Published online Apr 21, 2011. doi: 10.3748/wjg.v17.i15.2037
Revised: December 12, 2010
Accepted: December 19, 2010
Published online: April 21, 2011
AIM: To investigate the expression patterns of human differentiated embryo chondrocyte 1 (DEC1) in hepatocellular carcinoma (HCC) and corresponding adjacent non-tumor and the normal liver tissues, the association between DEC1 expression and histopathological variables and the role of DEC1 in hepatocarcinogenesis.
METHODS: The expression of DEC1 was detected immunohistochemically in 176 paraffin-embedded sections from 63 patients with HCC and 50 subjects with normal liver tissues.
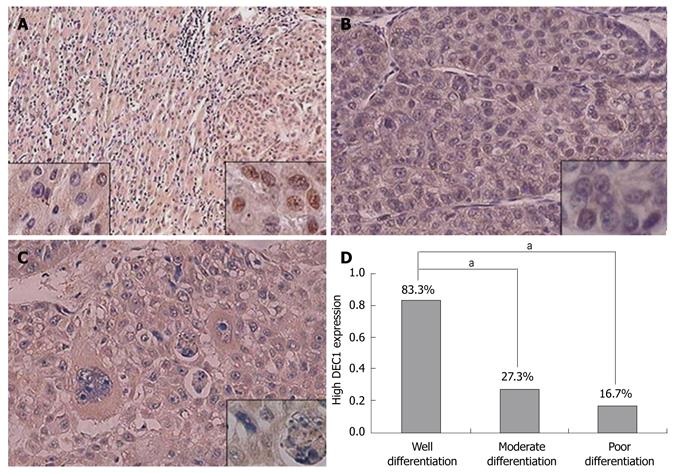
RESULTS: DEC1 protein was persistently expressed in the cytoplasm of hepatocytes in normal liver and HCC tissues. Compared with adjacent non-tumor liver tissues, HCC tissues showed high nuclear expression of DEC1 protein. However, high DEC1 nuclear expression was more frequently detected in well-differentiated (83.3%) than in moderately (27.3%) and poorly differentiated HCC (16.7%). Low DEC1 expression was associated with poor histological differentiation and malignancy progression. A correlation was found between the nuclear expression of DEC1 protein and histological differentiation (r = 0.376, P = 0.024).
CONCLUSION: DEC1 is expressed in the cytoplasm of hepatocytes and because nuclear DEC1 expression is decreased with decreasing differentiation status of HCC, nuclear DEC1 might be a marker of HCC differentiation.
- Citation: Shi XH, Zheng Y, Sun Q, Cui J, Liu QH, Qü F, Wang YS. DEC1 nuclear expression: A marker of differentiation grade in hepatocellular carcinoma. World J Gastroenterol 2011; 17(15): 2037-2043
- URL: https://www.wjgnet.com/1007-9327/full/v17/i15/2037.htm
- DOI: https://dx.doi.org/10.3748/wjg.v17.i15.2037
Hepatocellular carcinoma (HCC) is a major global health problem, with an estimated incidence of 500 000-1 000 000 cases and 600 000 deaths annually. It is the fifth most common cancer in the world and the third most common cause of cancer-related death[1]. The high morbidity and mortality of HCC is due to pre-existing primary chronic liver diseases, such as chronic viral hepatitis, aflatoxin B1, alcoholic liver disease and dysmetabolism, including hereditary haemochromatosis, obesity, diabetes and steatosis[2]. Each of these scenarios has its own genetic and epigenetic alterations, chromosomal aberrations, gene mutation and altered molecular pathways in the process of hepatocarcinogenesis[2,3]. Because of these varied background and heterogeneity, HCC is complex. Although dysregulation of signaling pathways such as Wnt/b-catenin, Ras, p14ARF/p53, p16INK4A/Rb, transforming growth factor-beta (TGF-beta) and PTEN/Akt has been reported in some HCC cases[4], the specific gene mutation(s) and exact molecular mechanism involved in hepatocarcinogenesis is not well known.
