Published online Dec 7, 2012. doi: 10.3748/wjg.v18.i45.6635
Revised: September 13, 2012
Accepted: October 22, 2012
Published online: December 7, 2012
AIM: To evaluate effects of UDP-glucuronosyltransferase1A1 (UGT1A1) and thymidylate synthetase (TS) gene polymorphisms on irinotecan in metastatic colorectal cancer (mCRC).
METHODS: Two irinotecan- and fluorouracil-based regimens, FOLFIRI and IFL, were selected as second-line therapy for 138 Chinese mCRC patients. Genomic DNA was extracted from peripheral blood samples before treatment. UGT1A1 and TS gene polymorphisms were determined by direct sequencing and restriction fragment length polymorphism, respectively. Gene polymorphisms of UGT1A1*28, UGT1A1*6 and promoter enhancer region of TS were analyzed. The relationship between genetic polymorphisms and clinical outcome, that is, response, toxicity and survival were assessed. Pharmacokinetic analyses were performed in a subgroup patients based on different UGT1A1 genotypes. Plasma concentration of irinotecan and its active metabolite SN-38 and inactive metabolite SN-38G were determined by high performance liquid chromatography. Differences in irinotecan and its metabolites between UGT1A1 gene variants were compared.
RESULTS: One hundred and eight patients received the FOLFIRI regimen, 29 the IFL regimen, and one irinotecan monotherapy. One hundred and thirty patients were eligible for toxicity and 111 for efficacy evaluation. One hundred and thirty-six patients were tested for UGT1A1*28 and *6 genotypes and 125 for promoter enhancer region of TS. Patients showed a higher frequency of wild-type UGT1A1*28 (TA6/6) compared with a Caucasian population (69.9% vs 45.2%). No significant difference was found between response rates and UGT1A1 genotype, although wild-type showed lower response rates compared with other variants (17.9% vs 24.2% for UGT1A1*28, 15.7% vs 26.8% for UGT1A1*6). When TS was considered, the subgroup with homozygous UGT1A1*28 (TA7/7) and non-3RG genotypes showed the highest response rate (33.3%), while wild-type UGT1A1*28 (TA6/6) with non-3RG only had a 13.6% response rate, but no significant difference was found. Logistic regression showed treatment duration was closely linked to clinical response. In toxicity comparison, UGT1A1*28 TA6/6 was associated with lower incidence of grade 2-4 diarrhea (27.8% vs 100%), and significantly reduced the risk of grade 4 neutropenia compared with TA7/7 (7.8% vs 37.5%). Wild-type UGT1A1*6 (G/G) tended to have a lower incidence of grade 3/4 diarrhea vs homozygous mutant (A/A) genotype (13.0% vs 40.0%). Taking UGT1A1 and TS genotypes together, lower incidence of grade 2-4 diarrhea was found in patients with non-3RG TS genotypes, when TA6/6 was compared with TA7/7 (35.3% vs 100.0%). No significant association with time to progression (TTP) and overall survival (OS) was observed with either UGT1A1 or TS gene polymorphisms, although slightly longer TTP and OS were found with UGT1A1*28 (TA6/6). Irinotecan PK was investigated in 34 patients, which showed high area under concentration curve (AUC) of irinotecan and SN-38, but low AUC ratio (SN-38G / SN-38) in those patients with UGT1A1*28 TA7/7.
CONCLUSION: A distinct distribution pattern of UGT1A1 genotypes in Chinese patients might contribute to relatively low toxicity associated with irinotecan and 5-fluorouracil in mCRC patients.
-
Citation: Wang Y, Shen L, Xu N, Wang JW, Jiao SC, Liu ZY, Xu JM.
UGT1A1 predicts outcome in colorectal cancer treated with irinotecan and fluorouracil. World J Gastroenterol 2012; 18(45): 6635-6644 - URL: https://www.wjgnet.com/1007-9327/full/v18/i45/6635.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i45.6635
The current management of metastatic colorectal cancer (mCRC) uses fluorouracil-based regimens in combination with either oxaliplatin or irinotecan. These regimens mainly differ in their toxicity profiles with neutropenia and late diarrhea being associated with irinotecan-based therapy versus neurotoxicity with oxaliplatin-based treatment.
In our previous studies on the relationship of UDP-glucuronosyltransferase1A1 (UGT1A1) polymorphisms and irinotecan-related diarrhea, we found that UGT1A1*28 genotype was significantly associated with the occurrence of diarrhea, while the polymorphisms of UGT1A7 and UGT1A9 variants were found to be unrelated. Mutated UGT1A1*28 genotype was seen infrequently in the Chinese population, therefore, this might explain the lower incidence of late diarrhea in our patients treated with irinotecan compared to that in the Caucasian population[1,2]. However, the role of UGT1A in clinical response to irinotecan- and 5-fluorouracil (5-FU)-based treatment, non-diarrhea toxicities and prognosis remains unclear. In addition, thymidylate synthetase (TS), the enzyme targeted by 5-FU, deserves more attention because most patients that receive irinotecan are treated with irinotecan and 5-FU in combination.
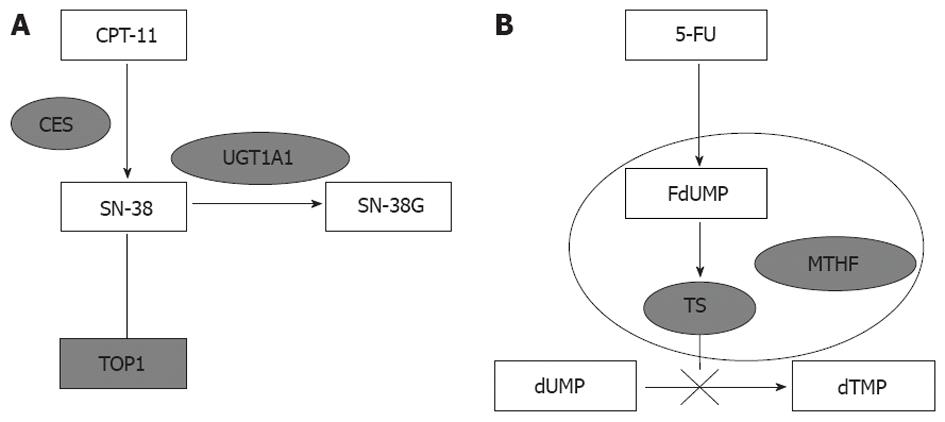
UGT1A polymorphisms have become the focus of irinotecan pharmacokinetics and toxicity research because they are involved in metabolism of cytotoxic SN-38 (an active metabolite of irinotecan) to inactive SN-38 glucuronide (SN-38G) (Figure 1). Di Paolo et al[3] have demonstrated that UGT1A1 polymorphisms are closely correlated with glucuronidation rates, and patients with higher concentration of SN-38G are less susceptible to irinotecan-induced toxicity. Although, several clinical trials have confirmed that patients carrying different genotypes of UGT1A1 had varied degrees of tolerance to irinotecan, it is still unclear whether UGT1A1 has any influence on treatment efficacy. Three studies tested if UGT1A1 isoforms had any impact on treatment outcome, however, their conclusions were inconsistent[4-6].
