Published online Jul 21, 2012. doi: 10.3748/wjg.v18.i27.3617
Revised: April 18, 2012
Accepted: April 20, 2012
Published online: July 21, 2012
AIM: To evaluate anti-hepatitis B virus (HBV) activity and cytotoxicity of interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α) following lamivudine treatment of HepG2.2.15 cells.
METHODS: HepG2.2.15 cells were treated with 2 μmol/L lamivudine for 16 d (lamivudine group), cultured for 10 d, followed by 5 ng/mL TNF-α and 1000 U/mL IFN-γ for 6 d (cytokine group), or treated with 2 μmol/L lamivudine for 10 d followed by 5 ng/mL TNF-α and 1000 U/mL IFN-γ for 6 d (sequential group), or cultured without additions for 16 d (control group). Intracellular DNA was extracted from 3 × 105 HepG2.2.15 cells from each group. The extracted DNA was further purified with mung bean nuclease to remove HBV relaxed circular DNA that may have remained. Both HBV covalently closed circular DNA (cccDNA) and HBV DNA were examined with real-time polymerase chain reaction. The titers of hepatitis B surface antigen (HBsAg) and hepatitis B e antigen (HBeAg) were quantified with enzyme-linked immunosorbent assay. Cell viability was measured with the cell counting kit-8 assay.
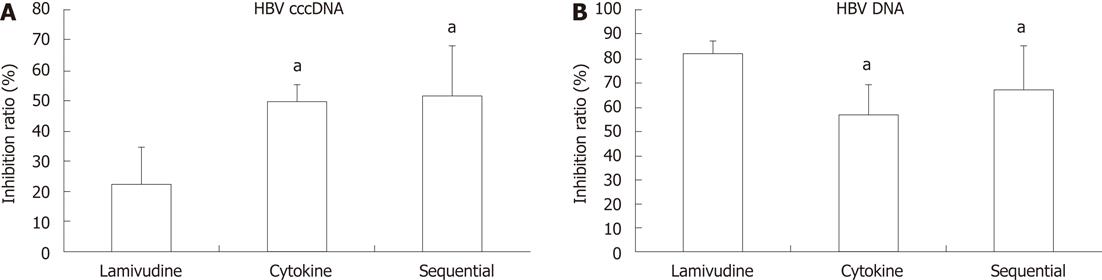
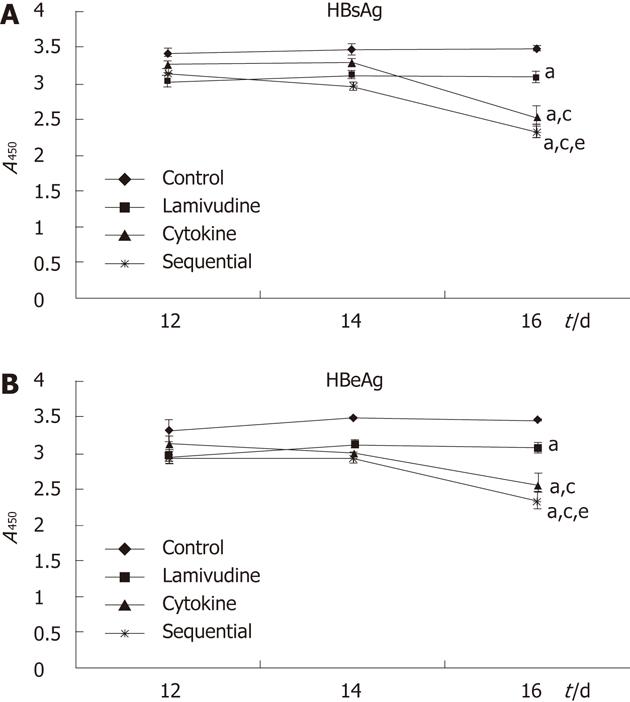
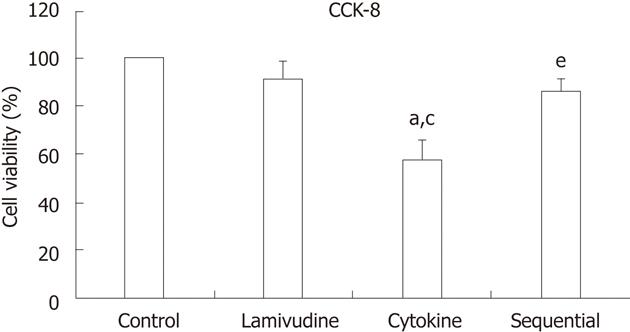
RESULTS: Compared to lamivudine alone (22.63% ± 0.12%), both sequential (51.50% ± 0.17%, P = 0.034) and cytokine treatment (49.66% ± 0.06%, P = 0.041) showed a stronger inhibition of HBV cccDNA; the difference between the sequential and cytokine groups was not statistically significant (51.50% ± 0.17% vs 49.66% ± 0.06%, P = 0.88). The sequential group showed less inhibition of HBV DNA replication than the lamivudine group (67.47% ± 0.02% vs 82.48% ± 0.05%, P = 0.014); the difference between the sequential and cytokine groups was not statistically significant (67.47% ± 0.02% vs 57.45% ± 