Published online Dec 14, 2010. doi: 10.3748/wjg.v16.i46.5815
Revised: August 17, 2010
Accepted: August 24, 2010
Published online: December 14, 2010
AIM: To investigated gene mutations and polymorphisms of TLR9 in Japanese ulcerative colitis (UC) patients.
METHODS: Three single nucleotide polymorphisms (SNPs) in TLR9 were identified in healthy controls, and were assessed in 48 UC patients and 47 healthy controls. Control subjects were matched for age, sex and date of blood sampling from among a subgroup of participants.
RESULTS: TLR9 -1486CC, 1174GG and 2848AA increase the risk of UC [odds ratio (OR) 2.64, 95% confidence interval (95% CI): 1.73-6.53, P = 0.042], and TLR9 -1486TT, 1174AA and 2848GG decrease the risk of UC (OR 0.30, 95% CI: 0.10-0.94, P = 0.039), although there were no correlations between SNPs and disease phenotype or TLR9 mRNA expression.
CONCLUSION: TLR9 polymorphisms are associated with increased susceptibility to UC.
- Citation: Fuse K, Katakura K, Sakamoto N, Ohira H. Toll-like receptor 9 gene mutations and polymorphisms in Japanese ulcerative colitis patients. World J Gastroenterol 2010; 16(46): 5815-5821
- URL: https://www.wjgnet.com/1007-9327/full/v16/i46/5815.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i46.5815
Inflammatory bowel diseases (IBDs), which include ulcerative colitis (UC) and Crohn’s disease (CD), are chronic inflammatory disorders of the digestive tract. Although the pathogenesis of IBDs is complex and remains unclear, it has been suggested that immunologic, environmental and genetic factors contribute to their etiology[1]. Both clinical studies of IBD and studies of animal colitis models implicate luminal bacteria as necessary for initiating and perpetuating intestinal inflammation[2,3].
Among several candidate IBD-related chromosomal regions and genes, the caspase recruitment domain 15 (CARD15) gene coding for the nucleotide oligomerization domain 2 (NOD2) gene was identified as having the strongest linkage for CD susceptibility[4,5]. NOD2/CARD15 is an intracytoplasmic receptor that binds bacterial peptidoglycan-derived muramyl dipeptide. While three common variants of this gene (R702W, G908R and L1007fsinsC) have been reported in CD patients in Western countries, there were no variants found in Japanese patients[6].
Similarly to NOD2, Toll-like receptors (TLRs) are essential components of innate immunity that recognize microbial compounds from bacteria, fungi and viruses[7-9]. While TLR activation leads to transcription of inflammatory and immunoregulatory genes, recent studies have demonstrated that TLR signaling in intestinal sites can inhibit inflammatory responses and maintain colonic homeostasis[10-12]. TLR9, which recognizes unmethylated CpG DNA in bacteria and viruses[13,14], and its signaling pathway protect mice from experimental colitis through production of type I interferons (IFN)[15]. Therefore, we need to know whether TLR9 is a protective molecule not only in a mouse model but also in human IBD.
As the relationships between UC and TLR9 gene variation have not been reported to date, we focused on TLR9 gene mutations or polymorphisms in UC. Our results demonstrate that TLR9 genetic polymorphisms are associated with an increased risk of UC in the Japanese population.
After obtaining written informed consent based on the Declaration of Helsinki of the World Medical Association, 48 UC patients and 47 healthy controls were enrolled in this study. All subjects were Japanese, and visited our hospital between December 2005 and January 2007. UC diagnoses were confirmed by review of medical charts by clinicians. Standard clinical, endoscopic and histological criteria were used[16-18]. The following clinical characteristics were analyzed: sex, age at diagnosis, disease location and disease severity. Control subjects were matched for age, sex and date of blood sampling from a subgroup of participants who were healthy volunteers, free from IBD and malignant tumors. Clinical characteristics of these subjects are shown in Table 1. This study was approved by the Ethics Committee of Fukushima Medical University.