Irinotecan is often combined with 5-FU in mCRC treatment, therefore, 5-FU should be taken into consideration for response and toxicity evaluation as well. In vivo, the active metabolite of 5-FU inhibits TS activity by forming complexes with TS and 5, 10-methylene-tetrahydrofolate (Figure 1)[7]. Evidence supports the determinant role of TS promoter region polymorphism in 5-FU treatment efficacy and tolerance[8,9]. Given higher accuracy and consistency in testing polymorphisms, tremendous efforts have been put into using TS polymorphism as a genetic marker for predicting clinical response, toxicity and prognosis in mCRC patients treated with 5-FU[10-13].
This study aimed to evaluate the effects of UGT1A1/TS polymorphisms in Chinese mCRC patients treated with irinotecan and fluorouracil including toxicity and clinical outcome.
Two regimens were selected for this study: (1) FOLFIRI: irinotecan (Camptosar; Pfizer, United States) 180 mg/m2 90-min i.v. infusion on day 1; leucovorin 200 mg/m2i.v. infusion on days 1 and 2; followed on days 1 and 2 by 5-FU 400 mg/m2i.v. bolus, then 600 mg/m2i.v. over 22 h continuous infusion; repeated every 2 wk; (2) IFL: irinotecan 125 mg/m2 as a 90-min i.v. infusion on days 1, 8, 15 and 22; leucovorin 20 mg/m2i.v. infusion on days 1, 8, 15 and 22; 5-FU 500 mg/m2i.v. bolus on days 1, 8, 15 and 22; every 6 wk.
The criteria for inclusion were: at least 18 years old; histologically confirmed mCRC; failed or intolerant to oxaliplatin-based regimens; the Eastern Cooperative Oncology Group (ECOG) performance status 0-2; no chemotherapy at least 4 wk before study enrollment; life expectancy > 3 mo; neutrophil count ≥ 1.5 × 109/L, platelet count ≥ 8 × 1010/L, serum creatinine ≤ 1.25 upper limit normal (ULN), total bilirubin ≤ 1.25 ULN, alanine aminotransferase and aspartate aminotransferase ≤ 2.5 ULN (≤ 5 ULN with liver metastasis); normal electrocardiogram. Written informed consent was required and the study was performed in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines. All patients’ information and samples were collected from trial participating centers.
Tumor response was assessed by RECIST 1.0[14] based on the results of computed tomography. Time to progression (TTP) was defined as the time from the start of treatment to the date of progression. Overall survival (OS) was defined as the time from the start of the treatment to the date of death. Toxicity was assessed according to the NCI-CTC 3.0[15].
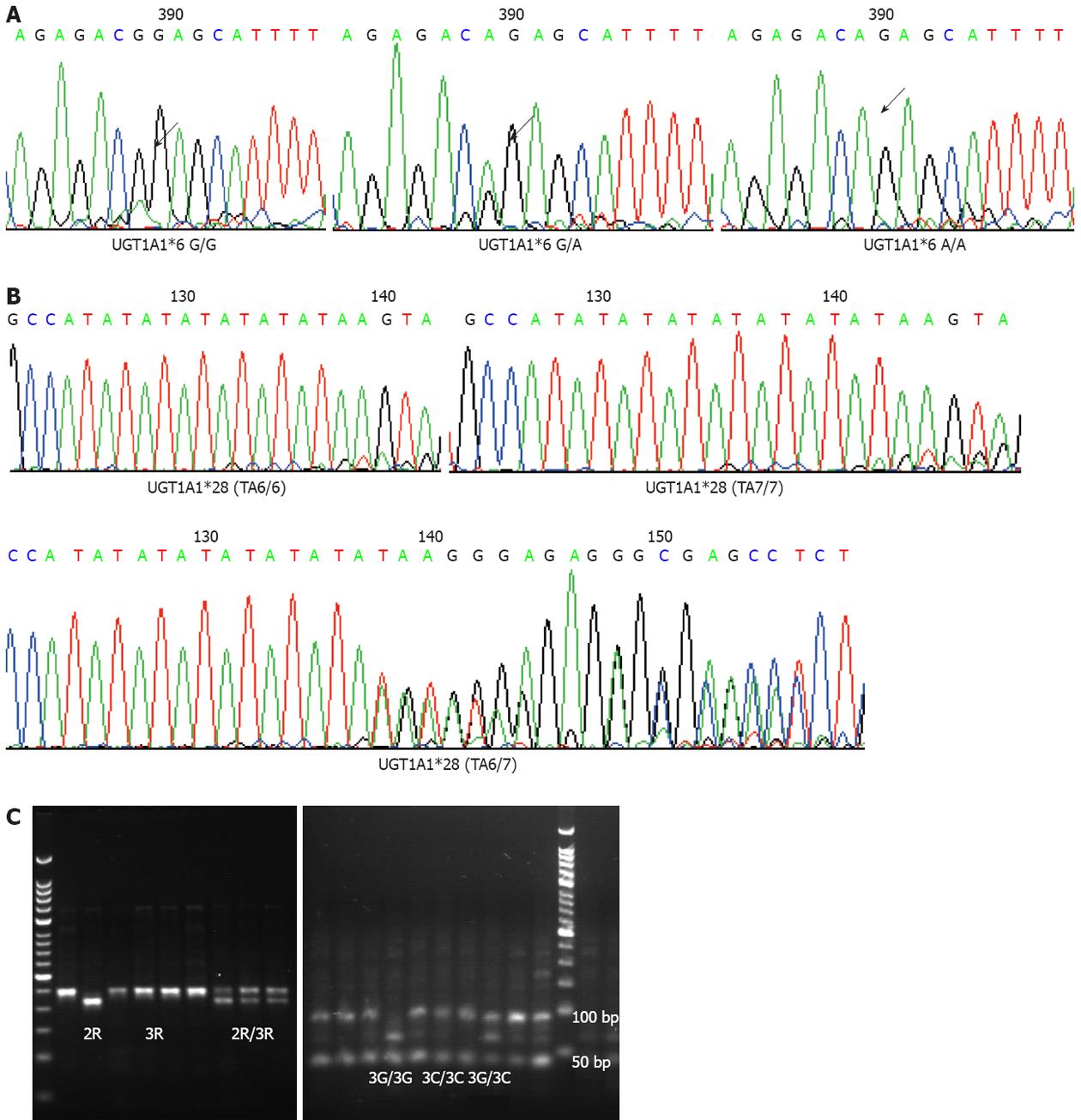
Two-milliliter peripheral blood samples were collected before starting treatment and frozen at -20 °C. Genomic DNA was extracted from these samples using the Peripheral Blood Extraction Kit (Tiangen, China). The fragments of UGT1A1*28 and UGT1A1*6 were amplified by polymerase chain reaction (PCR). The Primer 5.0 software was used to design the sense primer (5’-GCCAGTTCAACTGTTGTTGC-3’) and antisense primer (5’-GTCCGTCAGCATGACATCAA-3’). Each 25-μL PCR reaction mixture included 5 ng DNA template, 1 mmol/L dNTPs, 0.4 mmol/L MgSO4, 0.25 μmol/L primers, 0.5 U KOD-plus polymerase (TOYOBO, Japan) and 10 × KOD plus buffer. The PCR profile included 2.5 min denaturation at 94 °C, 35 cycles of 30 s at 94 °C, 60 s at 57 °C, 60 s at 68 °C, with a final 7-min extension at 68 °C. The PCR products were sequenced by ABI-3730 DNA analyzer and all single-nucleotide polymorphisms (SNPs) were analyzed by Polyphred 5.04 with additional manual proofreading.