0.07%, P = 0.071). The levels of HBsAg and HBeAg were significantly decreased in the sequential treatment group compared to the other groups [HBsAg: 3.48 ± 0.04 (control), 3.09 ± 0.08 (lamivudine), 2.55 ± 0.13 (cytokine), 2.32 ± 0.08 (sequential), P = 0.042 for each between-group comparison; HBeAg: 3.48 ± 0.01 (control), 3.08 ± 0.08 (lamivudine), 2.57 ± 0.15 (cytokine), 2.34 ± 0.12 (sequential), P = 0.048 for each between-group comparison]. Cell viability in the cytokine group was reduced to 58.03% ± 8.03% compared with control cells (58.03% ± 8.03% vs 100%, P = 0.000). Lamivudine pretreatment significantly reduced IFN-γ + TNF-α-mediated toxicity of HepG2.2.15 cells [85.82% ± 5.43% (sequential) vs 58.03% ± 8.03% (cytokine), P = 0.002].
CONCLUSION: Sequential treatment overcame the lower ability of lamivudine alone to inhibit cccDNA and precluded the aggressive cytotoxicity involving IFN-γ and TNF-α by decreasing the viral load.
- Citation: Shi H, Lu L, Zhang NP, Zhang SC, Shen XZ. Effect of interferon-γ and tumor necrosis factor-α on hepatitis B virus following lamivudine treatment. World J Gastroenterol 2012; 18(27): 3617-3622
- URL: https://www.wjgnet.com/1007-9327/full/v18/i27/3617.htm
- DOI: https://dx.doi.org/10.3748/wjg.v18.i27.3617
Hepatitis B virus (HBV) infection is a global health problem, and more than 350 million people are chronically infected with this virus worldwide[1]. Chronic hepatitis B infection is associated with an increased risk of cirrhosis, hepatic decompensation, and hepatocellular carcinoma[2]. Seven drugs are currently approved for the treatment for chronic hepatitis B, including conventional interferon (IFN)-α, pegylated IFN-α, and the nucleos(t)ide analogs lamivudine, adefovir, entecavir, telbivudine and tenofovir. However, none of the currently available drugs can eliminate viral covalently closed circular DNA (cccDNA) from the nucleus of infected hepatocytes[3,4].
