Published online Jan 21, 2012. doi: 10.3748/wjg.v18.i3.268
Revised: March 3, 2011
Accepted: March 8, 2011
Published online: January 21, 2012
AIM: To establish a multiple detection method based on comparative polymerase chain reaction (cPCR) and ligase detection reaction (LDR)/ligase chain reaction (LCR) to quantify the intestinal bacterial components.
METHODS: Comparative quantification of 16S rDNAs from different intestinal bacterial components was used to quantify multiple intestinal bacteria. The 16S rDNAs of different bacteria were amplified simultaneously by cPCR. The LDR/LCR was examined to actualize the genotyping and quantification. Two beneficial (Bifidobacterium, Lactobacillus) and three conditionally pathogenic bacteria (Enterococcus, Enterobacterium and Eubacterium) were used in this detection. With cloned standard bacterial 16S rDNAs, standard curves were prepared to validate the quantitative relations between the ratio of original concentrations of two templates and the ratio of the fluorescence signals of their final ligation products. The internal controls were added to monitor the whole detection flow. The quantity ratio between two bacteria was tested.
RESULTS: cPCR and LDR revealed obvious linear correlations with standard DNAs, but cPCR and LCR did not. In the sample test, the distributions of the quantity ratio between each two bacterial species were obtained. There were significant differences among these distributions in the total samples. But these distributions of quantity ratio of each two bacteria remained stable among groups divided by age or sex.
CONCLUSION: The detection method in this study can be used to conduct multiple intestinal bacteria genotyping and quantification, and to monitor the human intestinal health status as well.
- Citation: Tang ZR, Li K, Zhou YX, Xiao ZX, Xiao JH, Huang R, Gu GH. Comparative quantification of human intestinal bacteria based on cPCR and LDR/LCR. World J Gastroenterol 2012; 18(3): 268-274
- URL: https://www.wjgnet.com/1007-9327/full/v18/i3/268.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i3.268
Intestinal bacteria consist of more than 500 species with the concentrations ranging from 102-109/mL[1], and play important roles in human growth, immunity, drug metabolism, pathogenesis and health maintenance. Intestinal bacterial components can be divided into beneficial and pathogenic bacteria according to their effects on human health[2]. Previous research showed the quantity of Enterobacterim, Bifidobacterium, Enterococcus and Lactobacillus varied largely among the infants, middle/old-aged and diarrhea groups[3], which was helpful to know the status of human intestinal ecology. Quantitative assay of such species are of great value in clinical practice and scientific researches. However, most of the interested bacteria differ a lot in the amount between samples[4,5], which is a big challenge to quantitatively detect multiple intestinal bacteria simultaneously.
In this study, comparative polymerase chain reaction (cPCR) and ligase detection reaction (LDR)/ligase chain reaction (LCR) methods were first employed to quantify five intestinal bacterial species simultaneously based on their 16s rDNAs. The selected target bacteria consisted of three conditional pathogenic bacteria (Enterococcus, Enterobacterium, Eubacterium) and two beneficial bacteria Bifidobacterium and Lactobacillus.
cPCR has been used to quantify nucleic acids for many years[6]. In a cPCR reaction, the target and the internal reference templates have the same primer recognition sequence and length, and similar internal sequences, which guarantee the amplification efficiency between the templates[7]. Therefore, the amount of the amplified products can precisely reflect the initial concentrations of their own templates. The highly conserved sequences of the bacterial 16s rDNAs fit well with the requirement of cPCR. We applied one pair of universal primers to amplify multiple target bacterial DNAs simultaneously, each kind of bacterial 16s rDNA was a competitive template of the others.
LDR is an eminent method to genotype low-abundant DNA under high backgrounds with a high specificity[8,9]. LCR has even higher amplification efficiency than LDR, which was introduced to this study to detect the lower abundant templates. Given that the ligation efficiency of different probes kept constant among tubes under the same reaction conditions with the same running cycle number, the ratio of fluorescence signals of the LDR/LCR products could reflect their initial ratio of the template concentrations.
Standard curves were made to verify the feasibility of our method and were applied to subsequent quantification of samples. Two internal controls were added to monitor the whole detection flow. One was applied to the LDR detection, the other to the LCR detection. The internal controls were added into the mixed standard DNAs or sampled DNAs at a fixed concentration before PCR amplification.