Promoter enhancer region of 2R or 3R and G>C SNP in 3R were selected for testing. PCR-based restriction fragment length polymorphism was applied to detect these variants. The PCR conditions were same as described above with sense primer of 5’-GTGGCTCCTGCGTTTCCCCC-3’ and antisense primer of 5’-GCTCCGAGCCGGCCACAGGCATGGCGCGG-3’. The PCR profile included initial 15 cycles with annealing temperature at 63 °C, and another 30 cycles with annealing temperature at 62 °C. The PCR products were electrophoresized in 3% agarose gel to differentiate 215-bp 2R genotype and 243-bp 3R genotype. 3R gene polymorphism was detected via electrophoresis after enzyme digestion. After HaeIII treatment, 3RG genotype was separated to fragments of 66, 47, 45, 44, 28 and 13 bp, whereas 3RC genotype was separated to fragments of 94, 47, 45, 44 and 13 bp. The 20-μL reaction system contained 1 μL HaeIII Takara), 2 μL 10 × M buffer, 17 μL purified PCR products for 37 °C overnight incubation.
Five-milliliter heparinized blood samples were collected before administration of irinotecan, 1 and 1.5 h during infusion, and 0.25, 0.5, 1, 2, 4, 6, 8, 12, 24, 36, 48, 60 and 72 h after termination of infusion. Plasma samples were obtained by 3000 r/min centrifugation for 10 min and kept at -40 °C. The concentrations of the drug and its metabolites were determined by high performance liquid chromatography with postcolumn fluorescence derivatization (HPLC-FLD). Two hundred microliters plasma was added to 50 μL 1 ng/mL camptothecin, 150 μL methanol, and 200 μL acetonitrile, and vortexed for 2 min. The mixture was then centrifuged at 15 000 r/min for 10 min. Supernatant (200 μL) was placed with 100 μL 1 mol/L HCl before HPLC analysis. Agilent 1100 chromatographic system consisted of quaternary pumps, an automatic sample injection system, fluorescence detector, and Znertsil ODS-C18 Column. The mobile phases were eluted through the column and contained acetonitrile-sterile water (75:75 by volume) and phosphate buffer at pH 4. Irinotecan was kindly donated by Pfizer and camptothecin was provided by National Institute for the Control of Pharmaceutical and Biological Products. SN-38 was synthesized by Shanghai Zhangjiang Biochemical Company. Blank human plasma was obtained from Blood bank of 307 Hospital. The lowest limit of quantification of irinotecan, SN-38 and SN-38G were 50 ng/mL, 1.25 ng/mL and 5 ng/mL, respectively, with within-day and between-day imprecision < 15%. Cmax and Tmax were observed values. AUC0→t was acquired by linear trapezoidal approximation. AUC0→∞ = AUC0→t + Ct/ke.
The χ2 test was used to determine if the allele frequencies were matched to Hardy-Weinberg equilibrium. Associations between genotypes, clinical response and toxicity in Chinese and Caucasian populations were assessed by using Fisher’s exact test and Cochran-Armitage trend test. Kaplan-Meier estimates and the log-rank test were used in TTP and OS analysis. Cox regression and logistic regression models were examined in multivariate analysis with and without time variables, respectively. The linkage between genotypes and pharmacokinetic parameters was analyzed by Mann-Whitney U and Kruskal-Wallis tests. Two-sided tests were used to determine statistically significant P values (P < 0.05) using SPSS 13.0 software.
Between September 2005 and April 2009, 138 mCRC patients were enrolled. Of these, 130 patients were eligible for toxicity evaluation and 111 qualified for treatment efficacy evaluation. Of 138 patients, 108 received the FOLFIRI regimen, 29 the IFL regimen, and one irinotecan monotherapy. The median age of patients was 52 years (range: 26-81 years) with 63% being male. One hundred and eleven patients were followed until cancer progression or death. The overall response rate (complete + partial response) was 19.5%. The median TTP was 5.6 mo (95%CI: 3.9-7.3) and the median OS was 17 mo (95%CI: 11.9-22.1). The incidence of grade 3/4 neutropenia was 29.5%, and 17.4% participants developed grade 3/4 late diarrhea. However, the incidences in the FOLFIRI regimen were decreased, with 21.8% grade 3/4 neutropenia (95%CI: 13.7%-29.9%) and 10.9% grade 3/4 late diarrhea (95%CI: 4.8%-16.9%).
We tested UGT1A1 genotypes in 136 patients and TS promoter enhancer region in 125 (Figure 2). We observed that the response rates of UGT1A1*28 alleles TA6/6 and TA6/7 + TA7/7 were 17.9% and 24.2%, respectively, and those of UGT1A1*6 alleles G/G and G/A + A/A were 15.7% and 26.8%, respectively, without statistical significance observed.