During HBV infection, HBV cccDNA accumulates in cell nuclei where it persists as a stable episome and acts as a template for transcription of viral genes[5]. The elimination of cccDNA is a prerequisite for curing HBV infection[6]. Current knowledge suggests that clearance of HBV cccDNA occurs mainly through two pathways. The first is long-term and potent antiviral therapy, which effectively depletes the mature cytoplasmic nucleocapsid pool available for conversion into cccDNA. Based on mathematical models, the period needed to achieve complete clearance of intrahepatic cccDNA is 14.5 years[7]. Short-term antiviral therapy cannot completely exhaust the viral pool, which is stable and constitutes the source of renewal of viral production after cessation of therapy[8,9]. Nevertheless, long-term antiviral therapy can result in the development of antiviral resistance, and it is very expensive[10,11]. The second mechanism of cccDNA clearance involves two immune mechanisms: A cytotoxic T lymphocyte (CTL)-dependent cytolytic mechanism by which infected cells are eliminated and replaced with non-infected cells[12] and a non-cytolytic cytokine-dependent mechanism[13]. In addition to killing HBV-positive hepatocytes, HBV-specific CTLs can downregulate hepatocellular HBV gene expression and replication via a non-cytopathic, cytokine-induced process. These processes are mediated by inflammatory cytokines such as IFN-γ and tumor necrosis factor-α (TNF-α), which are secreted by CTLs following antigen recognition in the liver[14,15].
We previously reported that IFN-γ and TNF-α play a role in cell death of HBV- expressing HepG2.2.15 cells, a human hepatoblastoma cell line. Lamivudine treatment significantly reduces killing of HepG2.2.15 cells that is mediated by IFN-γ and TNF-α[16]. Lamivudine is the first oral nucleoside analog to be approved for the treatment of chronic hepatitis B patients, and it has been shown to suppress HBV replication by interfering with HBV DNA polymerase and disease activity, reducing the incidence of hepatocellular carcinoma and prolonging survival[17,18]. Lamivudine is potent and well tolerated, but its use is limited by the development of resistance. Viral breakthrough, which is defined as an abrupt increase in serum HBV DNA levels after a period of persistent suppression, may occur during lamivudine therapy[19]. Persistence of HBV cccDNA in hepatocytes plays a key role in viral persistence, reactivation of viral replication after cessation of antiviral therapy, and resistance to therapy[20]. To achieve effective suppression of HBV replication and elimination of HBV cccDNA and to avoid aggressive immune-mediated hepatitis and liver damage, we combined the above two established strategies of HBV cccDNA clearance. HBV-expressing HepG2.2.15 cells were initially given lamivudine to inhibit viral replication and reduce the level of HBV, and then the cells were given IFN-γ and TNF-α, two important immune mediators. We evaluated the antiviral potential of sequential treatment with lamivudine followed by IFN-γ and TNF-α in HepG2.2.15 cells, especially the potential to inhibit cccDNA amplification and eliminate its persistence.
HepG2.2.15 cells, which were derived from the stable transfection of HepG2 cells with a plasmid containing two head-to-tail dimers of the HBV genome, were used in this study. The HepG2.2.15 line supports persistent replication of HBV and produces intact HBV particles[21]. HBV cccDNA is detectable in the culture medium and intracellularly in HepG2.2.2.15 cells[22,23]. Cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 2 mmol/L l-glutamine, 50 IU/mL penicillin, 50 mg/L streptomycin, 500 mg/L G418, 5% (vol/vol) fetal bovine serum in 5 mL/L CO2 at 37 °C.
HepG2.2.15 cells were treated as follows: (1) with medium alone (control group); (2) 2 μmol/L lamivudine (GSK, London, United Kingdom) for 16 d (lamivudine group); (3) culture medium for 10 d, followed by 5 ng/mL recombinant human TNF-α (Invitrogen, CA, United State) and 1000 U/mL recombinant human IFN-γ (R and D Systems China, Shanghai, China) for 6 d (cytokine group); or (4) 2 μmol/L lamivudine for 10 d followed by 5 μg/L TNF-α and 1000 U/mL IFN-γ for 6 d (sequential group). The supernatant was replaced with fresh medium (with or without lamivudine and cytokines as per treatment protocols) every 2 d.
