Published online May 14, 2011. doi: 10.3748/wjg.v17.i18.2343
Revised: December 21, 2010
Accepted: December 28, 2010
Published online: May 14, 2011
AIM: To evaluate the association between xeroderma pigmentosum group D (XPD), genetic polymorphism Lys751Gln and esophageal cancer risk.
METHODS: We searched PubMed up to September 1, 2010 to identify eligible studies. A total of 10 case-control studies including 2288 cases and 4096 controls were included in the meta-analysis. Statistical analysis was performed with Review Manage version 4.2. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of the association.
RESULTS: The results suggested that there is no significant association between XPD Lys751Gln polymorphism and esophageal cancer susceptibility in the overall population. However, in subgroup analysis by histology type, a significant association was found between XPD Lys751Gln polymorphism and esophageal adenocarcinoma (for CC vs AA: OR = 1.25, 95% CI = 1.01-1.55, P = 0.05 for heterogeneity).
CONCLUSION: Our meta-analysis suggested that XPD Lys751Gln polymorphism may be associated with increased risk of esophageal adenocarcinoma.
- Citation: Yuan L, Cui D, Zhao EJ, Jia CZ, Wang LD, Lu WQ. XPD Lys751Gln polymorphism and esophageal cancer risk: A meta-analysis involving 2288 cases and 4096 controls. World J Gastroenterol 2011; 17(18): 2343-2348
- URL: https://www.wjgnet.com/1007-9327/full/v17/i18/2343.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i18.2343
Esophageal cancer, with a 5-year survival rate of < 20%, is considered as one of the most deadly malignancies[1,2]. It has already been identified that cigarette smoking, alcohol drinking, obesity, dietary factors, history of Barrett’s esophagus, and esophageal reflux disease can contribute to the development of esophageal cancer[3-6]. However, only a fraction of exposed individuals develop esophageal carcinoma, which suggests that genetic variations in sensitivity to carcinogen exposure and DNA repair capacity might be important inherited risk components in carcinogenesis[7,8]. DNA damage caused by exogenous, endogenous carcinogens or mutants is viewed as a crucial event in carcinogenesis. It can be repaired through activation of various pathways such as the nucleotide excision repair pathway (NER), base excision repair pathway (BER) and double-strand break pathway. The xeroderma pigmentosum group D (XPD) enzyme is involved in the NER pathway which plays an important role in the repair of bulky DNA adducts, such as pyrimidine dimmers, photo-products and cross-links[9]. Several single-nucleotide polymorphisms (SNPs) have been identified in the XPD gene. Among them, a polymorphism in the XPD gene, codon 751 A to C, resulting in an amino acid alteration from lysine (Lys) to glycine (Gln) has been reported to be associated with an increased susceptibility to lung cancer, and head and neck carcinoma[10-12]. Other malignancies such as esophageal cancer have also been investigated.
To date, many molecular epidemiological studies have explored the association between XPD Lys751Gln polymorphism and esophageal cancer risk[12-23]. However, results of these studies are controversial, which may be caused by the limitation of individual studies. Therefore, we performed a meta-analysis of 10 published case-control studies covering 6384 subjects in order to get a more precise evaluation of the relationship between the XPD Lys751Gln polymorphism and esophageal cancer risk.
We conducted a comprehensive search in the US National Library of Medicine’s PubMed database (as of September 1, 2010) using search terms including “XPD”, “xeroderma pigmentosum group D”, “ERCC2”, “excision repair cross-complementing rodent repair deficiency”, “polymorphism”, “esophageal”, “esophagus” and the combined phrases for all genetic studies on the relationship between XPD polymorphism and esophageal cancer. Moreover, we reviewed the references from original articles to search for more studies. No language restrictions were imposed. Two investigators conducted all searches independently. Studies were absorbed in this meta-analysis if they met the following criteria: (1) a case-control study of the XPD Lys751Gln polymorphism and esophageal cancer risk; and (2) the authors must offer the size of the sample, odds ratios (ORs) with 95% confidence intervals (CIs) or the information that can help infer the results in the articles. If data were reported in more than one study, the most recent and complete study was chosen for this analysis.