Based on mRNA transcriptional activity, TS was categorized into 3RG/3RG homozygous group with highest activity, 3RG heterozygous group (3RG/3RC and 3RG/2RG) with median activity, and non-3RG group (3RC/3RC, 3RC/2RG and 2RG/2RG) with lowest activity. Although not statistically significant, 3RG/3RG homozygous group was found with lowest response rate (17.6%) (Table 1). Taking UGT1A1 and TS genotypes together, the subgroup with TA7/7 and non-3RG genotypes showed the highest response rate of 33.3%. Furthermore, under the same non-3RG background, no significant difference in response rate was found between TA6/6 and TA7/7 (13.6% vs 33.3%, P = 0.154) (Table 2). A multivariate logistic regression model was fitted to our data. The covariates included: chemotherapy regimen, treatment duration, ECOG performance status, sex, age, UGT1A1 genotypes, and TS promoter enhancer region, but only treatment duration (odds ratio 1.268, 95%CI: 1.027-1.565, P = 0.027) was closely linked to clinical response.
| Genotypes | Tumor response | |
| n (%) | P value1 | |
| UGT1A1*28 | ||
| TA 6/6 | 14 (17.9) | 0.446 |
| TA 6/7, TA 7/7 | 8 (24.2) | |
| UGT1A1*6 | ||
| G/G | 11 (15.7) | 0.217 |
| G/A, A/A | 11 (26.8) | |
| TS promotor | ||
| 3RG/3RG | 3 (17.6) | 0.880 |
| 3RG/3RC, 3RG/2RG | 5 (23.8) | |
| 3RC/3RC, 3RC/2RG, 2RG/2RG | 12 (19.4) | |
| UGT1A1*28 | TS promoter | ||
| 3RG/3RG | 3RG/3RC, 3RG/2RG | 3RC/3RC, 3RC/2RG, 2RG/2RG | |
| TA6/6 | 2/11 (18.1) | 4/13 (30.8) | 6/44 (13.6) |
| TA6/7 | 1/4 (25) | 1/6 (16.7) | 5/15 (33.3) |
| TA7/7 | 0/2 (0) | 0/2 (0) | 1/3 (33.3) |
The enzyme activities of UGT1A1*28 alleles from highest to lowest were TA6/6, TA6/7 and TA7/7. TA7/7 showed the highest incidence of grade 2-4 late diarrhea (P < 0.0005). Compared with TA6/6, the risk ratio of TA6/7 and TA7/7 was 2.6 (95%CI: 1.20-5.63). Although not statistically significant, a trend of higher incidence of grade 3/4 diarrhea was observed in patients with lower activity genotypes. A similar finding was obtained in the UGT1A1*6 study. G/G with the highest activity was associated with lowest incidence of grade 3/4 diarrhea (trend test P = 0.057). Compared with G/G, the risk ratio of grade 3/4 diarrhea in patients with G/A and A/A was 2.18 (95%CI: 0.87-5.42) (Table 3). In addition, UGT1A1*28 showed a similar correlation pattern with neutropenia. Grade 4 neutropenia was dominantly observed in TA7/7 patients (P = 0.043, trend test, P = 0.017). Compared with TA6/6, the risk ratio of grade 3/4 neutropenia in patients carrying TA6/7 and TA7/7 was 1.75 (95%CI: 0.79-3.88). By contrast, no association between UGT1A1*6 and grade 3/4 neutropenia was found (Table 3). Among three TS groups, homozygous 3RG/3RG showed least incidence of grade 3/4 late diarrhea (15.8%) and neutropenia (26.3%), although not significantly different from the other two groups (P > 0.05). Further subgroup analysis of non-3RG TS genotypes found that patients with TA6/6 were less prone to grade 2-4 late diarrhea than TA7/7 (35.3% vs 100.0%, P = 0.019). Meanwhile, a trend of lower incidence of grade 3/4 neutropenia was observed in patients with TA6/6, although the association was not statistically significant (29.4% vs 50.0%, P = 0.582).
| Genotypes | Delayed diarrhea | Neutropenia | ||||||||||
| Grades 2-4 | Grades 3/4 | Grades 3/4 | Grade 4 | |||||||||
| n (%) | P value1 | P value2 | n (%) | P value1 | P value2 | n (%) | P value1 | P value2 | n (%) | P value1 | P value2 | |
| UGT1A1*28 | ||||||||||||
| TA 6/6 | 25 (27.8) | < 0.0005 | < 0.0005 | 14 (15.6) | 0.228 | 0.178 | 23 (25.6) | 0.088 | 0.055 | 7 (7.8) | 0.043 | 0.017 |
| TA 6/7 | 2 (37.5) | 6 (18.8) | 10 (31.3) | 2 (6.3) | ||||||||
| TA 7/7 | 8 (100) | 3 (37.5) | 5 (62.5) | 3 (37.5) | ||||||||
| UGT1A1*6 | ||||||||||||
| G/G | 24 (31.2) | 0.564 | 0.346 | 10 (13) | 0.12 | 0.057 | 23 (29.9) | 1 | 0.748 | 8 (10.4) | 0.857 | 0.473 |
| A/G | 19 (39.6) | 11 (22.9) | 14 (29.2) | 4 (8.3) | ||||||||
| A/A | 2 (40) | 2 (40) | 1 (20) | 0 (0) | ||||||||
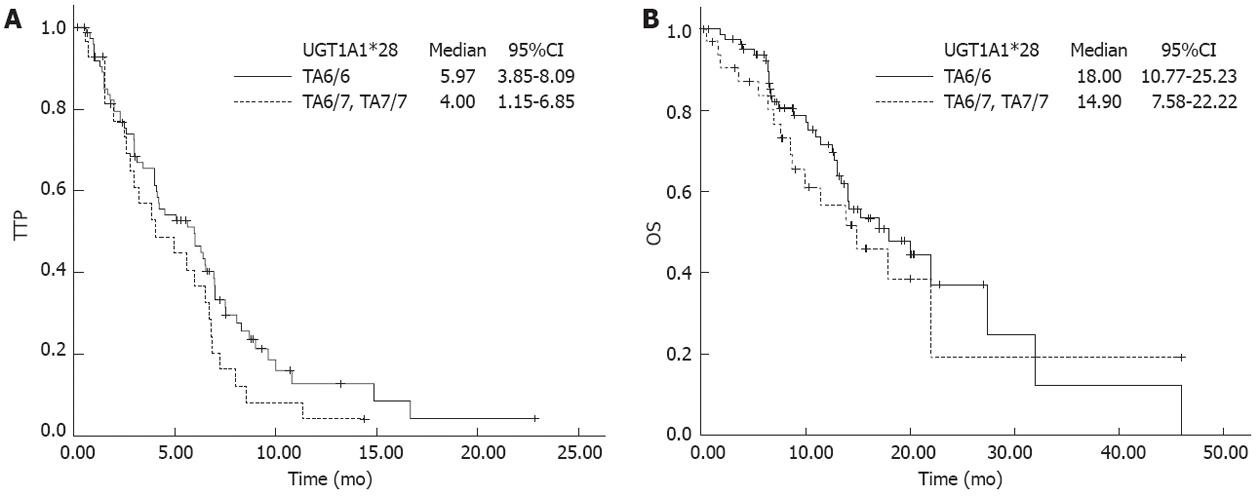
In 111 patients available for survival outcome analysis, no significant association with TTP and OS was observed with either UGT1A1*28 or UGT1A1*6, although slightly longer TTP and OS were found with UGT1A1*28 (TA6/6) (Figure 3). An additional study of 108 patients on TS revealed that there was no significant association between TS genotype and TTP/OS.