Intracellular DNA was extracted from 3 × 105 HepG2.2.15 cells from each group with the QIAamp Mini DNA kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The extracted product was further purified with mung bean nuclease (Gibco, Camarillo, CA, United State) to remove HBV relaxed circular DNA that may have remained. The purification reaction was carried out in a 50-μL volume containing 44 μL extracted DNA solution, 1 μL mung bean nuclease (10 000 U/mL), and 5 μL 10× mung bean nuclease buffer at 37 °C for 30 min. EGTA (2 μL; 100 mmol/L, pH 7.4) was then added to stop the reaction. HBV cccDNA was quantified with real-time polymerase chain reaction (PCR) using the ABI 7500 Real-Time PCR System (ABI, Foster City, CA, United States). Amplification was performed in a 20-μL reaction containing 2 μL isolated DNA and the Premix Ex Taq (Perfect Real-Time) kit (Takara, Dalian, China). The PCR primers were: forward 5’-TGAATCCYGCGGACGACC-3’ (nucleotides 1444-1461) and reverse 5’-CAGCTTGGAGGCTTGAACAG-3’ (nucleotides 1862-1881) (Y = C/T). The TaqMan probe was: 5’-FAM-CCTAATCATCTCWTGTTCATGTC-MGB-3’ (nucleotides 1836-1858) (W = A/T). For HBV cccDNA amplification, the cycling conditions were an initial incubation of 30 s at 95 °C followed by 40 cycles of 5 s at 95 °C and 34 s at 60 °C. The inhibition ratio of cccDNA was calculated as: (control-treatment)/control × 100%.
Intracellular DNA was extracted from 3 × 105 HepG2.2.15 cells from each group with the QIAamp Mini DNA kit. HBV DNA was quantified with real-time PCR using SYBR Premix Ex Taq (Perfect Real Time) (Takara) with the ABI 7500 Real-Time PCR System. The PCR primers were forward 5’-CCTCTTCATCCTGCTGCT-3’ and reverse 5’-AACTGAAAGCCAAACAGTG-3’. The PCR cycling program consisted of an initial denaturation step at 95 °C for 10 s, followed by 40 amplification cycles of 95 °C for 10 s, 60 °C for 10 s, 72 °C for 10 s, and 79 °C for 35 s, and then one cycle of 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s. The inhibition ratio of HBV DNA was calculated as: (control-treatment)/control × 100%.
HepG2.2.15 cell culture supernatants at days 12, 14 and 16 from each experimental group were collected and centrifuged at 118 ×g for 10 min to remove cellular debris and then transferred to clean tubes and stored at -20 °C until antigen measurement. The titers of hepatitis B surface antigen (HBsAg) and hepatitis B e antigen (HBeAg) were measured using a commercial enzyme-linked immunosorbent assay (ELISA) kit (Kehua, Shanghai, China) according to the manufacturer’s instructions.
After treatment for 10 d according to the protocols, cells were seeded at 3 × 104/well in 100 μL medium in 96-well plates and incubated overnight to allow cell adherence. Cells were then exposed to lamivudine and/or TNF-α + IFN-γ for 6 d. Ten microliters of 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt (Dojindo, Kumamoto, Japan) was added to each well, and the culture plate was incubated at 37 °C for 1 h. Absorbance was measured at 450 nm. The percent cell viability was calculated as: (value after treatment-blank)/(control-blank) × 100.
All data are expressed as the mean ± SD from three different experiments. Statistical analysis was performed with analysis of variance using SPSS l7.0 software. P < 0.05 was considered statistically significant.
The level of HBV cccDNA is an important parameter for determining the outcome of anti-HBV therapy. Both sequential treatment (51.50% ± 0.17%) and cytokine treatment (49.66% ± 0.06%) showed a stronger inhibition of HBV cccDNA compared to lamivudine alone (22.63% ± 0.12%) (51.50% ± 0.17% vs 22.63% ± 0.12%, P = 0.034; 49.66% ± 0.06% vs 22.63% ± 0.12%, P = 0.041). The difference between the sequential and cytokine groups was not statistically significant (P = 0.88) (Figure 1A).
The effect of the three different treatments on HBV DNA was investigated with real-time PCR. The sequential group (67.47% ± 0.02%) showed lower inhibition of HBV DNA than the lamivudine group (82.48% ± 0.05%) (P = 0.014). The inhibitory effect on HBV DNA between the sequential (67.47% ± 0.02%) and cytokine groups (57.45% ± 0.07%) was not significantly different (P = 0.071) (Figure 1B).