Two investigators independently extracted data and reached a consensus on all of the items. Information was collected from each article, including the first author’s name, year of publication, country of origin, racial descent of the subjects (categorized as Asian, European and mixed populations), sources of controls, genotyping method, histological type (categorized as esophageal adenocarcinoma and squamous cell carcinoma), number of different genotypes in cases and controls, Hardy-Weinberg equilibrium (HWE), and minor allele frequency in controls.
We assessed the strength of association between XPD Lys751Gln polymorphism and esophageal cancer risk by using ORs with 95% CIs which were obtained from the data given in the eligible studies. We evaluated the risk of codominant model (CC vs AA, CA vs AA), the dominant model (CA/CC vs AA), and recessive model (CC vs AA/CA), respectively. The between-study heterogeneity was investigated by Chi-square based Q-test[24], and it was considered significant if P < 0.05. The random-effects model (DerSimonian and Laird method) was then selected to pool the data[25]. Otherwise, the fixed-effects model (Mantel-Haenszel method) was used[26]. If heterogeneity was absent, these two models provided similar results. We used the funnel plot and the Egger weighted regression method (P < 0.05 was considered representative of statistical significance) to test possible publication bias in this meta-analysis[27]. All statistical analyses were performed in Statistical Analysis System software (v.9.13; SAS Institute, Cary, NC), and Review Manage (v.4.2; Oxford, England). All the tests were two-sided and the significant level was 0.05.