Human differentiated embryo chondrocyte 1 (DEC1), a basic helix-loop-helix (bHLH) transcription factor, has rat and mouse orthologs, named enhancer of split and hairy-related protein-2 (SHARP-2) and stimulation of retinoic acid 13 (Stra13), respectively[5-7]. The factors play important roles in regulation of gene expression in cell differentiation, proliferation, immune regulation and metabolism homeostatic control[8]. DEC1 is expressed ubiquitously in both embryonic and adult tissues with human and various extracellular stimuli such as growth factors, serum starvation, hypoxia, hormones, nutrients, cytokines, UV radiation, and infection, which regulate its expression[8,9]. The regulation of DEC1 is cell-type specific[5,9-11].
Several studies have described various DEC1 expression patterns in different tumor tissues, which suggest that it might contribute to oncogenesis. In human breast cancer, the overexpression of DEC1 contributes to a more aggressive phenotype[12]. The association between upregulation of DEC1 expression and differentiation of gastric cancer suggests its important role in the differentiation and progression of gastric cancer[13]. Linked to oncogenesis, DEC1 is highly expressed in colon carcinomas but not in the adjacent normal tissues[14]. It is involved in the UV signal transduction pathway and takes part in the process leading to skin cancer[15]. In combination with carbonic anhydrase-IX (CAIX) and carbonic anhydrase-XII (CAXII), DEC1 may help with a more accurate classification of all renal carcinomas[9]. However, in lung cancer, upregulated or downregulated DEC1 expression has been found[16,17]. The expression patterns and level of DEC1 protein in HCC have not been systematically investigated, and its potential role in hepatocarcinogenesis is unknown.
We aimed to investigate the expression of DEC1 in HCC. We evaluated the distribution and level of expression of DEC1 protein in 176 paraffin-embedded tissue sections from 63 patients with HCC and 50 subjects with normal liver tissues by immunohistochemistry. We also investigated the correlation of DEC1 expression with clinicopathological features and differentiation status of HCC to evaluate the functional characteristics of DEC1 in the development of HCC.
Three kinds of human liver sections (n = 176) were evaluated, including 126 HCC and adjacent non-tumor tissues from 63 patients with primary HCC, and 50 normal liver tissues from patients with hepatic hemangioma who underwent hepatectomy in Qianfoshan Hospital and Jinan Central Hospital, Shandong University, China. The 63 HCC patients included 52 males; the median age was 56 years (range, 35-77 years), and the 50 normal liver patients included 17 males. The formalin-fixed, paraffin-embedded tissue samples were retrospectively collected and randomly selected from the files of the Department of Pathology after the protocol was approved by the local research ethics committee. All HCC patients underwent surgery without prior radiotherapy or chemotherapy and other diseases such as viral hepatitis had been excluded in the hemangioma patients. All sections were reviewed independently by pathologists blinded to the clinicopathological characteristics, 63 HCC and 50 normal livers with the same pathological results were selected in our study. Among the 63 HCC samples, 18 were well differentiated, and 45 were moderately and poorly differentiated. We also collected data on sex, age, tumor size, hepatitis B virus infection, presence of cirrhosis, and α-fetoprotein (AFP) level in HCC.
The tissues were fixed in 10% neutral buffered formalin for 12 h and routinely processed. Paraffin wax-embedded tissue blocks were cut into 4-μm-thick sections. Briefly, formalin-fixed, paraffin-embedded sections were heated at 60°C for 60 min and placed into xylene to be deparaffinized and graded ethanol to be rehydrated, and then washed in phosphate-buffered saline (PBS). Antigen retrieval was performed in a prewarmed pressure cooker with a solution of antigen retrieval citrate buffer (pH 6.8) for 3 min. Following de-pressurization, cold water was poured into the cooker for 10 min, and then sections were rinsed well in warm water. Endogenous peroxide and oxidative compounds were quenched by incubation in 3% H2O2 in methanol for 10 min. Sections were washed 3 times with PBS, and incubated with rabbit polyclonal DEC1 antibody diluted in TBS-Tween20 (1:300 dilution) (Bethyl Laboratories Inc, Montgomery, TX) overnight in a moist chamber at 4°C. After a final wash, secondary antibody (KIT-5010, Max Vision, Maixin.Bio, China) was applied, and TBS-Tween20 was used in all the dilutions and intervening rinses involved. Diaminobenzidine (DAB) was the chromogenic substrate. Sections were allowed to develop in DAB for 5 min, and then counterstained with hematoxylin. Slides were reviewed under microscope. Sections incubated without primary antibody were used as negative controls, and breast cancer sections were used as positive controls. Positive and negative controls were included in each run.