Eighty-two fecal samples (45 from males and 37 from females) were used to test the method for specimen detection, and all target bacteria could be detected in these samples. The distributions of the quantity ratio between each two bacterial species were obtained. Samples were also divided into groups by age or gender, the distribution diversities between groups were analyzed by non-parametric test.
Bifidobacterium longum, Lactobacillus acidophilus and Lactobacillus casei strains were donated by Department of Medical Microbiology and Parasitology, Shanghai Jiao Tong University School of Medicine. Bifidobacterium longum was grown anaerobically in MRS broth with L-Cysteine hydrochloride at 37 °C for 48 h. Lactobacillus was grown anaerobically in MRS broth at 37 °C for 48 h.
Fecal samples were collected from healthy adults aged 20-86 years who had not received antibiotics or other intestinal drugs within 3 mo prior to sampling at the First Hospital of Suzhou University. Samples were collected in sterile bags, with the wet weight of 4 g. A fecal sample was added into 6 mL of sterile phosphate-buffered saline (PBS; 0.05 mmol/L, pH 7.4) and mixed by inverting and vortexed for 5-10 min. The sample was then centrifuged at 500 ×g for 5 min to collect the upper phase. The upper phase was then centrifuged to collect the bacterial cells in the pellets. The resulting pellets were washed in 20 μL pre-cooled ethanol for three times and then stored at 4 °C until use.
For cultured bacteria, genome DNA was extracted from 1 mL harvested culture using bacteria genome DNA extracting kit (HuaShun W6511, Shanghai, China). For fecal samples, 200 μL TE containing lysozyme (20 mg/mL) was added to each pretreated fecal sample, well mixed, and then incubated at room temperature for 40 min. The bacterial genome DNA was extracted using column bacterial genome DNA extraction kit (Sangon, Shanghai, China).
The specific 16s rDNA of each bacterial species was amplified by genus or species specific primers (Table 1) with the PCR product about 1500 bp in length. The amplified bacterial 16s rDNA was cloned in Escherichia coli (E. coli) DH5α by the pMD 18-T Vector system (Takara, Japan). Colonies carrying the specific inserts were cultured and their plasmid DNAs were extracted using a Spin Column Plasmid DNA Minipreps Kit (Sangon, Shanghai, China). The concentration of the extracted plasmid DNA was measured by ultraviolet spectrophotometer and then diluted to a fixed concentration as genus standard plasmid DNA (equivalent to 109/μL).
| Genus | Primer(5'→3') |
| Universal primer | F: CAGGATTAGATACCCTGGTAGT |
| R: TTGCGCTCGTTGCGGGACTT | |
| Enterococcus | F: CACCGGAGCTTGCTCCACCG |
| R: TGGCTCCAAAAGGTTACTTC | |
| Enterobacterium | F: AGAGCTTGCTCTCGGGTGAC |
| R:TAAGCTACCTACTTCTTTTGCAA | |
| Eubacterium | F: GCAACCCTCTCCGGAGGGAAGCG |
| R: TTCACCCCCCTCACCCTCCACAC | |
| Bifidobacterium | F: GGCTNGAGCTTGCTCCGGCT |
| R: GNCTCACCTTAGACGGCTCC | |
| Lactobacillus acidophilus | F: GAGTTTGATCCTGGCTCAGG |
| R: CTGTCCCACCTTAGRCGGCT | |
| Lactobacillus casei | F: GAGTTTGATCCTGGCTCAGG |
| R: CTGTCCCACCTTAGRCGGCT |
Universal primers and LDR probes were designed according to the DNA sequences of 16s rRNA gene available at GenBank.
For each kind of bacteria, more than fifty 16s rDNA sequences were used to align together (DNAsistant 2.0), a section containing three conserved regions and two variable regions was selected as the target segment. Universal primers were complementary to the conserved sequence flanking the target region; the variable region was the target of their specific LDR/LCR probes. Therefore, all the sequences of each bacterial species could be amplified by universal primers. LDR/LCR probes were genus-specific, except for Lactobacillus. As there was not a specific region matching with all Lactobacillus species, the Lactobacillus genus could be divided into two groups. Each group had a specific uniform sequence, and two specific probe sets were designed for typing the two groups of Lactobacillus genus.