A total of 12 potential relevant studies that described the association between the XPD genetic polymorphisms and esophageal cancer were retrieved through PubMed. After reading the full articles, one study by Liu et al[17] was excluded since the subjects had also been included in a study by Tse et al[20]. One other study was excluded because it did not list data clearly enough for further analysis[23]. Finally, we identified 10 eligible studies including 2288 cases and 4096 controls in total. As summarized in Table 1, four studies were conducted in Asians, four studies in Europeans, and two in mixed subjects. In terms of histology type, there were 4 studies of esophageal adenocarcinoma (EADC), 4 studies of esophageal squamous cell carcinoma (ESCC) and 2 of both EADC and ESCC. Diverse genotyping methods including PCR-RFLP, TaqMan and iPLEX™ were used. The classic PCR-RFLP assay was used in 60% (6/10) studies. Six studies mentioned the quality control. The genotype distributions in the controls of all the included studies were in accordance with HWE.
| First author | Year | Country | Racial descent | Source of controls | Genotyping method | Histological type | Genotype distribution | P for HWE1 | C | |||||
| Case | Control | |||||||||||||
| AA | AC | CC | AA | AC | CC | |||||||||
| 2002 | China | Asian | Age matched | PCR-RFLP | ESCC3 | 367 | 63 | 3 | 451 | 70 | 3 | 0.87 | 0.07 | |
| Yu | 2004 | China | Asian | Age matched | PCR-RFLP | ESCC | 108 | 16 | 11 | 133 | 17 | 2 | 0.11 | 0.07 |
| Casson | 2005 | Canada | European | Randomly selected | PCR-RFLP | EADC4 | 31 | 21 | 4 | 34 | 46 | 15 | 0.93 | 0.40 |
| Ye2 | 2006 | Sweden | European | Age matched | PCR-RFLP | EADC | 27 | 51 | 18 | 198 | 203 | 71 | 0.11 | 0.37 |
| Ye2 | 2006 | Sweden | European | Age matched | PCR-RFLP | ESCC | 23 | 44 | 14 | 198 | 203 | 71 | 0.11 | 0.37 |
| Sobti | 2007 | Indian | Asian | Age matched | PCR-RFLP | ESCC | 52 | 61 | 7 | 63 | 77 | 20 | 0.64 | 0.37 |