We further examined if the length of treatment duration was related to genotype. Patients with UGT1A1*28 (TA6/6) had a relatively longer treatment time compared with TA6/7 + TA7/7 genotype; near to statistical significance [9.57 ± 6.82 wk (range: 2-32 wk) vs 6.76 ± 5.05 wk (range: 1-19 wk), P = 0.078]. Although the 3RG homozygous group had longer treatment duration, it failed to display a significant correlation [3RG vs non-3RG, 9.03 ± 6.78 wk (range: 1-23 wk) vs 8.55 ± 6.16 wk (range: 2-32 wk), P = 0.895]. The longest TTP (6.9 mo) and OS (20 mo) were found in the subgroup with TA6/6 and 3RG genotypes, but the differences were still insignificant compared with other subgroups (P = 0.46 and 0.37, respectively). Independent variables that might affect OS were tested via Cox regression model analysis. These variables were ECOG performance status, age, sex, chemotherapy regimen, UGT1A1 and TS polymorphisms. Only chemotherapy regimen and ECOG were independent prognostic factors affecting OS. Higher ECOG score increased the death risk > 3 times [hazard ratio (HR): 3.325, 95 CI: 1.913-5.777, P < 0.0005). Compared with IFL, FOLFIRI showed a 69% decline in death risk (HR: 0.312, 95%CI: 0.132-0.738, P = 0.008). For TTP, the Cox model suggested that chemotherapy regimen and sex were independent prognostic factors. Treatment with FOLFIRI lowered the risk of disease progression by 78% (HR: 0.217, 95%CI: 0.104-0.451, P < 0.0005). Female sex reduced the progression risk to 60% (HR: 0.608, 95%CI: 0.387-0.968, P = 0.036).
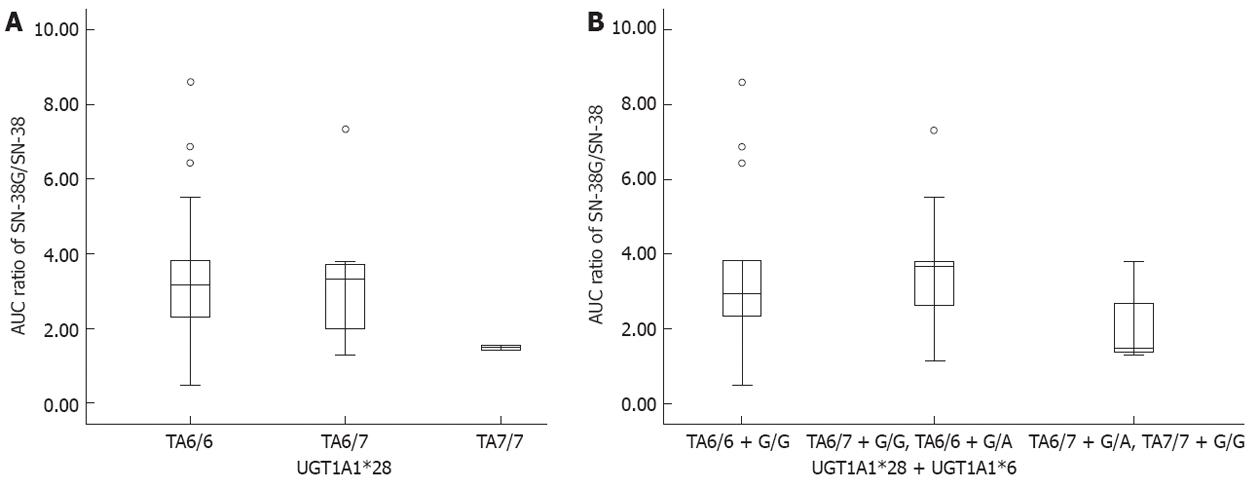
Thirty-four patients were enrolled in a pharmacokinetics study. Among UGT1A1*28 alleles, there were 22 with TA6/6, 10 with TA6/7, and two with TA7/7. Among UGT1A1*6 alleles, there were 25 with G/G and nine with G/A. Among them, two participants were found with a double heterozygous genetic profile; TA6/7 + G/A. The pharmacokinetic parameters showed that the area under concentration curve (AUC) values of irinotecan and SN-38 had an increasing trend among UGT1A1*28 alleles, and TA7/7 appeared to have the highest AUC values, although the AUC values of SN-38G in these three alleles showed no significant difference. Another indicator of SN-38 metabolic rate, the AUC ratio of SN-38G to SN-38 demonstrated that, compared with the other two alleles, the AUC ratio of TA7/7 was lower and close to statistical significance (P = 0.057). Due to no A/A allele being identified in UGT1A1*6, we were unable to study the effect of A/A on AUC. The combinations of UGT1A1*28 and UGT1A1*6 were divided into three groups: group 1 with wild type (TA6/6 + G/G); group 2 with one- mutated site variants (TA6/6 + G/A, TA6/7 + G/G); and group 3 with two-mutated site variants (TA6/7 + G/A, TA7/7 + G/A, TA7/7 + G/G). The higher AUC values of irinotecan and SN-38 and lower AUC ratio were found in group 3, and compared with the other two groups, the difference was near to statistical significance (P = 0.061 and 0.078, respectively) (Table 4). The plot of AUC ratios (Figure 4) clearly illustrated that group 3 had lower AUC ratio than the other two groups. Regarding UGT1A1*28, the AUC ratio of TA7/7 was less than half of the TA6/6 ratio (1.488 vs 3.178, P = 0.158).