The GenBank program Basic Local Alignment Search Tool was used to ensure that the proposed primers and probes were target specific. Ligation probes are listed in Table 2. Two sets of probe mixture for multiplex ligation reactions are shown in Table 1. Universal primers: up (5’-CAGGATTAGATACCCTGGTAGT-3’), down (5’-TTGCGCTCGTTGCGGGACTT-3’).
| Genus | Ligase detection reaction probes | Ligase chain reaction antisense probes | Product length (bp) |
| Enterococcus | F: (T)20-TTTGACCACTCTAGAGATAG | F: (T)10-TTGCCCCCGAAGGGGAAGCT | 80 |
| R: P-AGCTTCCCCTTCGGGGGCAA(T)20-FAM | R: CTATCTCTAGAGTGGTCAAA-(T)10 | ||
| Enterobacterium | F: (T)23-TTGGAGGTTGTGCCCTTGAG | F: CTCAAGGGCACAACCTCCAA-(T)10 | 85 |
| R: P-GCGTGGCTTCCGGAGCTAAC(T)22-FAM | R: (T)10-GTTAGCTCCGGAAGCCACGC | ||
| Eubacterium | F: (T)25-TTGACATATGGGTGAAGCGG | F: CCGCTTCACCCATATGTCAA-(T)10 | 90 |
| R: P-GGGAGACCCCGTGGCCGAGA(T)25-FAM | R: (T)10-TCTCGGCCACGGGGTCTCCC | ||
| Bifidobacterium | F: (T)28-GGATGTGGGGCCCGTTCCA | F: (T)10-TAGCTCCGACACGGAACCCG | 95 |
| R: P-CGGGTTCCGTGTCGGAGCTAT(T)27-FAM | R: TGGAACGGGCCCCACATCCA-(T)10 | ||
| Lactobacillus casei | F: (T)30-CAGGTCTTGACATCTTTTGA | F: (T)10-AAACCTGATCTCTCAGGTGA | 100 |
| R: P-TCACCTGAGAGATCAGGTTT(T)30-FAM | R: TCAAAAGATGTCAAGACCTG-(T)10 | ||
| Lactobacillus acidophilus | F: (T)33-GGTCTTGACATCTAGTGCAA | F: (T)10-GAACTCCGTATCTCTACGGA | 105 |
| R: P-TCCGTAGAGATACGGAGTTC(T)32-FAM | R: TTGCACTAGATGTCAAGACC-(T)10 | ||
| IC 1# | F: (T)30-CACAGGGCTTTCCACCATCCGTGTC | 110 | |
| R: P-GTAGCGGCCAAGCTGCCACGACAGG(T)30-FAM | |||
| IC 2# | F: (T)-33-GACATTCGGCAGGCAATCACAGCCT | F: (T)10-GTAAGGTCTTGCAAACGTTCACATC | 115 |
| R: P-GATGTGAACGTTTGCAAGACCTTAC(T)32-FAM | R: AGGCTGTGATTGCCTGCCGAATGTC-(T)10 |
Two oligonucleotides of internal control were synthesized to monitor the whole detecting flow. These oligonucleotides were approximately 310 bp in length, comparative to that of the bacterial 16s rDNA aplicon amplified with universal primers. Sequences at both ends were completely complementary to the universal primers, but the middle part was random. The internal control DNA was inserted into a plasmid and the concentration of internal control plasmid DNA was fixed to the same as bacterial standard plasmid DNA (equivalent to 109/μL).
Standard bacterial and internal control plasmid DNA was used to simulate the sample detection and verify the feasibility of the quantitative genotyping method. Each standard DNA was diluted to five series of concentrations. Six standard DNAs and two internal control DNAs were mixed together according to the symmetrically distributing test design[10] (Tables 2 and 3). The mixed templates were then applied to cPCR and LDR/LCR by two sets of mixed probes, and the fluorescence signal was collected by the ABI 377 sequencing system. Data of each two kinds of standard DNAs were used to obtain the linear equation. The ratio of two collected fluorescence signals of two templates and the ratio of their concentrations were used to simulate the linearity. The standard curves were prepared by combining all the equations.
The mixed standard DNA and sample DNA were processed as the same protocol. The cPCR reaction mixture consisted of 1 × PCR Buffer, 2 mmol/L MgCl2, 0.2 mmol/L deoxynucleoside triphosphate, 0.5 mmol/L universal primers, 1.0 U Taq DNA polymerase, and 1 μL of template DNA in a final volume of 10 μL. The PCR thermocycling program was: 95 °C for 15 min; 40 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s; and 72 °C for 7 min. The following LDR/LCR reaction mixture consisted of 1 × LDR Buffer, 0.1 mmol/L probe mixture No. 1 or No. 2 (Table 3), 4.0 U Taq ligase, and 1 μL of PCR products in a final volume of 10 μL. The LDR/LCR thermocycling program was: 94 °C for 2 min; 25 cycles of 94 °C for 30 s, and 60 °C for 2 min.
