-1 | CHN-XJ-SW36 (FJ775168) | 96 | 100 |
| GP-NJ-2 | swCH31 (DQ450072) | 96 | 100 |
| GP-NJ-3 | CHN-XJ-SW16 (GQ306000) | 99 | 100 |
| GP-NJ-4 | CHN-XJ-SW10 (FJ775167) | 93 | 100 |
| GP-NJ-5 | CHN-XJ-SW16 (GQ306000) | 99 | 100 |
| GP-NJ-6 | CHN-XJ-SW61 (FJ775170) | 93 | 100 |
| GP-NJ-7 | bjsw1 (GU206559) | 97 | 100 |
| GS-NJ-1 | CHN-XJ-SW36 (FJ775175) | 95 | 100 |
| GS-NJ-2 | swSH02 (DQ450074) | 96 | 99 |
| GS-NJ-3 | swIM8-4 (AB550626) | 95 | 97 |
| GS-NJ-4 | KMsw-3 (HQ008864) | 97 | 98 |
| GS-NJ-5 | CHN-XJ-SW16 (GQ306004) | 96 | 100 |
| GS-NJ-6 | CHN-XJ-SW10 (FJ775174) | 95 | 100 |
| GS-NJ-7 | CHN-XJ-SW16 (GQ306004) | 95 | 100 |
| GS-NJ-8 | swSH02 (DQ450074) | 95 | 98 |
| GS-NJ-9 | bjsw1 (GU206559) | 97 | 99 |
| GS-NJ-10 | KMsw-3 (HQ008864) | 98 | 98 |
| GS-NJ-11 | CHN-SJ-SW36 (FJ775175) | 95 | 100 |
| GS-NJ-12 | swIM6-43 (AB550624) | 100 | 97 |
| GS-NJ-13 | KMsw-3 (HQ008864) | 97 | 98 |
Among 62 serum specimens, 13 samples had positive result of HEV RNA using ORF2 primers. The ORF1 degenerate primer (P primer) set amplified 11 of 13 HEV sequences successfully, and 2 specimens positive for S primer were negative for P primer. While both primer sets could amplify human and animal HEV successfully[6,10,17,22], multiple nucleotide substitutions in the primer-binding regions or a few bases mismatch in 3’ end of primers, the narrow window of HEV incubation, low HEV RNA titer in sera and “false-positive” results from cross-reactivity with unknown antigens may result in a low detection rate of HEV RNA. Because the window for HEV diagnosis may be narrow, sample collection from the patients in due course is considered to be the key for HEV RNA detection.
Up until now, hepatitis E is diagnosed by detecting viral RNA in serum and/or feces during the incubation period or early acute phase of disease, or, more commonly, by demonstrating IgM anti-HEV or a rising titer of IgG anti-HEV in the serum during the late acute phase or convalescent phase of the illness[16]. However, there are still problems with the enzyme immunoassays used to detect current or previous HEV exposure. Commercial assays vary markedly in their sensitivity and specificity[4,23]. Recently, the Wantai kit used in this study was proved to be more sensitive than GeneLab kit (98% vs 56%)[24] , which has a high specificity for diagnosis of acute infection of HEV[25,26]. Detection of rising IgG or RNA was considered diagnostic with a specificity of 100%; the specificity of IgM was found to be 99.4%[15]. Yet, based on previous studies, it is still very hard for researchers to detect HEV RNA in all serum specimens positive for anti-HEV. In present study, because some patients did not seek medical care in the early stages of their illness or were treated at other hospitals at early period of illness, and the period of viraemia is short[3], only 5 of 10 cases with hepatic failure were confirmed by the presence of HEV RNA in serum samples. Although some cases negative for HEV RNA might be misdiagnosed as hepatitis E, 5 patients with liver failure (No. 4, 6, 7, 8, 10) were confirmed by detecting serum HEV RNA using S primers (designated as GS-NJ-11, GS-NJ-4, GS-NJ-6, GS-NJ-9, GS-NJ-5) and P primers (designated as GP-NJ-6, GP-NJ-7, GP-NJ-4, GP-NJ-7, GP-NJ-2). Patients 4 and 8 with no underlying liver diseases suffered from acute liver failure, and patient 4 died. Furthermore, 5 (No. 1, 5, 6, 7 and 9) out of 6 patients with CLD died of hepatic failure and relative complications, suggesting that it is imperative to develop a reliable hepatitis E vaccine.
In conclusion, this study presented the first finding that HEV genotype 4 could result in acute liver failure and acute-on-chronic liver failure. Genotype 4 was the dominant strain among the patients involved in this study, and there were at least 4 subgenotypes (4a, 4d and 2 new subgenotypes) prevalent in eastern China. Phylogenetic analysis showed a very close relationship between human and swine HEV, suggesting that swine may be a principal risk factor for occurrence of hepatitis E in eastern China. However, the implication for subgenotype classification and other issues such as the relationship between different genotypes/subtypes and different modes of transmission, pathogenicity, possibility of cross species transmission are still indistinct and remain to be understood.