| Genotype | AUC CPT-11 | AUC SN-38 | AUC SN-38G | AUC SN-38G/AUC SN-38 ratio | ||||
| Median | P value | Median | P value | Median | P value | Median | P value | |
| UGT1A1*28 | ||||||||
| TA6/6 | 5554.57 | 0.1631 | 176.40 | 0.1491 | 582.14 | 0.9881 | 3.178 | 0.1581 |
| TA6/7 | 6919.11 | 213.25 | 591.86 | 3.330 | ||||
| TA7/7 | 9049.03 | 390.00 | 584.63 | 1.488 | ||||
| UGT1A1*6 | ||||||||
| G/G | 5811.73 | 0.9532 | 189.62 | 0.3202 | 529.96 | 0.2342 | 2.924 | 0.5912 |
| G/A | 6582.17 | 231.50 | 679.17 | 3.759 | ||||
| UGT1A1*28+*6 | ||||||||
| TA6/6 + G/G | 5297.41 | 0.2821 | 169.62 | 0.0841 | 529.96 | 0.3861 | 2.924 | 0.1881 |
| TA6/7 + G/G, TA6/6 + G/A | 6649.50 | 217.87 | 679.17 | 3.680 | ||||
| TA7/7 + G/G TA6/7+ G/A | 6213.46 | 297.89 | 584.63 | 1.488 | ||||
| UGT1A1*28 | ||||||||
| TA6/6, TA6/7 | 6030.13 | 0.8842 | 191.92 | 0.0672 | 582.14 | 1.0002 | 3.177 | 0.0572 |
| TA7/7 | 9049.03 | 390.00 | 584.63 | 1.488 | ||||
| UGT1A1*28+*6 | ||||||||
| TA6/6 + G/G, TA6/7 + G/G, TA6/6 + G/A | 6030.13 | 0.7482 | 183.37 | 0.0612 | 582.14 | 0.9152 | 3.177 | 0.0782 |
| TA7/7 + G/G, TA6/7 + G/A | 6213.46 | 297.89 | 584.63 | 1.488 | ||||
Many western studies on the relationship of UGT1A polymorphisms and irinotecan toxicity and response have suggested that UGT1A1*28 is significantly associated with irinotecan-induced toxicity[16-18]. In particular, patients bearing UGT1A1*28 (TA7/7) have a high possibility to experience severe neutropenia and diarrhea. Based on this, a warning is labeled on irinotecan that patients with UGT1A1*28 (TA7/7) should start with a reduced dose of irinotecan, although the details of how to adjust the dose have not been specified[19]. By contrast, research in Asian countries has shown a lower incidence of UGT1A1*28 (TA7/7), while UGT1A1*6 (A/A) is more often found and may replace UGT1A1*28 as a key regulator in UGT1A1 expression[20-22].
Our early studies have found that few carrying UGT1A1*28 (TA7/7) might be the reason that less than expected diarrhea was observed in Chinese patients treated with irinotecan-based chemotherapy. Nevertheless, other UGT1A members, such as UGT1A1*6, UGT1A7*3 and UGT1A9*1 have a trend towards a high incidence of toxicity, and this association is racially different[1]. Although, recent data have implicated that UGT1A7 and UGT1A9 might be useful in predicting irinotecan-related toxicity[7], it might not be applicable for Asian patients because UGT1A1*6 is highly linked with UGT1A7 and UGT1A9 in the Asian population. In addition, Fujita et al[4] and Toffoli et al[5] have proposed that the homozygous UGTlAl*28 (TA7/7) is a predictive marker for better treatment outcome and longer survival time. The results of our study confirmed that only 6.6% of Chinese patients had UGT1A1*28 (TA7/7), whereas a higher frequency of TA6/6 was identified compared with that in Caucasians (69.9% vs 45.2%, P = 0.002). The allele distribution difference might account for lower incidence of severe late diarrhea and grade 3/4 neutropenia in Chinese patients treated with irinotecan. At the same time, we discovered that the association between UGT1A1*6 and grade 3/4 late diarrhea was close to statistically significance. Furthermore, we did not find that treatment efficacy was discounted by either UGT1A1*28 or UGT1A1*6 variants. Besides that, multivariate logistic regression analysis failed to show that UGT1A1/TS polymorphisms and chemotherapy regimen had any effect on clinical response, but indicated that longer treatment time increased response rate by 26%. No OS benefit was obtained in patients with high incidence of toxicity. This could be caused by shorter treatment duration due to drug intolerance.
Several studies have indicated that TS polymorphism is associated with 5-FU toxicity and prognosis. Although the results are inconclusive, it is well accepted that the allele 3RG/3RG is correlated with long OS and low toxicity[23,24]. We introduced TS polymorphism into treatment outcome and toxicity evaluation of irinotecan/5-FU chemotherapy and found that patients carrying UGT1A1*28 (TA6/6) and 3RG/3RG seemed to experience mild toxicity, but relatively low tumor response. Similar to UGT1A1*28, the OS of patients with 3RG/3RG was extended because therapy was well tolerated and its duration was prolonged. However, these observed differences did not reach statistical significance. We conjectured that the differences of treatment response and toxicity between TS genotypes could be narrowed by the synergistic effect of 5-FU combined with irinotecan regimen.
Unfortunately, the combination of two genetic markers was unable to increase the predictive accuracy. In our study, blood samples were used for polymorphism testing. Possibly, examining TS polymorphism and expression level in tumor tissues might yield more convincing data.
The in vivo metabolism of irinotecan is complicated. Our data showed that the plasma concentrations of irinotecan and its metabolites varied significantly in each patient. These variations are strongly related to the polymorphisms of many metabolic enzymes and transport proteins. We found that the AUC values of irinotecan and SN-38 were gradually elevated from TA6/6 to TA7/7, whereas the AUC ratios of SN-38G/SN-38 were decreased from TA6/6 to T7/7 (P = 0.057). A similar trend was found for UGT1A1*6. If UGT1A1*28 and UGT1A1*6 were taken together, the AUC ratios of group 1 (wild type), group 2 (one-mutated site) and group 3 (two mutated sites) were shifted from the highest to the lowest. Clearly, both alleles had effects on irinotecan metabolism.