In hepatocytes, DEC1 protein was persistently expressed in all cytoplasms. We used a scoring standard for nuclear DEC1 expression according to our former paper[13]: negative expression, no nuclear staining; low expression, nuclear staining < 10% of cancer cells; and high expression, nuclear staining > 10% of cancer cells. The staining was evaluated by two independent observers.
Categorical variables were compared by χ2 test or Fisher’s exact test as appropriate. Spearman analysis was used to assess the correlation between DEC1 expression and tumor differentiation status. All statistical analyses were performed using SPSS v11 for Windows (SPSS, Inc., Chicago, IL). P < 0.05 was considered statistically significant.
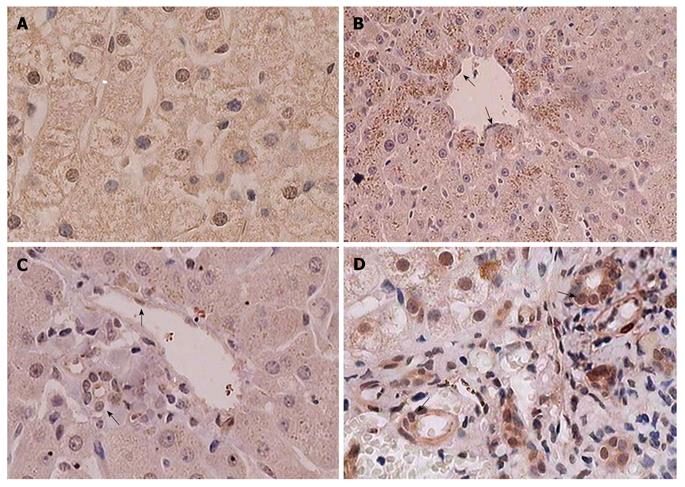
In normal liver tissues, DEC1 expression was diffuse in the cytoplasm of hepatocytes accompanied by a varying degree of nuclear immunoreactivity (Figure 1A). A strong granular pattern of DEC1 staining was seen within the cytoplasm of the hepatocytes around the central vein in the intact hepatic lobule, and this special staining strength became gradually weakened away from the central vein (Figure 1B). However, no granular cytoplasmic staining was seen around the portal area (Figure 1C). The two different cytoplasmic staining patterns may result from the special anatomic structure and the particular blood supplication of the hepatic lobule. The specific conditions around the central vein, such as hypoxia, low nutrition and acidity, could affect DEC1 expression status and lead to the granular pattern, and the diffusive expression pattern may be hypoxia independent. In addition, endothelial and bile duct epithelial cells showed nuclear and/or cytoplasmic DEC1 immunoreactivity (Figure 1A, B, D).
Diffuse cytoplasm expression of DEC1 protein was observed in all 126 HCC and adjacent non-tumor liver tissues, except in 2 poorly differentiated HCC samples with weak cytoplasmic staining. Nuclear DEC1 expression was detected in 36 (57.1%) of 63 samples of primary HCC, with 26 (41.3%) samples showing high nuclear expression (Table 1). Nuclear DEC1 expression was observed in only 17 (27.0%) of the 63 matched adjacent non-tumor tissues. Compared with HCC tissue, adjacent non-tumor tissues showed reduced nuclear DEC1 expression (P = 0.001). The proportion of positive DEC1 nuclear staining was higher in normal (66.7%) than in HCC tissues, but was not significantly different (P = 0.650). In LO-2 cells (normal hepatocytes), nuclear DEC1 expression was higher than in HepG-2 cells (data not shown). Compared with the normal control, adjacent non-tumor tissues showed a significantly low positive staining rate (P = 0.002), which may result from extrusion around the tumor or the response of hepatocytes to extracellular stimulation from the tumor. In addition, in HCC tissues with negative nuclear staining, only two cases showed positive nuclear staining in the corresponding adjacent non-tumor tissues. Therefore, the specific microenvironment around the HCC tumor and the present chronic liver disease such as viral hepatitis may affect DEC1 expression in adjacent non-tumor tissues.