| UC (n = 48) | Controls (n = 47) | |
| Age (yr) | 43.49 ± 15.48 | 41.21 ± 15.36 |
| Sex (M/F) | 21 (43.8)/27 (56.2) | 21 (44.7)/26 (55.3) |
| Age at diagnosis (yr) | 35.07 ± 14.39 | |
| Disease duration (yr) | 8.42 ± 8.49 | |
| Familial disease | 0 (0) | 0 (0) |
| Disease location | ||
| Proctitis | 3 (6.25) | |
| Left colitis | 27 (56.3) | |
| Right colitis | 1 (2.1) | |
| Pancolitis | 17 (35.4) | |
| Disease severity | ||
| Light | 17 (35.4) | |
| Mild | 18 (37.5) | |
| Severe | 13 (27.1) | |
| History of colectomy | 5 (10.4) | |
| History of immunosuppressant | 6 (12.5) | |
| History of colon cancer | 3 (6.3) |
Peripheral blood mononuclear cells (PBMC) from UC patients and healthy controls were isolated from heparinized blood by Ficoll Paque density-gradient centrifugation (Lymphoprep, Axis-Shield PoC AS, Oslo, Norway). Total cellular RNA was extracted from isolated PBMC using TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA).
cDNA was generated by using the HotStarTaq Master Mix Kit (QIAGEN GmbH, Hilden, Germany). Quantitative real-time reverse transcriptase-polymerase chain reaction (RT-PCR) was performed using a TaqMan® fast universal PCR master mix (Applied Biosystems, Foster City, CA, USA) and on-demand gene-specific primers, assessed using StepOne™ real time PCR system (Applied Biosystems). The primers were as follows: TLR9 (Hs00370913_s1) and GAPDH (Hs02786624_g1). Both primers were purchased from Applied Biosystems. Relative quantification was achieved by normalizing to the values of the GAPDH gene.
Genomic DNA from UC patients and healthy controls was extracted from heparinized whole blood samples using the PUREGENE® DNA purification kit (Gentra Systems, Minneapolis, MN, USA).
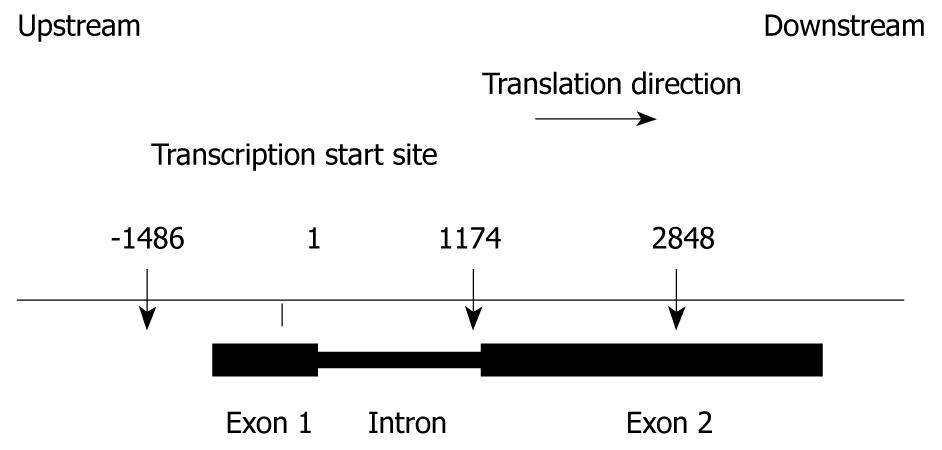
All exons, introns and approximately 1500 bases of the 5’-flanking region and 1500 bases of the 3’-flanking region of the TLR9 gene were amplified from the genomic DNA of one healthy control. The genomic sequences were based on the GenBank reference sequence NM_017442 and AC097637.2 for TLR9. The subject was selected at random. Thirteen TLR9 gene segments were amplified by PCR with the primer pairs shown in Table 2, Taq DNA polymerase (QIAGEN GmbH, Hilden, Germany) and genomic DNA from a healthy control as a template. Amplified products were purified using the GFX™ PCR DNA and Gel Band Purification Kit (Amersham Biosciences UK Limited, Buckinghamshire, UK), and were subjected to direct sequencing with the GenomeLab™ DTCS-Quick Start Kit (Beckman Coulter, Fullerton, CA, USA) on a CEQ2000 DNA Sequencer (Beckman Coulter). SNP offsets were calculated relative to the A base of the TLR9 ATG start codon, such that SNPs in the promoter region upstream of the first intron have negative position numbers.