| Doecke | 2008 | Australia | Mixed | Age matched | iPLEXTM | EADC | 108 | 124 | 31 | 575 | 588 | 174 | 0.22 | 0.35 |
| Ferguson | 2008 | Ireland | European | Randomly selected | TaqMan | EADC | 80 | 94 | 34 | 91 | 121 | 35 | 0.61 | 0.39 |
| Tse | 2008 | America | Mixed | Age matched | TaqMan | EADC | 104 | 159 | 49 | 193 | 208 | 52 | 0.72 | 0.34 |
| Pan2 | 2009 | America | European | Age matched | TaqMan | EADC | 137 | 153 | 56 | 187 | 216 | 53 | 0.43 | 0.24 |
| Pan2 | 2009 | America | European | Age matched | TaqMan | ESCC | 17 | 18 | 3 | 187 | 216 | 53 | 0.43 | 0.24 |
| Zhai | 2009 | China | Asian | Age matched | PCR-RFLP | ESCC | 167 | 31 | 2 | 148 | 51 | 1 | 0.12 | 0.13 |
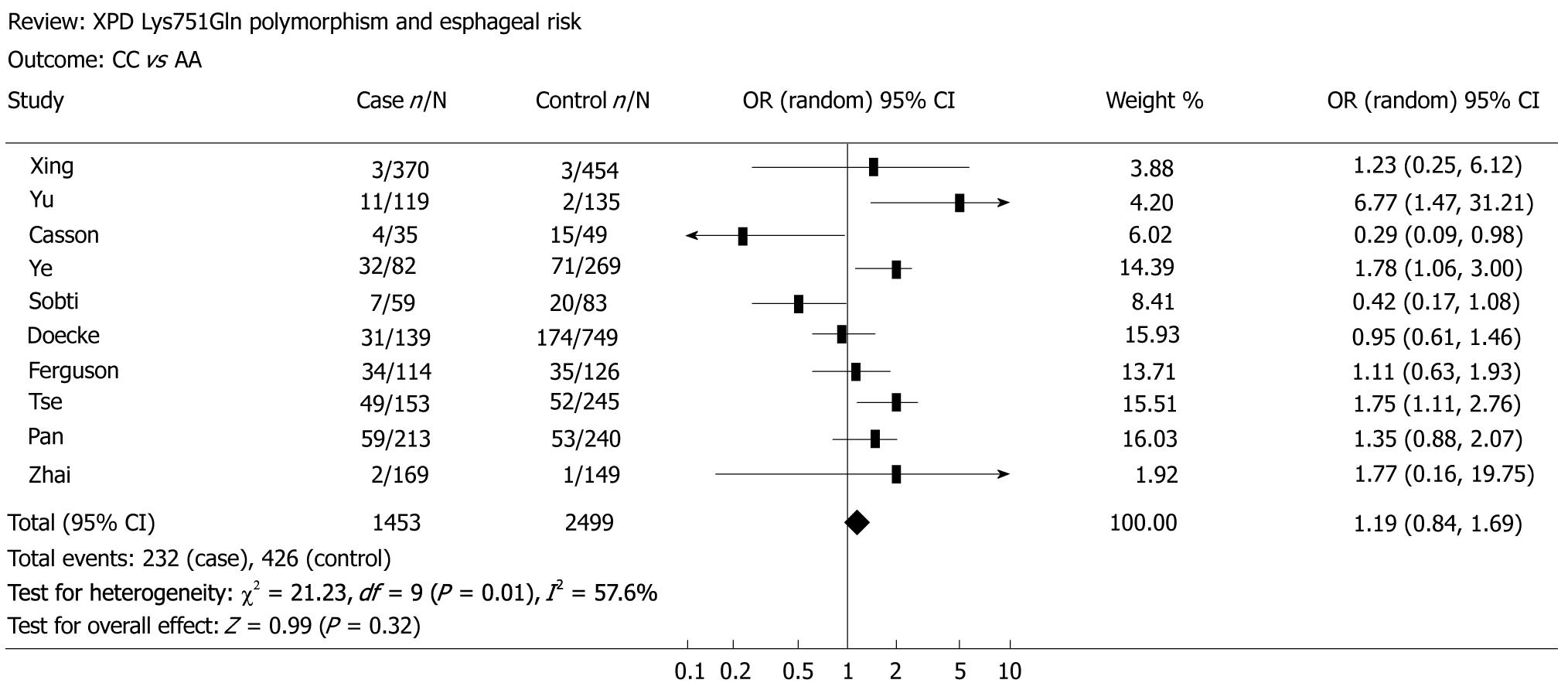
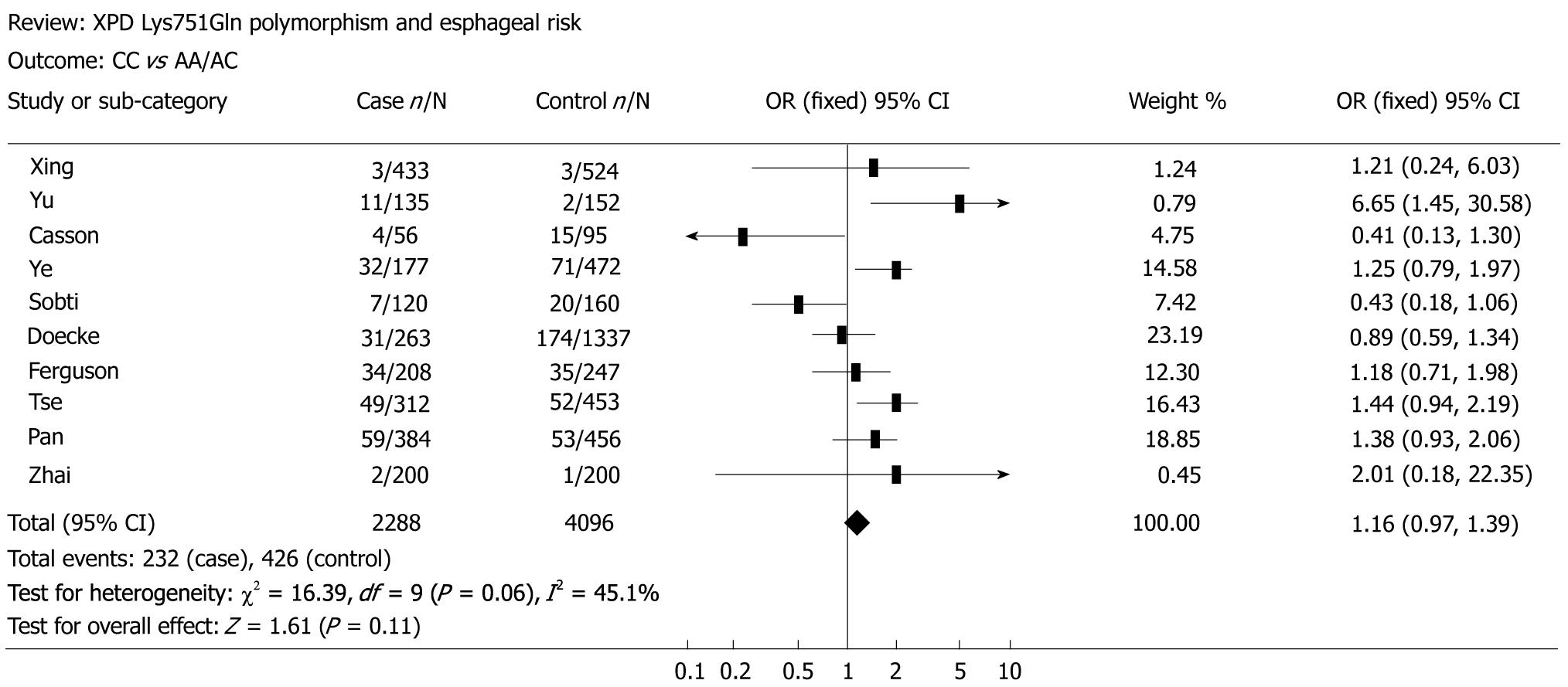
The main results of the meta-analysis on the association between XPD Lys751Gln polymorphism and esophageal cancer risk are shown in Table 2. Overall, no significant association was found between XPD Lys751Gln polymorphism and esophageal cancer risk (for CC vs AA: OR = 1.19, 95% CI = 0.84-1.69, P = 0.01 for heterogeneity, Figure 1; for CA vs AA: OR = 1.03, 95% CI = 0.83-1.27, P = 0.01 for heterogeneity; for the dominant model CA/CC vs AA: OR = 1.05, 95% CI = 0.85-1.32, P = 0.01 for heterogeneity; for the recessive model CC vs CA/AA: OR = 1.16, 95% CI = 0.97-1.39, P = 0.06 for heterogeneity, Figure 2). In subgroup analysis by ethnicity, we also did not detect any significant association in all genetic models. However, further analysis by histological type revealed that individuals carrying the variant homozygote CC genotype showed an elevated risk to EADC compared to those with the wild-type AA genotype (OR = 1.25, 95% CI = 1.01-1.55, P = 0.05 for heterogeneity).