For the 36 cases with positive DEC1 nuclear staining, the proportion of DEC1 staining in well, moderately and poorly differentiated HCC tissues was 94.4% (17/18), 42.4% (14/33) and 41.7% (5/12), respectively. Negative DEC1 nuclear protein staining was associated with high histological HCC grade (Table 2). Well-differentiated tissue was significantly different from moderate and poorly differentiated tissues (P <0.001). Factors such as sex (P = 0.886), age (P = 0.383), tumor size (P = 0.571), hepatitis B virus infection (P = 0.842) and cirrhosis (P = 0.616) had no significant association with DEC1 nuclear expression.
| Clinicopathologic features | n | Nuclear DEC1 protein expression | P value | |
| Negative | Positive | |||
| Sex | ||||
| Male | 52 | 23 | 29 | 0.8861 |
| Female | 11 | 4 | 7 | |
| Age (yr) | ||||
| < 56 | 31 | 15 | 16 | 0.383 |
| ≥ 56 | 32 | 12 | 20 | |
| Tumor size (cm) | ||||
| ≤ 5 | 28 | 11 | 17 | 0.571 |
| > 5 | 30 | 14 | 16 | |
| Pathological grade | ||||
| Grade (I) | 18 | 1 | 17 | 0.000 |
| Grade (II-III) | 45 | 26 | 19 | |
| Venous infiltration | ||||
| Absent | 9 | 2 | 7 | 0.0702 |
| Present | 10 | 7 | 3 | |
| No. tumor nodules | ||||
| < 3 | 49 | 22 | 27 | 0.7811 |
| ≥ 3 | 9 | 3 | 6 | |
| Hepatitis B virus infection | ||||
| Yes | 51 | 22 | 29 | 0.8421 |
| No | 3 | 2 | 1 | |
| Cirrhosis | ||||
| Yes | 47 | 21 | 26 | 0.616 |
| No | 16 | 6 | 10 | |
| α-fetoprotein level (ng/mL) | ||||
| > 20 | 40 | 18 | 22 | 0.713 |
| ≤ 20 | 20 | 8 | 12 | |
As a transcription factor, DEC1 plays its role in the nucleus. We found strong nuclear-positive staining and high nuclear DEC1 expression in well-differentiated HCC samples. In poorly differentiated tumors with large and pleomorphic cells, DEC1 nuclear expression was weak, even negative. We investigated the nuclear immunoreactivity of DEC1 related to differentiation status (Figure 2A-C). The proportion of high DEC1 nuclear expression in well, moderately and poorly differentiated HCC tissues was 83.3%, 27.3% and 16.7%, respectively (Figure 2D). We found a correlation between nuclear DEC1 protein expression and histological differentiation in HCC (r = 0.376, P = 0.024) (Table 3).
HCC is a complex and heterogeneous cancer, and the key drivers are not well known[3,4]. Human DEC1 and the homologs rat SHARP-2 and mouse Stra13 belong to the bHLH family[8]. In many tumor-derived cell lines and tumor tissues, DEC1 mRNA levels are upregulated[9,18]. Its protein is also widely expressed with restricted patterns in many tissues[9,10]. Nevertheless, the distribution and role of DEC1 in carcinogenesis and in the differentiation of human HCC are unknown. A previous study showed 5 of 6 cases of HCC with positive cytoplasmic and nuclear staining[9]. However, the number of samples in this study was too small. In the current research, we studied the expression of DEC1 in 176 samples of HCC and normal liver tissues by immunohistochemistry.