Toffoli et al[5] concluded that for Caucasians, UGT1A1*28 not only affected irinotecan-related toxicity, but also changed the drug metabolic rate. On the contrary, reports from Asia suggested that UGT1A1*6 was involved in irinotecan pharmacokinetics and toxicity. Han et al[21] discovered that few UGT1A1*28 variants were found in the Korean population, while more UGT1A1*6 variants were observed. They claimed that the role of UGT1A1*28 in predicating irinotecan pharmacokinetics and toxicity could be replaced by UGT1A1*6 for Koreans. The research from Jada et al[24] also downplayed the role of UGT1A1*28 in the irinotecan metabolic pathway. However, Minami et al[20] contradicted this hypothesis by showing that both UGT1A1*28 and UGT1A1*6 were involved in irinotecan pharmacokinetics. Thus, for the Japanese population, the homozygous (UGT1A1*28/*28 and UGT1A1*6/*6) and the heterozygous (UGT1A1*6/*28) reduced the AUC ratios of SN-38G/SN-38. Our data favored Minami’s conclusion that both UGT1A1*28 and UGT1A1*6 participated in glucuronidation.
Derived from the AUC ratios of UGT1A1*28 and UGT1A1*6 variants, we estimated that genetic alterations could inactive the metabolic efficiency of SN-38 by 50%. Therefore we recommend a 50% dose reduction of irinotecan to treat group 3 patients with double-site mutations. Minami et al[20] described that the ratio of SN-38 AUC to irinotecan dose was 2.4 in group 3, while the ratio was 1.4 in group 1. Based on their findings, it would be rational to lower the initial dose of irinotecan by 50% for group 3 patients. Another study by Innocenti et al[25] investigated the association between the pharmacokinetics of UGT1A1*28 variants and neutropenia. They recommended that a 20% decrease in drug dose should be considered in patients with UGT1A1*28 (TA7/7).
To date, no clinical trial has used genotypes for determining the proper dose for initial irinotecan-based therapy. We recommend a 50% dose reduction of irinotecan for patients with double-site mutations to avoid severe toxicity, and ensure better efficacy with sufficient treatment given. However, accurate dose modification for patients with different UGT1A1 genotype is difficult. The limitations of our pharmacokinetics study were lack of UGT1A1*6 A/A information and small sample size. A large-scale prospective trial focused on dose modification of irinotecan will be our next work.
We thank Guang-Tao Hao, MD, for his contribution to the PK analysis.
Irinotecan is a prodrug that is hydrolyzed by carboxylesterase in vivo to form an active metabolite SN-38. SN-38 is further conjugated and detoxified by UDP-glucuronosyltransferase (UGT) to yield its β-glucuronide. Remarkable inter-individual variations in the pharmacokinetics and clinical outcomes have been reported. Genetic polymorphisms of UDP-glucuronosyltransferase1A1 (UGT1A1), especially UGT1A1*28 and UGT1A1*6 are important determinants of individual variations in Asian patients. 5-Fluorouracil (5-FU) is a fundamental component of all chemotherapic combinations for treatment of colorectal cancer (CRC). Thymidylate synthetase (TS) is the main intracellular target of fluoropyrimidines. The TS gene polymorphisms are known to influence the activity of TS, which are related to 5-FU clinical response. The allele containing the triple repeat (3R) in 5’-UTR of the TS is associated with 3-4-fold translational efficiency compared with the double repeat allele (2R), and a G>C base change in 3R alleles makes the transcriptional activity of the 3R allele as low as that of the 2R allele.
Current studies have revealed the relationship of UGT1A polymorphisms and irinotecan related toxicity and pharmacokinetics. The polymorphisms of UGT1A1*28 are considered to be important in the Caucasian population, while UGT1A1*6 seems to be more important than UGT1A1*28 in Asian studies. It is still unclear whether UGT1A1 has any influence on treatment efficacy. Three studies tested if UGT1A1 isoforms had any impact on treatment outcome, however, their conclusions were inconsistent. TS polymorphisms are of the most broadly studied genetic variants in CRC. It is accepted that the 3R allele is associated with increased expression levels of TS and poorer outcome in patients treated with 5-FU-based regimens. However, the role of gene polymorphisms of UGT1A1 and TS on clinical response to irinotecan and 5-FU-based treatment, non-diarrhea toxicities and prognosis remains unclear.
Only 6.6% of Chinese patients had UGT1A1*28 (TA7/7), whereas higher frequency of TA6/6 was identified compared with that in Caucasians (69.9% vs 45.2%, P = 0.002). Mutant variants of UGT1A1*28 and UGT1A1*6 were associated with increased toxicity and decreased SN-38 disposition, but the response rate did not increase accordingly, although this group of patients surely had a trend towards a better response. This probably related to shorter treatment time in patients carrying double-site mutations. Derived from the area under concentration curve ratios of UGT1A1*28 and UGT1A1*6 variants, authors estimated that genetic alterations could inactive the metabolic efficiency of SN-38 by 50%. Therefore, the authors recommend a 50% dose reduction of irinotecan to treat patients with double-site mutations.
To date, no clinical trial has used genotypes for determining the proper dose for initial irinotecan-based therapy. The authors recommend a 50% dose reduction of irinotecan for patients with double-site mutations to avoid severe toxicity, and ensure better efficacy with sufficient treatment given. However, accurate dose modification for patients with different UGT1A1 genotypes is difficult. A large-scale prospective trial focusing on dose modification of irinotecan will be next work.
Polymorphisms predict response and toxicity in patients with metastatic CRC treated with irinotecan and 5-FU. This study is interesting, and the manuscript is worthy of publication.