| CC vs AA | CA vs AA | CA/CC vs AA | CC vs CA/AA | |||||
| OR (95% CI) | P1 | OR (95% CI) | P1 | OR (95% CI) | P1 | OR (95% CI) | P1 | |
| Total | 1.19 (0.84-1.69) | 0.01 | 1.03 (0.83-1.27) | 0.01 | 1.05 (0.85-1.32) | 0.01 | 1.16 (0.97-1.39) | 0.062 |
| Ethnicity | ||||||||
| European | 1.26 (0.95-1.65) | 0.052 | 1.00 (0.64-1.54) | 0.01 | 1.01 (0.65-1.56) | 0.01 | 1.20 (0.94-1.55) | 0.282 |
| Asian | 1.44 (0.37-5.66) | 0.02 | 0.91 (0.71-1.15) | 0.122 | 0.96 (0.63-1.45) | 0.03 | 1.47 (0.38-5.71) | 0.02 |
| Histological type | ||||||||
| EADC | 1.25 (1.01-1.55) | 0.052 | 1.13 (0.85-1.52) | 0.02 | 0.98 (0.68-1.41) | 0.01 | 1.18 (0.97-1.44) | 0.212 |
| ESCC | 1.24 (0.56-2.70) | 0.04 | 1.02 (0.73-1.41) | 0.04 | 1.05 (0.74-1.50) | 0.01 | 1.06 (0.71-1.58) | 0.062 |
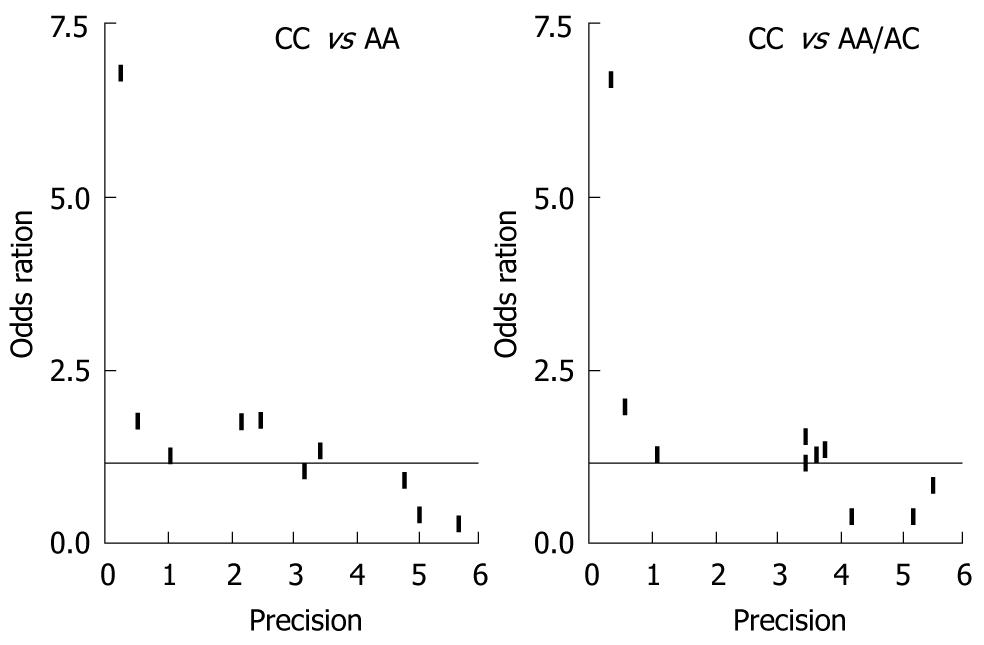
Funnel plot and the Egger’s test were performed to assess possible publication bias. As shown in Figure 3, no publication bias was revealed by the funnel plots, which was approximately symmetrical for the codominant model CC vs AA and the recessive model CC vs CA/AA. Statistical evidence from the results of Egger’s test confirmed the funnel plot symmetry (for CC vs AA: t = 2.23, P = 0.06; for CA vs AA: t = 2.03, P = 0.08; for CA/CC vs AA: t = 1.48, P = 0.18; for CC vs CA/AA: t = 2.33, P = 0.05).