We found that DEC1 protein was persistently expressed in the cytoplasm of both normal and malignant hepatocytes, with varying degrees of nucleic immunoreactivity and two staining patterns, granular and diffusive, in the cytoplasm of hepatocytes. Compared with the adjacent non-tumor tissues, HCC tissue showed predominant DEC1 nuclear protein expression, but the nuclear expression of DEC1 was decreased from well to moderately and poorly differentiated HCC. It seems that low DEC1 expression is associated with poor histological differentiation and malignancy progression in HCC. However, the mechanism of this distribution of DEC1 expression in HCC needs further investigations.
The intracellular distribution of DEC1 protein shows cell specificity: in Hela cells, DEC1 is equally present in both the nucleus and cytoplasm; in HepG2 cells, DEC1 is located mostly in the cytoplasm; and in the 786-0 cell line, the nuclear expression of DEC1 is predominant[9]. In the present study, DEC1 protein was persistently expressed in almost all cytoplasms of hepatocytes, with a varying degree of nuclear immunoreactivity. This expression pattern implies that DEC1 may take part in the normal vital functions of the liver such as metabolism and detoxification. DEC1-positive granular staining gradually weakened outward from the central vein in the cytoplasm of hepatocytes. The mechanism underlying this phenomenon might be the hypoxic environment around the central vein. Hypoxia-induced factor-1α (HIF-1α), the most important hypoxia effecter, can upregulate the expression of DEC1 by binding hypoxia response element (HRE) on DEC1 gene promoter[19]. We also found that the DEC1 expression level in HCC was higher than in normal liver tissues. The oxygen tension in HCC was similar to that in normal liver tissues, so the overexpressed HIF-1α in HCC was independent of hypoxia[20]. Thus, the diffuse staining pattern of DEC1 in normal liver tissues and HCC was independent of hypoxia, and the granular staining pattern of DEC1 may represent a low oxygen concentration. Whether the different patterns of staining imply a different function of DEC1 in the cytoplasm of hepatocytes awaits further investigations. Additional studies are required to unravel the mechanism by which DEC1 performs its function in cytoplasm. Furthermore, the subcellular localization of DEC1 may be helpful in identifying the primary origin of certain tissues and cells.
DEC1 usually acts as a transcription repressor, but it also can function as a transcriptional activator under particular circumstances. It has been shown that signal transducers and activators of transcription 3 (STAT3), which are essential for the carcinogenesis of HCC, are a protein partner of DEC1[21,22]. DEC1 can bind with phosphorylated STAT3β and/or STAT3α isoforms, thus activating the downstream transcription from STAT-dependent cis-elements[22]. For example, co-expression of STAT3α or STAT3β with DEC1 can modify the transcription status of Fas, which can regulate cell survival and apoptosis[22]. DEC1, together with STAT3, is involved in complex regulation of STAT downstream transcriptional targets. Thus, inappropriate co-activation of DEC1 and STAT3 may lead to oncogenic transformation in hepatocytes. Through this network, DEC1 may contribute to the regulation of critical processes of cell survival and growth in hepatocellular carcinogenesis.