| Fragment (product) | Forward | Reverse |
| TLR9 polymorphisms | ||
| -2109 to -1293 (817 bp) | 5'-CCAAGGGACTCTGGGAAAG-3' | 5'-CATGTCACCCTCTCAACAGGG-3' |
| -1626 to -1095 (532 bp) | 5'-CAGCCTTCACTCAGAAATACCC-3' | 5'-GGCCAACAAGGCCCTATG-3' |
| -1296 to -553 (662 bp) | 5'-CATGGGAGCAGAGACATAATG-3' | 5'-GCCAGGGTGTAGCTTGA-3' |
| -795 to -134 (662 bp) | 5'-GAGTCTCCTCACCTAGATCAG-3' | 5'-TATACCAGCCTAGTAGC-3' |
| 79 to 962 (884 bp) | 5'-CTGCAAGCAACAGTGACGG-3' | 5'-AGCTTTCACTTAACCAATCCC-3' |
| -393 to 1698 (2092 bp) | 5'-TACCCGCTACTGGTGCTATC-3' | 5'-TGGCAGAGTCTAGCATCAGG-3' |
| 996 to 1687 (692 bp) | 5'-CTGGTTCTGAAGCCTAATTC-3' | 5'-AGCATGAGGATGTTGGTATGG-3' |
| 976 to 1899 (924 bp) | 5'-CTGGATTCTAGGTCTCAGTCC-3' | 5'-CTGGATTCTAGGTCTCAGTCC-3' |
| 1540 to 2368 (829 bp) | 5'-CCTGCCACATGACCATCGAG-3' | 5'-CCTGCCACATGACCATCGAG-3' |
| 1812 to 2520 (709 bp) | 5'-AACCTCACCCACCTGTCAC-3' | 5'-AACCTCACCCACCTGTCAC-3' |
| 2407 to 3350 (944 bp) | 5'-TGCAGATGAACTTCATCAACC-3' | 5'-GCTGTTGCAGCTGACATC-3' |
| 3267 to 4057 (791 bp) | 5'-CAGGAAACCAGCTGAAGG-3' | 5'-CAGGAAACCAGCTGAAGG-3' |
| 3867 to 4701 (835 bp) | 5'-GACTGGGTGTACAACGAGCTT-3' | 5'-TTCTGCATGGGAAAGGTAGG-3' |
| SNP | ||
| -1486 | 5'-CAGCCTTCACTCAGAAATACCC-3' | 5'-GGCCAACAAGGCCCTATG-3' |
| 1174 | 5'-CTGGTTCTGAAGCCTAATTC-3' | 5'-AGCATGAGGATGTTGGTATGG-3' |
| 2848 | 5'-TGCAGATGAACTTCATCAACC-3' | 5'-GCTGTTGCAGCTGACATC-3' |
Three common SNPs (-1486T/C, 1174A/G, 2848G/A) were analyzed for genotyping. The -1486T/C SNP was genotyped by restriction fragment length polymorphism (RFLP). Briefly, genomic DNA fragments containing -1486T/C SNP were amplified by PCR. PCR products were digested with the restriction enzyme Afl II (TAKARA SHUZO Co., Ltd., Shiga, Japan), run on a 3% agarose gel and subsequently stained with ethidium bromide to visualize the bands. The other two SNPs (1174A/G and 2848G/A) were confirmed by direct sequencing. The primers used for PCR or sequencing are shown in Table 2.
Statistical analyses were performed using SPSS version 11.0.1 and SNPAlyze version 6.0 (Dynacom Co., Ltd. Yokohama, Japan). Cases and controls were compared using Fisher’s exact test (Table 3, Table 4, Table 5, Table 6,) for categorical items. Simple logistic regression models were used to analyze Genotype-Diplotype associations. The results are expressed as odds ratio (OR) with corresponding 95% confidence intervals (95% CI). The level of significance was set at 5%. We examined Lewontin’s D’ (|D’|) and the linkage disequilibrium coefficient r2 between each pair of SNPs using the expectation-maximization (EM) algorithm in SNPAlyze software.