Before the detection, 105 copies of internal control template were added to each sample. The mixed samples were subject to cPCR and LDR (with probe mixture No. 1) along with designed mixed standard DNAs, 377 sequencer was used to collect the signals. If the fluorescence signal of any specific probe was not obtained by LDR, the PCR product was subject to LCR by probe mixture No. 2.
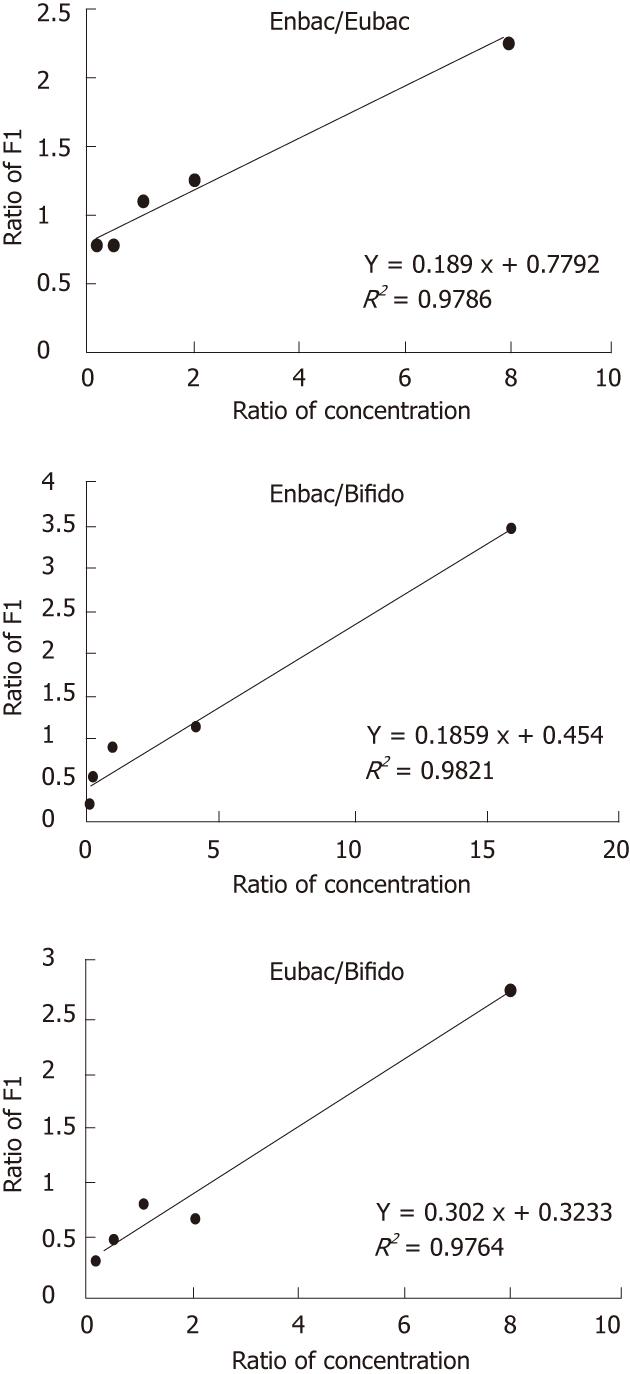
Standard DNAs were mixed at different concentrations (Table 4) and applied to LDR and LCR test simultaneously. The results showed that the fluorescence intensity of each two templates was correlated with their initial concentrations in good linearity by LDR probes (R2 > 0.97) (Figure 1) while the LCR probes could not contribute to a satisfactory linearity (R2 ranged from 0.57 to 0.96).