Secretion of HBsAg and HBeAg into cell media at days 12, 14 and 16 during the treatment period was detected with a commercial ELISA kit. After 16 d of treatment, we observed statistically significant decreases in the levels of HBsAg and HBeAg with sequential treatment compared to the other groups [HBsAg: 3.48 ± 0.04 (control), 3.09 ± 0.08 (lamivudine), 2.55 ± 0.13 (cytokine), 2.32 ± 0.08 (sequential), P = 0.042 for each between-group comparison; HBeAg: 3.48 ± 0.01 (control), 3.08 ± 0.08 (lamivudine), 2.57 ± 0.15 (cytokine), 2.34 ± 0.12 (sequential), P = 0.048 for each between-group comparison] (Figure 2).
The effect of each treatment on HepG2.2.15 cell viability was quantified with a CCK-8 assay. The viability in the cytokine group was reduced to 58.03% ± 8.03% compared with control cells (100%, P = 0.000). Lamivudine pretreatment significantly reduced TNF-α + IFN-γ-mediated toxicity of HepG2.2.15 cells [85.82% ± 5.43% (sequential) vs 58.03% ± 8.03% (cytokine), P = 0.002] (Figure 3).
HBV infection affects about 350 million people globally and is a leading cause of end-stage liver disease, hepatocellular carcinoma, and mortality[24]. HBV primarily infects hepatocytes by a single pathway, although the exact mechanism remains poorly understood. After endocytosis, nucleocapsids are released into the cytoplasm, and the relaxed circular DNA genome is transported to the nucleus where it is converted into cccDNA[25]. Persistence of HBV cccDNA in hepatocytes plays a key role in viral persistence, reactivation of viral replication after cessation of antiviral therapy, and resistance to therapy[20,26]. Thus, prevention of the formation of cccDNA and elimination of cccDNA to prevent reactivation of viral replication after withdrawal of antiviral therapy are of special clinical interest[27]. Although many new therapies have been developed against HBV infection[11], therapeutic elimination of HBV cccDNA remains a major challenge in curing chronic HBV infections. Novel approaches to improving current therapy for HBV infection are demanding but remain a global health priority.
IFN-γ and TNF-α are important immune mediators in host defense against HBV infection[28,29]. Synergistic antiviral activity of murine IFN-γ and murine TNF-α on HBV gene expression was previously demonstrated in an HBV-Met transgenic hepatocyte cell line[30]. IFN-γ and TNF-α abolished HBV gene expression and replication without killing the hepatocytes[31,32]. Nevertheless, IFN-γ and TNF-α also play a role in apoptotic cell death. IFN-γ inhibits large HBV surface protein storage disease and the ground-glass hepatocyte appearance, but it exacerbates inflammation and apoptosis in HBV surface protein-accumulating transgenic livers[33]. IFN-γ has been proposed to act as a pro-apoptotic regulator, triggering death receptors and other mediators[34]. HBV X protein (HBx) sensitizes cells to apoptosis by TNF-α[35]. We previously reported that IFN-γ and TNF-α play a role in cell death of HBV-expressing HepG2.2.15 cells. Expression of HBV sensitizes the cells to IFN-γ + TNF-α-mediated apoptosis. Increased expression of genes encoding interferon regulatory factor 1, c-myc and caspase 7 may be responsible for the synergistic induction of apoptosis by IFN-γ and TNF-α[16]. Cell death mediated by both IFN-γ and TNF-α may help eradicate the virus by eliminating residually infected cells. In patients with high levels of HBV DNA, however, aggressive immune-mediated processes involving IFN-γ and TNF-α may contribute to HBV-associated fulminant hepatic failure[16].
To achieve effective suppression of HBV replication and elimination of HBV cccDNA and also avoid aggressive immune-mediated hepatitis and liver damage, we combined the two established strategies of clearance of HBV cccDNA. We found that sequential treatment with lamivudine followed by IFN-γ and TNF-α had a stronger suppressive effect on HBV cccDNA replication and antigen expression compared to the lamivudine-only group. Although the differences between the sequential and cytokine groups in the inhibition ratio of both HBV cccDNA and HBV DNA were not statistically significant, sequential treatment showed stronger inhibition of antigen expression than cytokines only. More importantly, lamivudine pretreatment significantly reduced IFN-γ + TNF-α-induced cytotoxicity of HepG2.2.15 cells.