Through analyzing data from the 10 eligible studies on relationship between XPD Lys751Gln polymorphism and esophageal cancer risk, we found no significant association between XPD Lys751Gln polymorphism and esophageal cancer risk in overall population. However, in the stratified analysis according to histological type, positive association were observed between XPD Lys751Gln polymorphism and elevated susceptibility to EADC.
The XPD gene has been mapped in chromosome 19q13.3. It spans over 20 kb, contains 23 exons and encodes the 761-amino acid protein. The XPD protein possesses both single-strand DNA-dependant ATPase and 5'-3' DNA helicase activities, which is essential for NER pathway and transcription[28]. The NER pathway generally removes bulky adducts caused by exogenous carcinogens, especially from cigarette smoking which is a well defined risk factor for EADC[29]. Any functional variation in NER pathway such as SNPs of key repair genes may lead to a deficiency in the DNA repair capacity (DRC) which is associated with a higher risk of cancer[28,30-32]. Benhamou et al[33] found that the single nucleotide substitution from A to C at codon 751 in the XPD gene leads to a complete change in the electronic configuration of the resulting amino acid, and reduces DNA repair efficiency. Many epidemiological studies have investigated the association between XPD Lys751Gln polymorphism and esophageal cancer, but the results were inconclusive. Xing et al[12] first explored the polymorphisms of DNA repair gene XPD and their associations with risk of esophageal squamous cell carcinoma in a Chinese population, but a Lys751Gln polymorphism in the XPD gene did not influence risk of ESCC in this study. However, two other studies on the relationship between XPD Lys751Gln polymorphism and ESCC revealed a contradictive result which suggested an increased risk of ESCC in association with the XPD 751 Gln/Gln genotype[13,15]. The more interesting finding revealed by Zhai et al[22] suggested an inverse association, which indicated that the XPD codon 751Gln allele was a protective factor rather than a risk factor to ESCC (OR = 0.628, 95% CI = 0.400-0.986). The first study on the association between XPD 751 codon polymorphism and EADC was conducted by Casson, who observed the protective effect of the homozygous variant of XPD Lys751Gln for EADC (OR = 0.24, 95% CI = 0.07-0.88)[14]. However, this result has not been supported by more studies. Both studies of Ye et al and Tse et al suggested that the XPD 751Gln allele was associated with an elevated risk for esophageal adenocarcinoma which is consistent with the result of our meta-analysis.
Some limitations of our meta-analysis should be acknowledged. Firstly, though it is known that the XPD gene has more polymorphisms than just Lys751Gln, we focused our meta-analysis on the most studied Lys751Gln polymorphism due to limited evidence on others. Secondly, some studies on this relationship were modified by some other potentially suspected factors such as BMI, smoking status, alcohol consumption, history of gastroesophageal reflux disease and lifestyle; however, our results were based on unadjusted estimates due to a lack of the original data. Finally, the XPD gene may influence susceptibility to esophageal cancer with other genes, but we did not conduct the gene-gene interactions analysis in our study.
In conclusion, our meta-analysis suggested that XPD Lys751Gln polymorphism may be a risk factor for esophageal adenocarcinoma. Large and well designed epidemiological studies will be necessary to combine genetic factors together with other potential risk factor such as smoking status, alcohol consumption and history of gastroesophageal reflux disease in order to validate the relationship between XPD Lys751Gln polymorphism and esophageal cancer risk.
Esophageal cancer is one of the most deadly malignancies. Many studies have explored the association between the Xeroderma pigmentosum group D (XPD) genetic polymorphism Lys751Gln and esophageal cancer risk, but the results are inconclusive and even controversial. It is, therefore, necessary to perform a meta-analysis in order to get a more precise evaluation of the relationship between the XPD Lys751Gln polymorphism and esophageal cancer risk.
The XPD gene is responsible for bulky adducts and repair of UV-induced DNA damage. To date, there have been many case-control studies on the association between XPD Lys751Gln and esophageal cancer risk, but few meta-analyses were conducted on this topic.
Our meta-analysis suggested that XPD Lys751Gln polymorphism might alter individuals’ susceptibility to esophageal adenocarcinoma. Further studies are needed to confirm it.