| Trial | Factor | |||||||
| Enterobacterium c | Eubacterium | Bifidobacterium | Enterococcus | Lactobacillus casei | Lactobacillus acidophilus | IC 1# | IC 2# | |
| Detected with probe mixture No. 1 | ||||||||
| 1 | 1 | 0.5 | 0.0625 | 0.01 | 0.005 | 0.000625 | 0.25 | 0.0025 |
| 2 | 0.5 | 0.0625 | 0.125 | 0.005 | 0.000625 | 0.00125 | 0.0625 | 0.000625 |
| 3 | 0.25 | 0.25 | 0.25 | 0.0025 | 0.0025 | 0.0025 | 1 | 0.01 |
| 4 | 0.125 | 1 | 0.5 | 0.00125 | 0.01 | 0.005 | 0.125 | 0.00125 |
| 5 | 0.0625 | 0.125 | 1 | 0.000625 | 0.00125 | 0.1 | 0.5 | 0.005 |
| Detected with probe mixture No. 2 | ||||||||
| 1 | 1 | 0.005 | 0.000625 | 0.01 | 0.005 | 0.000625 | 0.25 | 0.0025 |
| 2 | 0.5 | 0.000625 | 0.00125 | 0.005 | 0.000625 | 0.00125 | 0.0625 | 0.000625 |
| 3 | 0.25 | 0.0025 | 0.0025 | 0.0025 | 0.0025 | 0.0025 | 1 | 0.01 |
| 4 | 0.125 | 0.01 | 0.005 | 0.00125 | 0.01 | 0.005 | 0.125 | 0.00125 |
| 5 | 0.0625 | 0.00125 | 0.01 | 0.000625 | 0.00125 | 0.1 | 0.5 | 0.005 |
Eighty-two samples were detected for the six bacterial strains. The detected rates ranked as: Lactobacillus casei > Enterobacterium = Lactobacillus acidophilus > Bifidobacterium > Eubacterium > Enterococcus (Table 5). Eubacterium and Bifidobacterium had the largest quantities among the six bacteria, which could be detected by LDR probes. In Bifidobacterium, 46 of the 82 samples could be detected by LDR probe, the other 23 samples were detected by LCR probe. Lactobacillus and Enterococcus were able to be detected only by LCR probes, while the detection rate of the former was much higher than the latter (> 90% vs 35.37%).
| Bacterium | Enterococcus | Enterobacterium | Eubacterium | Bifidobacterium | Lactobacillus casei | Lactobacillus acidophilus | |
| Probe | LCR | LDR | LDR | LDR | LCR | LCR | LCR |
| No. of detected sample | 29 | 77 | 42 | 46 | 23 | 80 | 77 |
| Detection rate(%) | 35.37 | 93.90 | 51.22 | 84.15 | 97.56 | 93.90 | |
The ratio of the initial template concentrations between two bacteria were calculated by combining the LDR/LCR fluorescence signals and the standard curves. Eubacterium was selected as the comparing standard because of its highest detection rate and relatively steady quantity. The other bacteria were compared with Eubacterium except for Enterococcus because of its insufficient statistic data.
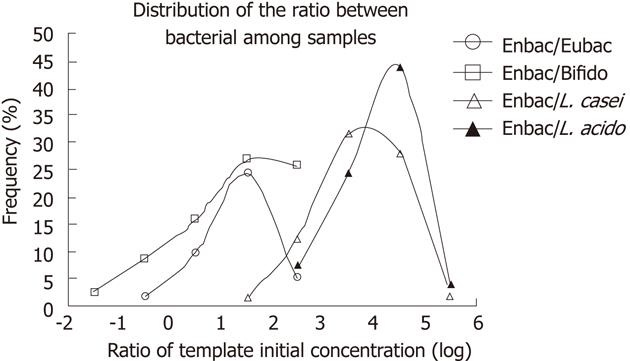
The ratio of two bacteria templates ranged from < 0.01 to > 104 deduced from the fluorescence signals and standard curves. In order to compensate the unsatisfactory linearity of the LCR products, the ratio of the initial template concentration was divided into several sections with 10-fold each. Frequencies of ratios in each section among all the samples were determined. The distributions of the quantity ratio between each two bacteria were plotted (Figure 2). Variations between distributions were analyzed by nonparametric test. There was on significant difference in the distributions between Enterobacterium/Eubacterium and Enterobacterium/Bifidobacterium (P = 0.605), which indicated that the quantity of Bifidobacterium and Enterobacterium kept constant in all the samples, while the distributions varied significantly among the other samples (P < 0.05).
These samples were divided into different groups by gender or age (young, 20-39 years; middle, 40-59; and old, above 60), and the bacterial ratio distribution was compared among groups through nonparametric test, without significant difference.