Peer reviewers: Takuya Watanabe, MD, PhD, Associate Professor, Department of Internal Medicine and Gastroenterology, Medical Hospital, The Nippon Dental University school of Life Dentistry at Niigata, 1-8 Hamauracho, Chu-o-ku, Niigata 951-8580, Japan; Dr. Luca Morelli, MD, UO, Anatomy and Histology, Ospedale S Chiara, Largo Medaglie d’Oro 9, 38100 Trento, Italy
S- Editor Gou SX L- Editor Kerr C E- Editor Li JY
| 1. | Wang Y, Xu JM, Shen L, Xu N, Wang JW, Jiao SC, Zhang JS, Song ST, Li J, Bao HY. Polymorphisms of UGT1A gene and irinotecan toxicity in Chinese colorectal cancer patients. Zhonghua Zhongliu Zazhi. 2007;29:913-916. [PubMed] |
| 2. | Braun MS, Richman SD, Quirke P, Daly C, Adlard JW, Elliott F, Barrett JH, Selby P, Meade AM, Stephens RJ. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: results from the UK MRC FOCUS trial. J Clin Oncol. 2008;26:2690-2698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 221] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 3. | Di Paolo A, Bocci G, Polillo M, Del Re M, Di Desidero T, Lastella M, Danesi R. Pharmacokinetic and pharmacogenetic predictive markers of irinotecan activity and toxicity. Curr Drug Metab. 2011;12:932-943. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 4. | Fujita K, Ando Y, Nagashima F, Yamamoto W, Eodo H, Araki K, Kodama K, Miya T, Narabayashi M, Sasaki Y. Genetic linkage of UGT1A7 and UGT1A9 polymorphisms to UGT1A1*6 is associated with reduced activity for SN-38 in Japanese patients with cancer. Cancer Chemother Pharmacol. 2007;60:515-522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 5. | Toffoli G, Cecchin E, Corona G, Russo A, Buonadonna A, D'Andrea M, Pasetto LM, Pessa S, Errante D, De Pangher V. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24:3061-3068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 259] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 6. | Cecchin E, Innocenti F, D'Andrea M, Corona G, De Mattia E, Biason P, Buonadonna A, Toffoli G. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol. 2009;27:2457-2465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 169] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 7. | Popat S, Matakidou A, Houlston RS. Thymidylate synthase expression and prognosis in colorectal cancer: a systematic review and meta-analysis. J Clin Oncol. 2004;22:529-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 344] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 8. | Martinez-Balibrea E, Abad A, Martínez-Cardús A, Ginés A, Valladares M, Navarro M, Aranda E, Marcuello E, Benavides M, Massutí B. UGT1A and TYMS genetic variants predict toxicity and response of colorectal cancer patients treated with first-line irinotecan and fluorouracil combination therapy. Br J Cancer. 2010;103:581-589. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 9. | Ren DN, Kim IY, Koh SB, Chang SJ, Eom M, Yi SY, Seong SH, Kim MD, Bronner MP, Cho MY. Comparative analysis of thymidylate synthase at the protein, mRNA, and DNA levels as prognostic markers in colorectal adenocarcinoma. J Surg Oncol. 2009;100:546-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 10. | Karlberg M, Ohrling K, Edler D, Hallström M, Ullén H, Ragnhammar P. Prognostic and predictive value of thymidylate synthase expression in primary colorectal cancer. Anticancer Res. 2010;30:645-651. [PubMed] |
| 11. | Park CM, Lee WY, Chun HK, Cho YB, Yun HR, Heo JS, Yun SH, Kim HC. Relationship of polymorphism of the tandem repeat sequence in the thymidylate synthase gene and the survival of stage III colorectal cancer patients receiving adjuvant 5-flurouracil-based chemotherapy. J Surg Oncol. 2010;101:22-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 12. | Kristensen MH, Pedersen PL, Melsen GV, Ellehauge J, Mejer J. Variants in the dihydropyrimidine dehydrogenase, methylenetetrahydrofolate reductase and thymidylate synthase genes predict early toxicity of 5-fluorouracil in colorectal cancer patients. J Int Med Res. 2010;38:870-883. [PubMed] |
| 13. | Gibson TB. Polymorphisms in the thymidylate synthase gene predict response to 5-fluorouracil therapy in colorectal cancer. Clin Colorectal Cancer. 2006;5:321-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 14. | Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer. 2009;45:228-247. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15860] [Cited by in RCA: 21602] [Article Influence: 1350.1] [Reference Citation Analysis (1)] |
| 15. | National Cancer Institute. Common Terminology Criteria for Adverse Events, version 3.0. National Institutes of Health, National Cancer Institute. 2006;. |
| 16. | Strassburg CP, Kalthoff S, Ehmer U. Variability and function of family 1 uridine-5'-diphosphate glucuronosyltransferases (UGT1A). Crit Rev Clin Lab Sci. 2008;45:485-530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 47] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 17. | Shulman K, Cohen I, Barnett-Griness O, Kuten A, Gruber SB, Lejbkowicz F, Rennert G. Clinical implications of UGT1A1*28 genotype testing in colorectal cancer patients. Cancer. 2011;117:3156-3162. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 18. | Gagné JF, Montminy V, Belanger P, Journault K, Gaucher G, Guillemette C. Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7-ethyl-10-hydroxycamptothecin (SN-38). Mol Pharmacol. 2002;62:608-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 264] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 19. | Palomaki GE, Bradley LA, Douglas MP, Kolor K, Dotson WD. Can UGT1A1 genotyping reduce morbidity and mortality in patients with metastatic colorectal cancer treated with irinotecan? An evidence-based review. Genet Med. 2009;11:21-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 101] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 20. | Minami H, Sai K, Saeki M, Saito Y, Ozawa S, Suzuki K, Kaniwa N, Sawada J, Hamaguchi T, Yamamoto N. Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet Genomics. 2007;17:497-504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 230] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 21. | Han JY, Lim HS, Park YH, Lee SY, Lee JS. Integrated pharmacogenetic prediction of irinotecan pharmacokinetics and toxicity in patients with advanced non-small cell lung cancer. Lung Cancer. 2009;63:115-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 82] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 22. | Gusella M, Frigo AC, Bolzonella C, Marinelli R, Barile C, Bononi A, Crepaldi G, Menon D, Stievano L, Toso S. Predictors of survival and toxicity in patients on adjuvant therapy with 5-fluorouracil for colorectal cancer. Br J Cancer. 2009;100:1549-1557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 74] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 23. | Fernández-Contreras ME, Sánchez-Hernández JJ, Guijarro M, Gisbert JP, Rivas N, García de Paredes ML, Hinojar-Gutiérrez A, Gamallo C. Influence of thymidylate synthase DNA polymorphisms and gender on the clinical evolution of patients with advanced colorectal cancer. Oncol Rep. 2010;23:1393-1400. [PubMed] |
| 24. | Jada SR, Lim R, Wong CI, Shu X, Lee SC, Zhou Q, Goh BC, Chowbay B. Role of UGT1A1*6, UGT1A1*28 and ABCG2 c.421C& gt; A polymorphisms in irinotecan-induced neutropenia in Asian cancer patients. Cancer Sci. 2007;98:1461-1467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 96] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 25. | Innocenti F, Undevia SD, Iyer L, Chen PX, Das S, Kocherginsky M, Karrison T, Janisch L, Ramírez J, Rudin CM. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol. 2004;22:1382-1388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 767] [Cited by in RCA: 723] [Article Influence: 34.4] [Reference Citation Analysis (0)] |