Unlike other bHLH proteins, such as c-Myc and ID, which exhibit intrinsically growth-promoting activity, DEC1 can cause proliferation inhibition and differentiation promotion. In NIH 3T3 cells, induced Stra13 strongly represses the expression of the cell proliferation-associated gene c-Myc by interacting with the basal transcription factor TFIIB[18]. The proximal promoter region of ID1 gene contains several potential DEC1 responsive elements by which DEC1 can inhibit ID1 expression, thus promoting cell differentiation[23]. DEC1 has been involved in various differentiation processes, such as neurogenesis[6,7], chondrogenesis[5] and myogenesis[24]. The differentiation inducer hydroxyurea can markedly induce the expression of DEC1[13]. The promotion or inhibition function of DEC1 in cell differentiation is cell-type specific and differs in different original tissues. Overexpression of Stra13 inhibits mesodermal but promotes neuronal differentiation in P19 cells[6]. In rats, the expression of sharp-2 mRNA was slightly higher in the well-differentiated than in the poorly differentiated malignant hepatoma cell lines[25]. Similarly, in our study, high DEC1 nuclear expression was more frequently detected in well-differentiated HCC than in poorly differentiated HCC. Thereby, DEC1 may serve as a marker for the degree of HCC differentiation. As a transcription repressor, DEC1 can regulate expression of many other transcription factors and maintain the homeostasis of the tissues and cells[8]. The reduced expression of DEC1 in poorly differentiated HCC might lead to exacerbation through impairing the homeostasis of the liver.
Epidemiological studies indicated that metabolic disturbance, especially abnormal lipid and glucose metabolism, is a major characteristic of the cause of HCC[4]. Liver X receptor (LXR), together with sterol-regulatory-element-binding protein 1c (SREBP1c) and fatty acid synthase (FAS), enhances the development of HCC caused by chronic viral infection[26]. And insulin resistance increases the risk of HCC[27]. Recent reports have proved that DEC1 was closely associated with LXR, SREBP1c, FAS and insulin[28-31]. However, further researches are needed about the relationship between the function of DEC1 interacted with these genes in liver metabolism and the differentiation grade of HCC.
In summary, we demonstrated DEC1 protein expression patterns in the liver and characterized the relationship between nuclear DEC1 protein expression and histological differentiation of HCC. DEC1 might be a marker of HCC differentiation. However, DEC1 expression can be regulated through several pathways to contribute to its specific functions in different tissues. In the liver, DEC1 may be situated at the crossroads of a complex transcriptional network that is able to modulate metabolism, energy, physiological function and tumor genesis. Dysregulation of DEC1 gene causes alteration of homeostasis in tissues and cells, leading to abnormal cell proliferation, differentiation and death as well as subsequent development of cancer in a cell type-dependent manner.
Hepatocellular carcinoma (HCC) is one of the common fatal cancers worldwide. The aggressive phenotype and resistance to therapy makes HCC particularly dangerous. Recent reports indicate that the transcription factor differentiated embryo chondrocyte 1 (DEC1) contributes to tumorigenesis. The expression of DEC1 and its role in human HCC are unknown. DEC1 may take part in the vital metabolism function of liver and in the procession of hepatocarcinogenesis.
DEC1 is a new transcriptional factor with helix-loop-helix (HLH) domains. It plays an important role in the maintenance of the homeostatus of metabolism and energy, oncogenesis, cell growth and apoptosis, immune balance and circadian rhythm.
This is the first study to describe the expression of DEC1 in HCC and normal liver tissues. In this study, an inverse relationship was found between the nuclear DEC1 protein expression and the histological grade of HCC. DEC1 might be a marker for HCC differentiation. Based on all these results, DEC1 may be situated at the crossroads of a complex transcriptional network that is able to modulate metabolism, energy, physiological function and tumor genesis in the liver.
By understanding the distribution of DEC1 protein expression and the function of this bHLH molecule, this study may represent a future strategy for therapeutic intervention in patients with HCC.
Dec1 and Hif are transcriptional factors with HLH domains. They are important in oncogenesis. Sterol-regulatory-element-binding protein 1c (SREBP1c), fatty acid synthase (FAS) and liver X receptor (LXR) play vital functions in metabolism and tumor genesis in the liver. All these proteins take part in the maintenance of the homeostatus of metabolism and energy, cell growth and apoptosis, oncogenesis.
The morbidity and mortality of HCC are high in China. In the procession of hepatocarcinogenesis, there are already pre-existing chronic liver diseases. In this paper, the authors demonstrated for the first time that DEC1 protein expression patterns in normal liver tissue and HCC systematically. It is important for further studying the development of HCC and the function of DEC1.