The Bifidobacterium standard DNA was selected and was serially diluted by 10-fold. The LDR and LCR detection could both obtain signals at a concentration of 10-9 of the original products (1-10 copies/μL) after 40 cycles of PCR and 25 cycles of LDR.
There is a detection limit of the concentration range of templates because of the amplification limit of PCR and LDR and the signal detection range of 377 sequencer.
The detection limit of LDR probes was tested by mixing standard DNAs of Eubacterium and internal control 1# at a serial ratio. The concentrations and detection results of the two components are shown in Table 6. The results showed that signals could be obtained by LDR probes simultaneously at the concentration varying between the two templates within 30 times.
| Eubac | LDR | Dilution times | 64 | 32 | 16 | 8 | 4 | 2 | 11 | 1 | 1 | 1 | 1 | 1 | 1 | ||
| Detection result | - | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| IC 1# | LDR | Dilution times | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | ||
| Detection result | + | + | + | + | + | + | + | + | + | + | + | + | + | ||||
| Bifido | LCR | Dilution times | 100 | 200 | 400 | 800 | 1600 | 3200 | 6400 | 12 800 | 25 600 | ||||||
| Detection result | + | + | + | + | + | + | + | + | - | ||||||||
| IC 1# | LDR | Dilution times | 11 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||
| Detection result | + | + | + | + | + | + | + | + | + | ||||||||
| Bifido | LCR | Detecion result | 128 | 64 | 32 | 16 | 8 | 4 | 2 | 12 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Dilution times | - | + | + | + | + | + | + | + | + | + | + | + | + | + | + | ||
| L. casei | LCR | Detection result | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 |
| Dilution times | + | + | + | + | + | + | + | + | + | + | + | + | + | + | - |
The detection limit between LDR and LCR probes was tested by Bifidobacterium and internal control 1# standard DNAs. The concentrations and detection results of the two components are listed in Table 6. The results showed that both LDR and LCR signals could be detected simultaneously, even at template concentrations varying within 10 000 times. The detection limit between LCR probes was tested by Bifidobacterium and Lactobacillus.casei standard DNAs. The concentrations and detection results of the two components are shown in Table 6.
LDR and LCR were both introduced to this study to genotype and comparatively quantify the final cPCR products, which extended the quantitative detection range significantly. LDR is a linear amplification process, while LCR is an exponential one. The templates of high quantity samples were detected by LDR probes, while those of low quantity by LCR probes. Therefore, the ligation products of different templates could be detected on one platform simultaneously.
The universality of universal primers was validated as they could direct the amplification of all the investigated bacterial standard DNAs. By mixed LDR/LCR probes, only one fluorescence signal of the target product was obtained after the LDR/LCR reaction templated by all bacterial standard PCR products. PCR products of other bacterial 16s rDNA including E. coli, Clostridium coccoides, Bacteroides and Clostridium leptum were not able to produce fluorescence signals (data not shown).
According to the standard curves, the ratio of LDR products and that of the initial template concentrations have perfect linear relationship because of the stable ligation efficiency in the linear amplification process. However, the relationship between the ratio of signals from LCR probes and that of the initial template concentrations did not show good linearity, which implied that the efficiency of the exponentially amplifying process could not keep stable in every cycle and between different reaction tubes. Nevertheless, on a large scale (10-fold), the amount of the LCR product was able to reflect the concentration of the templates, because of the limited number of reaction cycles (25 cycles) and the same reaction condition.
The quantity differences were shown between bacterial species, while the distribution of quantity ratio among species showed no difference in healthy human population whether grouped by sex or by age.
In this study, a precise genotyping and a stable comparative quantification were achieved by combining cPCR and LDR, and the detection throughout could be developed to tens of target bacteria at one time. Although the template concentration range of quantification needs to be extended and optimized, the cPCR and LDR/LCR method has shown its potential in intestinal bacteria detection in both basic research and clinical practice.
We would like to thank our colleagues from the Department of Medical Microbiology and Parasitology, Shanghai Jiao Tong University School of Medicine for donating the bacterial strains and those from the First Hospital of Suzhou University for their help in sample collection, and Chen B for her experimental assistance and cooperation.
Increasing researches suggested that the human health status was tightly correlated with the quantitative variation of intestinal bacterial components. However, it is a big challenge to quantify multiple intestinal bacterial components simultaneously, for the significantly quantitative variations between different components and individual samples.