Our in vitro findings suggest that sequential treatment with IFN-γ and TNF-α following lamivudine not only overcame the lower ability of lamivudine alone to inhibit HBV cccDNA by cytokine treatment but also precluded the aggressive immune-mediated cytotoxicity involving IFN-γ and TNF-α by decreasing the viral load with lamivudine pretreatment. This novel treatment suggests a new strategy for treating HBV infection and may shorten the course of antiviral therapy, minimize the emergence of drug-resistant mutants, and reduce the financial burden of patients. Because our present work was carried out in vitro, and HBV cccDNA was not eliminated in the relative short study period, further in vivo studies are warranted to evaluate the sequential treatment protocol to combat HBV infection.
The authors thank Professor Zheng-Hong Yuan (Key Laboratory of Medical Molecular Virology, Fudan University) for providing HepG2.2.15 cells.
Hepatitis B virus (HBV) infection affects about 350 million people globally and is a leading cause of end-stage liver disease, hepatocellular carcinoma, and mortality. Although many new therapies have been developed against HBV infection, therapeutic elimination of HBV covalently closed circular DNA (cccDNA) remains a major challenge in curing chronic HBV infections. Novel approaches to improve current therapy for HBV infection are demanding but remain a global health priority.
Current knowledge suggests that clearance of HBV cccDNA occurs mainly through two pathways: The first is long-term and potent antiviral therapy, which effectively depletes the mature cytoplasmic nucleocapsid pool available for conversion into cccDNA. The second involves two immune mechanisms: A cytotoxic T lymphocyte (CTL)-dependent cytolytic mechanism by which infected cells are eliminated and replaced with non-infected cells and a non-cytolytic cytokine-dependent mechanism. These processes are mediated by inflammatory cytokines such as interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α), which are secreted by CTLs following antigen recognition in the liver. Nevertheless, IFN-γ and TNF-α have been reported to play a role in cell death of HBV-expressing HepG2.2.15 cells. Increased expression of genes encoding interferon regulatory factor 1, c-myc, and caspase 7 may be responsible for the synergistic induction of apoptosis by IFN-γ and TNF-α.
To achieve effective suppression of HBV replication and elimination of HBV cccDNA and also to avoid aggressive immune-mediated hepatitis and liver damage, the authors combined the two established strategies of clearance of HBV cccDNA. HBV-expressing HepG2.2.15 cells were initially treated with lamivudine to inhibit viral replication and reduce HBV levels, and then the cells were treated with IFN-γ and TNF-α, two important immune mediators, to achieve a synergistic effect on elimination of HBV. Based on the results, the authors found that sequential treatment with lamivudine followed by IFN-γ and TNF-α not only overcame the lower ability of lamivudine alone to inhibit HBV cccDNA by cytokine treatment but also precluded the aggressive immune-mediated cytotoxicity involving IFN-γ and TNF-α by decreasing the viral load with lamivudine pretreatment.
This novel treatment suggests a new strategy for treating HBV infection and may shorten the course of antiviral therapy, minimize the emergence of drug-resistant mutants, and reduce the financial burden of patients. Further in vivo studies are warranted to evaluate the sequential treatment protocol for combating HBV infection.
HBV cccDNA: During HBV infection, HBV cccDNA accumulates in cell nuclei where it persists as a stable episome and acts as a template for the transcription of viral genes. Persistence of HBV cccDNA in hepatocytes plays a key role in viral persistence, reactivation of viral replication after cessation of antiviral therapy, and resistance to therapy. The elimination of cccDNA is necessary for curing HBV infection.