Peer reviewer: Filip Braet, Associate Professor, Australian Key Centre for Microscopy and Microanalysis, Madsen Building (F09), The University of Sydney, Sydney NSW 2006, Australia
S- Editor Sun H L- Editor Ma JY E- Editor Ma WH
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. |
| 2. | Lodato F, Mazzella G, Festi D, Azzaroli F, Colecchia A, Roda E. Hepatocellular carcinoma prevention: a worldwide emergence between the opulence of developed countries and the economic constraints of developing nations. World J Gastroenterol. 2006;12:7239-7249. |
| 4. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. |
| 5. | Shen M, Kawamoto T, Yan W, Nakamasu K, Tamagami M, Koyano Y, Noshiro M, Kato Y. Molecular characterization of the novel basic helix-loop-helix protein DEC1 expressed in differentiated human embryo chondrocytes. Biochem Biophys Res Commun. 1997;236:294-298. |
| 6. | Boudjelal M, Taneja R, Matsubara S, Bouillet P, Dolle P, Chambon P. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix-loop-helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 1997;11:2052-2065. |
| 7. | Rossner MJ, Dörr J, Gass P, Schwab MH, Nave KA. SHARPs: mammalian enhancer-of-split- and hairy-related proteins coupled to neuronal stimulation. Mol Cell Neurosci. 1997;10:460-475. |
| 8. | Yamada K, Miyamoto K. Basic helix-loop-helix transcription factors, BHLHB2 and BHLHB3; their gene expressions are regulated by multiple extracellular stimuli. Front Biosci. 2005;10:3151-3171. |
| 9. | Ivanova A, Liao SY, Lerman MI, Ivanov S, Stanbridge EJ. STRA13 expression and subcellular localisation in normal and tumour tissues: implications for use as a diagnostic and differentiation marker. J Med Genet. 2005;42:565-576. |
| 10. | Turley H, Wykoff CC, Troup S, Watson PH, Gatter KC, Harris AL. The hypoxia-regulated transcription factor DEC1 (Stra13, SHARP-2) and its expression in human tissues and tumours. J Pathol. 2004;203:808-813. |
| 11. | Shi X, Zheng Y, Ma W, Wang Y. Possible involvement of DEC1 on the adverse effects of quinolone antibiotics. Toxicology. 2010;271:1-4. |
| 12. | Chakrabarti J, Turley H, Campo L, Han C, Harris AL, Gatter KC, Fox SB. The transcription factor DEC1 (stra13, SHARP2) is associated with the hypoxic response and high tumour grade in human breast cancers. Br J Cancer. 2004;91:954-958. |
| 13. | Zheng Y, Jia Y, Wang Y, Wang M, Li B, Shi X, Ma X, Xiao D, Sun Y. The hypoxia-regulated transcription factor DEC1 (Stra13, SHARP-2) and its expression in gastric cancer. OMICS. 2009;13:301-306. |
| 14. | Li Y, Zhang H, Xie M, Hu M, Ge S, Yang D, Wan Y, Yan B. Abundant expression of Dec1/stra13/sharp2 in colon carcinoma: its antagonizing role in serum deprivation-induced apoptosis and selective inhibition of procaspase activation. Biochem J. 2002;367:413-422. |
| 15. | Li Y, Bi Z, Yan B, Wan Y. UVB radiation induces expression of HIF-1alpha and VEGF through the EGFR/PI3K/DEC1 pathway. Int J Mol Med. 2006;18:713-719. |
| 16. | Falvella FS, Colombo F, Spinola M, Campiglio M, Pastorino U, Dragani TA. BHLHB3: a candidate tumor suppressor in lung cancer. Oncogene. 2008;27:3761-3764. |
| 17. | Giatromanolaki A, Koukourakis MI, Sivridis E, Turley H, Wykoff CC, Gatter KC, Harris AL. DEC1 (STRA13) protein expression relates to hypoxia- inducible factor 1-alpha and carbonic anhydrase-9 overexpression in non-small cell lung cancer. J Pathol. 2003;200:222-228. |