We are grateful to Professor Malcolm A McCrae of Warwick University, UK for proofreading the manuscript.
Hepatitis E, caused by the hepatitis E virus (HEV), is the most important cause of acute viral hepatitis in adults throughout Asia, the Middle East and Africa where the sanitation conditions are usually substandard. HEV genotypes 1 and 2, which have similar epidemiological and sporadic features, can result in acute hepatitis, acute liver failure, and acute-on-chronic liver failure. However, HEV genotype 3 and 4, which were generally considered to cause acute, self-limiting illness followed by a complete recovery, seems to be less virulent for humans than genotypes 1 and 2 and have not been implicated in causing severe liver diseases. However, there were few reports on the association between genetic characteristics and pathogenicity of HEV infection. In addition, whether genotype 3 and 4 HEV could induce liver failure in normal population and patients with chronic liver disease (CLD) is still unclear.
HEV is widespread in swine and is likely to be endemic in many developed/developing countries. There is a very close relationship between human and swine HEV by phylogenetic analysis of partial/full-length genomic sequence. Accumulated evidences support the hypothesis that hepatitis E is a zoonosis, and swine may be a principal risk factor for occurrence of hepatitis E in many areas. HEV infection in healthy individuals are associated with a mortality rate of 0.04%-4%. Some studies have reported a high prevalence of HEV antibodies in patients with CLDs and others have suggested that cirrhotics were prone to HEV infection, and superinfection with HEV in patients with underlying CLD can cause severe hepatic decompensation, leading to increased morbidity and mortality.
In China, superinfection with genotype 4 HEV in patients with underlying CLD, especially in patients with chronic hepatitis B, can cause severe hepatic decompensation, leading to increased morbidity and mortality. This study presented the first finding that genotype 4 HEV can induce liver failure in normal population and patients with CLD. The GS-NJ-12 strain isolated from a case of sporadic hepatitis E shared a 100% nucleotide identity with two swine HEVs (swIM6-43 and swIM6-41) isolated from Inner Mongolia, China. Moreover, all swine HEVs showing the highest nucleotide identities comparable with human HEVs were isolated from different areas of mainland China, indicating that sporadic hepatitis E is acquired domestically, and swine may be a principal risk factor for occurrence of sporadic hepatitis E in eastern China.
This study found that the dominant genotype of HEV prevalent in eastern China was genotype 4, which can induce liver failure in normal population and patients with CLD and swine may be a principal risk factor for occurrence of sporadic hepatitis E. Therefore, a standardized management of swine in stock farm is the key to prevent HEV transmission from swine to human.
The work investigated the genetic characteristics of the HEV in Eastern China and assessed the potential risk factors. Among acute infected patients, they found a seroprevalence of HEV RNA in 21%. The isolated HEV strains shared a high percentage of nucleotide homology with a HEV strain isolated from swine. The manuscript demonstrates a well performed study which hints one more time the direction that HEV may be acquired by the contact to swines/wild boars.
Peer reviewers: Kilian Weigand, MD, Department of Gastroenterology, Infectious Diseases and Intoxications, University Hospital Heidelberg, Im Neuenheimer Feld 410, 69120 Heidelberg, Germany; Deepak Narayan Amarapurkar, Consultant Gastroenterologist and Hepatologist, Department of Gastroenterology, Bombay Hospital and Medical Research Centre, Mumbai 400 020, India
S- Editor Gou SX L- Editor Ma JY E- Editor Xiong L
| 1. | Zhao C, Ma Z, Harrison TJ, Feng R, Zhang C, Qiao Z, Fan J, Ma H, Li M, Song A. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J Med Virol. 2009;81:1371-1379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 253] [Cited by in RCA: 271] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 2. | Lu L, Li C, Hagedorn CH. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. 2006;16:5-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 582] [Cited by in RCA: 592] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 3. | Purcell RH, Emerson SU. Hepatitis E: an emerging awareness of an old disease. J Hepatol. 2008;48:494-503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 508] [Cited by in RCA: 494] [Article Influence: 29.1] [Reference Citation Analysis (0)] |
| 4. | Acharya SK, Panda SK. Hepatitis E: water, water everywhere - now a global disease. J Hepatol. 2011;54:9-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 5. | Zheng Y, Ge S, Zhang J, Guo Q, Ng MH, Wang F, Xia N, Jiang Q. Swine as a principal reservoir of hepatitis E virus that infects humans in eastern China. J Infect Dis. 2006;193:1643-1649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 103] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 6. | Fu H, Li L, Zhu Y, Wang L, Geng J, Chang Y, Xue C, Du G, Li Y, Zhuang H. Hepatitis E virus infection among animals and humans in Xinjiang, China: possibility of swine to human transmission of sporadic hepatitis E in an endemic area. Am J Trop Med Hyg. 2010;82:961-966. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 7. | Sonoda H, Abe M, Sugimoto T, Sato Y, Bando M, Fukui E, Mizuo H, Takahashi M, Nishizawa T, Okamoto H. Prevalence of hepatitis E virus (HEV) Infection in wild boars and deer and genetic identification of a genotype 3 HEV from a boar in Japan. J Clin Microbiol. 2004;42:5371-5374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 132] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 8. | Okamoto H. Genetic variability and evolution of hepatitis E virus. Virus Res. 2007;127:216-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 255] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 9. | Johne R, Plenge-Bönig A, Hess M, Ulrich RG, Reetz J, Schielke A. Detection of a novel hepatitis E-like virus in faeces of wild rats using a nested broad-spectrum RT-PCR. J Gen Virol. 2010;91:750-758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 259] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 10. | Geng J, Wang L, Wang X, Fu H, Bu Q, Zhu Y, Zhuang H. Study on prevalence and genotype of hepatitis E virus isolated from Rex Rabbits in Beijing, China. J Viral Hepat. 2011;18:661-667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 11. | Meng XJ. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet Microbiol. 2010;140:256-265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 396] [Cited by in RCA: 364] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 12. | Xiong LS, Cui SF, Zhou JG, Xing Y. [Detection and analysis of HAV-HEV, HGV infection in patients with viral hepatitis]. Zhonghua Ganzangbing Zazhi. 2004;12:395-396. [PubMed] |
| 13. | Hamid SS, Atiq M, Shehzad F, Yasmeen A, Nissa T, Salam A, Siddiqui A, Jafri W. Hepatitis E virus superinfection in patients with chronic liver disease. Hepatology. 2002;36:474-478. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 14. | Zhang X, Ke W, Xie J, Zhao Z, Xie D, Gao Z. Comparison of effects of hepatitis E or A viral superinfection in patients with chronic hepatitis B. Hepatol Int. 2010;4:615-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 48] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 15. | Huang S, Zhang X, Jiang H, Yan Q, Ai X, Wang Y, Cai J, Jiang L, Wu T, Wang Z. Profile of acute infectious markers in sporadic hepatitis E. PLoS One. 2010;5:e13560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 109] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 16. | Dalton HR, Bendall R, Ijaz S, Banks M. Hepatitis E: an emerging infection in developed countries. Lancet Infect Dis. 2008;8:698-709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 498] [Cited by in RCA: 483] [Article Influence: 28.4] [Reference Citation Analysis (0)] |
| 17. | Chang Y, Wang L, Geng J, Zhu Y, Fu H, Ren F, Li L, Wang X, Zhuang H. Zoonotic risk of hepatitis E virus (HEV): A study of HEV infection in animals and humans in suburbs of Beijing. Hepatol Res. 2009;39:1153-1158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 18. | Teshale EH, Grytdal SP, Howard C, Barry V, Kamili S, Drobeniuc J, Hill VR, Okware S, Hu DJ, Holmberg SD. Evidence of person-to-person transmission of hepatitis E virus during a large outbreak in Northern Uganda. Clin Infect Dis. 2010;50:1006-1010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 19. | Howard CM, Handzel T, Hill VR, Grytdal SP, Blanton C, Kamili S, Drobeniuc J, Hu D, Teshale E. Novel risk factors associated with hepatitis E virus infection in a large outbreak in northern Uganda: results from a case-control study and environmental analysis. Am J Trop Med Hyg. 2010;83:1170-1173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 20. | Khuroo MS, Kamili S, Yattoo GN. Hepatitis E virus infection may be transmitted through blood transfusions in an endemic area. J Gastroenterol Hepatol. 2004;19:778-784. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 138] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 21. | Mushahwar IK. Hepatitis E virus: molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. J Med Virol. 2008;80:646-658. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 178] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 22. | Ji Y, Zhu Y, Liang J, Wei X, Yang X, Wang L, Li L, Chang Y, Tang R, Zhuang H. Swine hepatitis E virus in rural southern China: genetic characterization and experimental infection in rhesus monkeys (Macaca mulatta). J Gastroenterol. 2008;43:565-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 23. | Ghabrah TM, Tsarev S, Yarbough PO, Emerson SU, Strickland GT, Purcell RH. Comparison of tests for antibody to hepatitis E virus. J Med Virol. 1998;55:134-137. [PubMed] |
| 24. | Bendall R, Ellis V, Ijaz S, Ali R, Dalton H. A comparison of two commercially available anti-HEV IgG kits and a re-evaluation of anti-HEV IgG seroprevalence data in developed countries. J Med Virol. 2010;82:799-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 247] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 25. | Bendall R, Ellis V, Ijaz S, Thurairajah P, Dalton HR. Serological response to hepatitis E virus genotype 3 infection: IgG quantitation, avidity, and IgM response. J Med Virol. 2008;80:95-101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 78] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 26. | Legrand-Abravanel F, Thevenet I, Mansuy JM, Saune K, Vischi F, Peron JM, Kamar N, Rostaing L, Izopet J. Good performance of immunoglobulin M assays in diagnosing genotype 3 hepatitis E virus infections. Clin Vaccine Immunol. 2009;16:772-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 66] [Article Influence: 4.1] [Reference Citation Analysis (0)] |