The finding that individuals carrying the variant homozygote CC genotype showed an elevated risk to esophageal adenocarcinoma indicates that genetic variations in the DNA repair protein may contribute to the risk of EADC. This meta-analysis gave a structured and systematic integration of information on the etiology of esophageal cancer, and the result may provide valuable information for researchers and clinicians.
This is a review of the ten previously published association studies and extracted the importance of esophageal adenocarcinoma. This information is worthwhile.
Peer reviewer: Haruhiko Sugimura, First Department of Pathology,Hamamatsu University School of Medicine, 1-20-1 Handayama, Higashi-ku, Hamamatsu, 431-3192, Japan
S- Editor Tian L L- Editor Rutherford A E- Editor Ma WH
| 1. | Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71-96. |
| 2. | Yang SJ, Yokoyama A, Yokoyama T, Huang YC, Wu SY, Shao Y, Niu J, Wang J, Liu Y, Zhou XQ. Relationship between genetic polymorphisms of ALDH2 and ADH1B and esophageal cancer risk: a meta-analysis. World J Gastroenterol. 2010;16:4210-4220. |
| 3. | Vaughan TL, Davis S, Kristal A, Thomas DB. Obesity, alcohol, and tobacco as risk factors for cancers of the esophagus and gastric cardia: adenocarcinoma versus squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 1995;4:85-92. |
| 4. | Drewitz DJ, Sampliner RE, Garewal HS. The incidence of adenocarcinoma in Barrett’s esophagus: a prospective study of 170 patients followed 4.8 years. Am J Gastroenterol. 1997;92:212-215. |
| 5. | Mayne ST, Risch HA, Dubrow R, Chow WH, Gammon MD, Vaughan TL, Farrow DC, Schoenberg JB, Stanford JL, Ahsan H. Nutrient intake and risk of subtypes of esophageal and gastric cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:1055-1062. |
| 6. | Lagergren J, Bergström R, Lindgren A, Nyrén O. Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N Engl J Med. 1999;340:825-831. |
| 7. | Poulsen HE, Loft S, Wassermann K. Cancer risk related to genetic polymorphisms in carcinogen metabolism and DNA repair. Pharmacol Toxicol. 1993;72 Suppl 1:93-103. |
| 8. | Ishibe N, Kelsey KT. Genetic susceptibility to environmental and occupational cancers. Cancer Causes Control. 1997;8:504-513. |
| 10. | Sturgis EM, Zheng R, Li L, Castillo EJ, Eicher SA, Chen M, Strom SS, Spitz MR, Wei Q. XPD/ERCC2 polymorphisms and risk of head and neck cancer: a case-control analysis. Carcinogenesis. 2000;21:2219-2223. |
| 11. | Stern MC, Johnson LR, Bell DA, Taylor JA. XPD codon 751 polymorphism, metabolism genes, smoking, and bladder cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1004-1011. |
| 12. | Xing D, Tan W, Wei Q, Lin D. Polymorphisms of the DNA repair gene XPD and risk of lung cancer in a Chinese population. Lung Cancer. 2002;38:123-129. |
| 13. | Yu HP, Wang XL, Sun X, Su YH, Wang YJ, Lu B, Shi LY, Xiong CL, Li YY, Li F. Polymorphisms in the DNA repair gene XPD and susceptibility to esophageal squamous cell carcinoma. Cancer Genet Cytogenet. 2004;154:10-15. |
| 14. | Casson AG, Zheng Z, Evans SC, Veugelers PJ, Porter GA, Guernsey DL. Polymorphisms in DNA repair genes in the molecular pathogenesis of esophageal (Barrett) adenocarcinoma. Carcinogenesis. 2005;26:1536-1541. |
| 15. | Ye W, Kumar R, Bacova G, Lagergren J, Hemminki K, Nyrén O. The XPD 751Gln allele is associated with an increased risk for esophageal adenocarcinoma: a population-based case-control study in Sweden. Carcinogenesis. 2006;27:1835-1841. |