| 18. | Sun H, Taneja R. Stra13 expression is associated with growth arrest and represses transcription through histone deacetylase (HDAC)-dependent and HDAC-independent mechanisms. Proc Natl Acad Sci USA. 2000;97:4058-4063. |
| 19. | Miyazaki K, Kawamoto T, Tanimoto K, Nishiyama M, Honda H, Kato Y. Identification of functional hypoxia response elements in the promoter region of the DEC1 and DEC2 genes. J Biol Chem. 2002;277:47014-47021. |
| 20. | Tanaka H, Yamamoto M, Hashimoto N, Miyakoshi M, Tamakawa S, Yoshie M, Tokusashi Y, Yokoyama K, Yaginuma Y, Ogawa K. Hypoxia-independent overexpression of hypoxia-inducible factor 1alpha as an early change in mouse hepatocarcinogenesis. Cancer Res. 2006;66:11263-11270. |
| 21. | Li WC, Ye SL, Sun RX, Liu YK, Tang ZY, Kim Y, Karras JG, Zhang H. Inhibition of growth and metastasis of human hepatocellular carcinoma by antisense oligonucleotide targeting signal transducer and activator of transcription 3. Clin Cancer Res. 2006;12:7140-7148. |
| 22. | Ivanova AV, Ivanov SV, Zhang X, Ivanov VN, Timofeeva OA, Lerman MI. STRA13 interacts with STAT3 and modulates transcription of STAT3-dependent targets. J Mol Biol. 2004;340:641-653. |
| 23. | Qian Y, Chen X. ID1, inhibitor of differentiation/DNA binding, is an effector of the p53-dependent DNA damage response pathway. J Biol Chem. 2008;283:22410-22416. |
| 24. | Sun H, Li L, Vercherat C, Gulbagci NT, Acharjee S, Li J, Chung TK, Thin TH, Taneja R. Stra13 regulates satellite cell activation by antagonizing Notch signaling. J Cell Biol. 2007;177:647-657. |
| 25. | Hirano S, Yamada K, Kawata H, Shou Z, Mizutani T, Shigematsu Y, Mayumi M, Miyamoto K. The rat enhancer of split- and hairy-related protein-2 gene: hepatic expression, genomic structure, and promoter analysis. Arch Biochem Biophys. 2004;422:81-90. |
| 26. | Na TY, Shin YK, Roh KJ, Kang SA, Hong I, Oh SJ, Seong JK, Park CK, Choi YL, Lee MO. Liver X receptor mediates hepatitis B virus X protein-induced lipogenesis in hepatitis B virus-associated hepatocellular carcinoma. Hepatology. 2009;49:1122-1131. |
| 27. | Siegel AB, Zhu AX. Metabolic syndrome and hepatocellular carcinoma: two growing epidemics with a potential link. Cancer. 2009;115:5651-5661. |
| 28. | Noshiro M, Usui E, Kawamoto T, Sato F, Nakashima A, Ueshima T, Honda K, Fujimoto K, Honma S, Honma K. Liver X receptors (LXRalpha and LXRbeta) are potent regulators for hepatic Dec1 expression. Genes Cells. 2009;14:29-40. |
| 29. | Choi SM, Cho HJ, Cho H, Kim KH, Kim JB, Park H. Stra13/DEC1 and DEC2 inhibit sterol regulatory element binding protein-1c in a hypoxia-inducible factor-dependent mechanism. Nucleic Acids Res. 2008;36:6372-6385. |
| 30. | Kawamoto T, Noshiro M, Furukawa M, Honda KK, Nakashima A, Ueshima T, Usui E, Katsura Y, Fujimoto K, Honma S. Effects of fasting and re-feeding on the expression of Dec1, Per1, and other clock-related genes. J Biochem. 2006;140:401-408. |
| 31. | Yamada K, Kawata H, Shou Z, Mizutani T, Noguchi T, Miyamoto K. Insulin induces the expression of the SHARP-2/Stra13/DEC1 gene via a phosphoinositide 3-kinase pathway. J Biol Chem. 2003;278:30719-30724. |