The authors assessed the effect of sequential treatment with lamivudine followed by IFN-γ and TNF-α on HBV in HepG2.2.15 cells. It was observed that sequential treatment not only overcame the lower ability of lamivudine alone to inhibit HBV cccDNA but also precluded the aggressive immune-mediated cytotoxicity involving IFN-γ and TNF-α. The methodology is sound and the results obtained in cell culture studies have useful potential to be used in humans.
Peer reviewers: Dr. BS Anand, Professor, Digestive Diseases Section (111D), VA Medical Center, 2002 Holcombe Blvd., Houston, TX 77030, United State; Runu Chakravarty, PhD, ICMR Virus Unit, Kolkata, GB 4, 1st Floor ID and BG Hospital Campus, Kolkata 700010, India
S- Editor Lv S L- Editor A E- Editor Xiong L
| 1. | Huang LM, Lu CY, Chen DS. Hepatitis B virus infection, its sequelae, and prevention by vaccination. Curr Opin Immunol. 2011;23:237-243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 2. | Aspinall EJ, Hawkins G, Fraser A, Hutchinson SJ, Goldberg D. Hepatitis B prevention, diagnosis, treatment and care: a review. Occup Med (Lond). 2011;61:531-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 3. | European Association For The Study Of The Liver. EASL Clinical Practice Guidelines: management of chronic hepatitis B. J Hepatol. 2009;50:227-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1152] [Cited by in RCA: 1155] [Article Influence: 72.2] [Reference Citation Analysis (0)] |
| 4. | Krastev ZA. The "return" of hepatitis B. World J Gastroenterol. 2006;12:7081-7086. [PubMed] |
| 5. | Zoulim F. New insight on hepatitis B virus persistence from the study of intrahepatic viral cccDNA. J Hepatol. 2005;42:302-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 191] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 6. | Gao W, Hu J. Formation of hepatitis B virus covalently closed circular DNA: removal of genome-linked protein. J Virol. 2007;81:6164-6174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 166] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 7. | Tsiang M, Gibbs CS. Analysis of hepatitis b virus dynamics and its impact on antiviral development. Hepatitis B and D protocols. Vol. 2. Immunology, model systems, and clinical studies. Totowa, NJ: Humana Press Inc 2004; 361-377. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 8. | Zhu Y, Yamamoto T, Cullen J, Saputelli J, Aldrich CE, Miller DS, Litwin S, Furman PA, Jilbert AR, Mason WS. Kinetics of hepadnavirus loss from the liver during inhibition of viral DNA synthesis. J Virol. 2001;75:311-322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 194] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 9. | Abdelhamed AM, Kelley CM, Miller TG, Furman PA, Isom HC. Rebound of hepatitis B virus replication in HepG2 cells after cessation of antiviral treatment. J Virol. 2002;76:8148-8160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 10. | Ghany MG, Doo EC. Antiviral resistance and hepatitis B therapy. Hepatology. 2009;49:S174-S184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 175] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 11. | Hoofnagle JH, Doo E, Liang TJ, Fleischer R, Lok AS. Management of hepatitis B: summary of a clinical research workshop. Hepatology. 2007;45:1056-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 440] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 12. | Thimme R, Wieland S, Steiger C, Ghrayeb J, Reimann KA, Purcell RH, Chisari FV. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol. 2003;77:68-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 705] [Cited by in RCA: 763] [Article Influence: 34.7] [Reference Citation Analysis (1)] |
| 13. | Jung MC, Pape GR. Immunology of hepatitis B infection. Lancet Infect Dis. 2002;2:43-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 177] [Cited by in RCA: 194] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 14. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1700] [Cited by in RCA: 1712] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 15. | Guidotti LG, Chisari FV. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu Rev Immunol. 2001;19:65-91. [PubMed] |