Comparative quantification polymerase chain reaction (cPCR) has been used to quantify nucleic acids for many years. Due to the similarity of 16S rDNAs of different intestinal bacteria, it is hard to quantify multiple intestinal bacteria by cPCR alone. ligase detection reaction/ ligase chain reaction (LDR/LCR) is an eminent method to genotype low-abundant DNA under high backgrounds and its merit of high specificity fits for multiple detections. In this study, the authors tried to combine the advantages of both cPCR and LDR/LCR to fulfill multiple quantitative detections.
Using the universal primer target at the 16S rDNA of intestinal bacteria, cPCR could get amplifying product of nearly all species with similar efficiency. LDR could achieve precise genotyping and quantification of different bacteria simultaneously. This study demonstrated the possibility to realize accurate multiple quantification of intestinal bacteria by the combined cPCR and LDR/LCR.
By the detection method used in this study, the authors could quantify tens of intestinal bacteria simultaneously after test optimization. And it could be used to monitor the intestinal health status of human as well.
To quantify multiple intestinal bacterial components simultaneously, the authors described a multiple detection method for comparative quantification of 16S rDNA from different intestinal bacterial components based on cPCR and LDR/LCR methods. The results show differences between bacterial species, while the distribution of quantity ratio among species made no difference in healthy human population. This study indicates potential combination of cPCR and LDR/LCR method in measuring multiple intestinal bacterial components simultaneously.
Peer reviewer: Guang-Cun Huang, MD, PhD, Center for Clinical and Translational Research, The Research Institute at Nationwide Children's Hospital, Research II, WA 2112, Columbus, OH 43205, United States
S- Editor Tian L L- Editor Ma JY E- Editor Xiong L
1The standard concentration of standard DNA equivalent to 109/μL; 2The 100 times dilution of the standard concentration.+: the positive result; -: the negative result. LDR: Ligase detection reaction; LCR: Ligase chain reaction; Bifido:
| 1. | Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2194] [Cited by in RCA: 2114] [Article Influence: 96.1] [Reference Citation Analysis (0)] |
| 2. | Hart AL, Stagg AJ, Frame M, Graffner H, Glise H, Falk P, Kamm MA. The role of the gut flora in health and disease, and its modification as therapy. Aliment Pharmacol Ther. 2002;16:1383-1393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 3. | Hopkins MJ, Sharp R, Macfarlane GT. Age and disease related changes in intestinal bacterial populations assessed by cell culture, 16S rRNA abundance, and community cellular fatty acid profiles. Gut. 2001;48:198-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 399] [Cited by in RCA: 378] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 4. | Sghir1 A, Doré1 J, Mackie RI; Molecular diversity and phylogeny of human colonic bacteria. In: Bell CR, Brylinsky M, Johnson-Green. Molecular Ecology in Gastrointestinal Systems Microbial Biosystems: New Frontiers Proceedings of the 8th International Symposium on Microbial Ecology. Halifax, Canada: Atlantic Canada Society for Microbial Ecology. . |
| 5. | Wang M, Ahrné S, Antonsson M, Molin G. T-RFLP combined with principal component analysis and 16S rRNA gene sequencing: an effective strategy for comparison of fecal microbiota in infants of different ages. J Microbiol Methods. 2004;59:53-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 76] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 6. | Menzo S, Bagnarelli P, Giacca M, Manzin A, Varaldo PE, Clementi M. Absolute quantitation of viremia in human immunodeficiency virus infection by competitive reverse transcription and polymerase chain reaction. J Clin Microbiol. 1992;30:1752-1757. |
| 7. | Clementi M, Bagnarelli P, Manzin A, Menzo S. Competitive polymerase chain reaction and analysis of viral activity at the molecular level. Genet Anal Tech Appl. 1994;11:1-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 15] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 8. | Xiao Z, Xiao J, Jiang Y, Zhang S, Yu M, Zhao J, Wei D, Cao H. A novel method based on ligase detection reaction for low abundant YIDD mutants detection in hepatitis B virus. Hepatol Res. 2006;34:150-155. [PubMed] [DOI] [Full Text] |
| 9. | McNamara DT, Thomson JM, Kasehagen LJ, Zimmerman PA. Development of a multiplex PCR-ligase detection reaction assay for diagnosis of infection by the four parasite species causing malaria in humans. J Clin Microbiol. 2004;42:2403-2410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 54] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 10. | Test design and data process; Beijng, China: Beijing-Chemical Industry Press, 2005. . |