| 16. | Sobti RC, Singh J, Kaur P, Pachouri SS, Siddiqui EA, Bindra HS. XRCC1 codon 399 and ERCC2 codon 751 polymorphism, smoking, and drinking and risk of esophageal squamous cell carcinoma in a North Indian population. Cancer Genet Cytogenet. 2007;175:91-97. |
| 17. | Liu G, Zhou W, Yeap BY, Su L, Wain JC, Poneros JM, Nishioka NS, Lynch TJ, Christiani DC. XRCC1 and XPD polymorphisms and esophageal adenocarcinoma risk. Carcinogenesis. 2007;28:1254-1258. |
| 18. | Ferguson HR, Wild CP, Anderson LA, Murphy SJ, Johnston BT, Murray LJ, Watson RG, McGuigan J, Reynolds JV, Hardie LJ. No association between hOGG1, XRCC1, and XPD polymorphisms and risk of reflux esophagitis, Barrett’s esophagus, or esophageal adenocarcinoma: results from the factors influencing the Barrett’s adenocarcinoma relationship case-control study. Cancer Epidemiol Biomarkers Prev. 2008;17:736-739. |
| 19. | Doecke J, Zhao ZZ, Pandeya N, Sadeghi S, Stark M, Green AC, Hayward NK, Webb PM, Whiteman DC. Polymorphisms in MGMT and DNA repair genes and the risk of esophageal adenocarcinoma. Int J Cancer. 2008;123:174-180. |
| 20. | Tse D, Zhai R, Zhou W, Heist RS, Asomaning K, Su L, Lynch TJ, Wain JC, Christiani DC, Liu G. Polymorphisms of the NER pathway genes, ERCC1 and XPD are associated with esophageal adenocarcinoma risk. Cancer Causes Control. 2008;19:1077-1083. |
| 21. | Pan J, Lin J, Izzo JG, Liu Y, Xing J, Huang M, Ajani JA, Wu X. Genetic susceptibility to esophageal cancer: the role of the nucleotide excision repair pathway. Carcinogenesis. 2009;30:785-792. |
| 22. | Zhai XD, Mo YN, Xue XQ, Zhao GS, Gao LB, Ai HW, Ye Y. XRCC1 codon 280 and ERCC2 codon 751 polymorphisms and risk of esophageal squamous cell carcinoma in a Chinese population. Bull Cancer. 2009;96:E61-E65. |
| 23. | Ma WJ, Lv GD, Zheng ST, Huang CG, Liu Q, Wang X, Lin RY, Sheyhidin I, Lu XM. DNA polymorphism and risk of esophageal squamous cell carcinoma in a population of North Xinjiang, China. World J Gastroenterol. 2010;16:641-647. |
| 24. | Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820-826. |
| 25. | Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719-748. |
| 26. | DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177-188. |
| 27. | Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629-634. |
| 28. | Lunn RM, Helzlsouer KJ, Parshad R, Umbach DM, Harris EL, Sanford KK, Bell DA. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis. 2000;21:551-555. |
| 29. | Gammon MD, Schoenberg JB, Ahsan H, Risch HA, Vaughan TL, Chow WH, Rotterdam H, West AB, Dubrow R, Stanford JL. Tobacco, alcohol, and socioeconomic status and adenocarcinomas of the esophagus and gastric cardia. J Natl Cancer Inst. 1997;89:1277-1284. |
| 30. | Benhamou S, Tuimala J, Bouchardy C, Dayer P, Sarasin A, Hirvonen A. DNA repair gene XRCC2 and XRCC3 polymorphisms and susceptibility to cancers of the upper aerodigestive tract. Int J Cancer. 2004;112:901-904. |
| 31. | Hu JJ, Hall MC, Grossman L, Hedayati M, McCullough DL, Lohman K, Case LD. Deficient nucleotide excision repair capacity enhances human prostate cancer risk. Cancer Res. 2004;64:1197-1201. |
| 32. | Li C, Hu Z, Liu Z, Wang LE, Strom SS, Gershenwald JE, Lee JE, Ross MI, Mansfield PF, Cormier JN. Polymorphisms in the DNA repair genes XPC, XPD, and XPG and risk of cutaneous melanoma: a case-control analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:2526-2532. |
| 33. | Benhamou S, Sarasin A. ERCC2/XPD gene polymorphisms and cancer risk. Mutagenesis. 2002;17:463-469. |