| 16. | Shi H, Guan SH. Increased apoptosis in HepG2.2.15 cells with hepatitis B virus expression by synergistic induction of interferon-gamma and tumour necrosis factor-alpha. Liver Int. 2009;29:349-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 17. | Liaw YF, Sung JJ, Chow WC, Farrell G, Lee CZ, Yuen H, Tanwandee T, Tao QM, Shue K, Keene ON, Dixon JS, Gray DF, Sabbat J. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N Engl J Med. 2004;351:1521-1531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1739] [Cited by in RCA: 1740] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 18. | Papatheodoridis GV, Dimou E, Dimakopoulos K, Manolakopoulos S, Rapti I, Kitis G, Tzourmakliotis D, Manesis E, Hadziyannis SJ. Outcome of hepatitis B e antigen-negative chronic hepatitis B on long-term nucleos(t)ide analog therapy starting with lamivudine. Hepatology. 2005;42:121-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 160] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 19. | Leung N. Viral breakthrough during lamivudine therapy for chronic hepatitis B. Intervirology. 2003;46:344-349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 20. | Caruntu FA, Molagic V. CccDNA persistence during natural evolution of chronic VHB infection. Rom J Gastroenterol. 2005;14:373-377. [PubMed] |
| 21. | Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005-1009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 868] [Cited by in RCA: 939] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 22. | Liu MC, Wang GQ, Piao WH, Xi HL, Lu HY, Wang Y, Wang QH. [Dynamic expression of hepatitis B virus covalently closed circular DNA in 2.2.15 cell]. Zhonghua Shiyan He Linchuangbing Duxue Zazhi. 2005;19:391-394. [PubMed] |
| 23. | Liu MC, Yu M, Zhang NL, Gong WB, Wang Y, Piao WH, Wang QH, Wang GQ. Dynamic analysis of hepatitis B virus DNA and its antigens in 2.2.15 cells. J Viral Hepat. 2004;11:124-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 24. | Dienstag JL. Hepatitis B virus infection. N Engl J Med. 2008;359:1486-1500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 689] [Article Influence: 40.5] [Reference Citation Analysis (0)] |
| 25. | Schädler S, Hildt E. HBV life cycle: entry and morphogenesis. Viruses. 2009;1:185-209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 84] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 26. | Zoulim F. Antiviral therapy of chronic hepatitis B: can we clear the virus and prevent drug resistance? Antivir Chem Chemother. 2004;15:299-305. [PubMed] |
| 27. | Levrero M, Pollicino T, Petersen J, Belloni L, Raimondo G, Dandri M. Control of cccDNA function in hepatitis B virus infection. J Hepatol. 2009;51:581-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 398] [Cited by in RCA: 431] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 28. | Puro R, Schneider RJ. Tumor necrosis factor activates a conserved innate antiviral response to hepatitis B virus that destabilizes nucleocapsids and reduces nuclear viral DNA. J Virol. 2007;81:7351-7362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 29. | Bertoletti A, D'Elios MM, Boni C, De Carli M, Zignego AL, Durazzo M, Missale G, Penna A, Fiaccadori F, Del Prete G. Different cytokine profiles of intraphepatic T cells in chronic hepatitis B and hepatitis C virus infections. Gastroenterology. 1997;112:193-199. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 225] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 30. | Pasquetto V, Wieland SF, Uprichard SL, Tripodi M, Chisari FV. Cytokine-sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J Virol. 2002;76:5646-5653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 110] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 31. | Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity. 1996;4:25-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 840] [Cited by in RCA: 840] [Article Influence: 29.0] [Reference Citation Analysis (1)] |
| 32. | Guidotti LG, Chisari FV. To kill or to cure: options in host defense against viral infection. Curr Opin Immunol. 1996;8:478-483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 182] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 33. | Reifenberg K, Hildt E, Lecher B, Wiese E, Nusser P, Ott S, Yamamura K, Rutter G, Löhler J. IFNgamma expression inhibits LHBs storage disease and ground glass hepatocyte appearance, but exacerbates inflammation and apoptosis in HBV surface protein-accumulating transgenic livers. Liver Int. 2006;26:986-993. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 34. | Herzer K, Sprinzl MF, Galle PR. Hepatitis viruses: live and let die. Liver Int. 2007;27:293-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 35. | Su F, Schneider RJ. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor alpha. Proc Natl Acad Sci USA. 1997;94:8744-8749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 251] [Article Influence: 9.0] [Reference Citation Analysis (0)] |