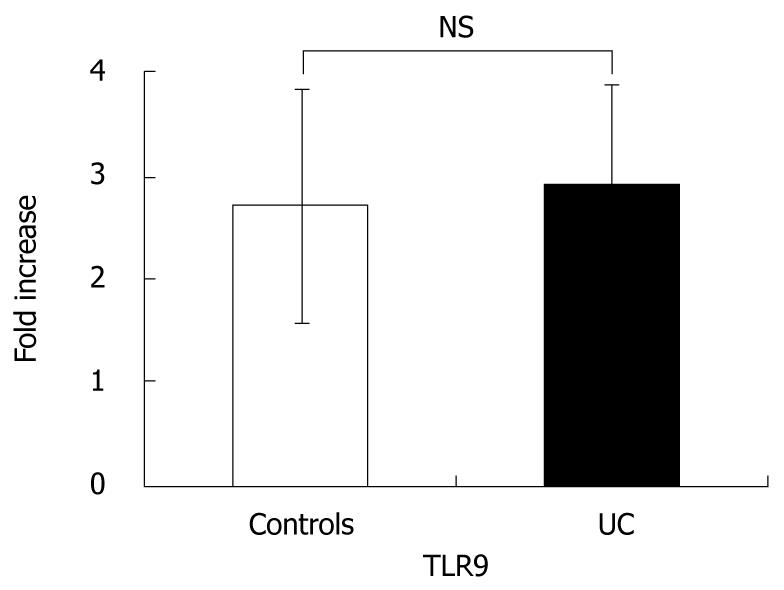
In order to confirm whether there were differences in TLR9 expression, we performed quantitative RT-PCR of TLR9 using PBMC from UC patients and healthy controls. There were no significant differences in TLR9 mRNA expression (Figure 1).
We sequenced both exons and approximately 1500 bases of the 5’- and 3’-regions of TLR9 in one healthy control. Offsets for each SNP were calculated relative to the transcription start site of TLR9 mRNA (GenBank Accession No. NM_017442). We identified three SNPs in the regions sequenced. Their locations in chromosome 3 and in the TLR9 gene are illustrated in Figure 2. These SNPs were confirmed in dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and JSNP (http://snp.ims.u-tokyo.ac.jp/index_ja.html). The remainder of the analysis will focus on these three relatively common SNPs.
We found a marginally significant association between the SNPs at -1486, 1174, 2848 and UC. Elevated risk for UC was associated with a C allele at -1486 (Fisher’s exact test, P = 0.013), a G allele at 1174 (Fisher’s exact test, P = 0.013), and an A allele at 2848 (Fisher’s exact test, P = 0.013) (Table 3).
We then analyzed genotype frequencies between UC patients and healthy controls. As shown in Table 4, a pattern of increased prevalence of CC genotypes at -1486 (Fisher’s exact test, P = 0.047), GG genotypes at 1174 (Fisher’s exact test, P = 0.047), AA genotypes at 2848 (Fisher’s exact test, P = 0.042), and a decreased prevalence of TT at -1486 (Fisher’s exact test, P = 0.039), AA genotypes at 1174 (Fisher’s exact test, P = 0.039), GG genotypes at 2848 (Fisher’s exact test, P = 0.039) was observed in UC patients, as compared with controls.
We observed absolutely identical allele frequencies for the three common SNPs. Table 7 shows that three SNPs are in strong linkage disequilibrium (LD) in both UC patients and healthy controls. We also inferred three SNP haplotypes by using an expectation-maximization algorithm. The number of statistically inferred haplotypes is eight (i.e. 23). A total of three of the eight possible haplotypes are observed in Table 5. The usage of haplotype C-G-A was more frequent in UC patients when compared with healthy controls, and the difference was statistically significant. (Fisher’s exact test, P = 0.013, Table 5).
| SNPs | UC (n = 48) | Controls (n = 47) | |||
| |D’| | r2 | |D’| | r2 | ||
| -1486 T/C | 1174 A/G | 1 | 1 | 1 | 1 |
| -1486 T/C | 2848 G/A | 1 | 0.9522 | 1 | 0.9581 |
| 1174 A/G | 2848 G/A | 1 | 0.9522 | 1 | 0.9581 |
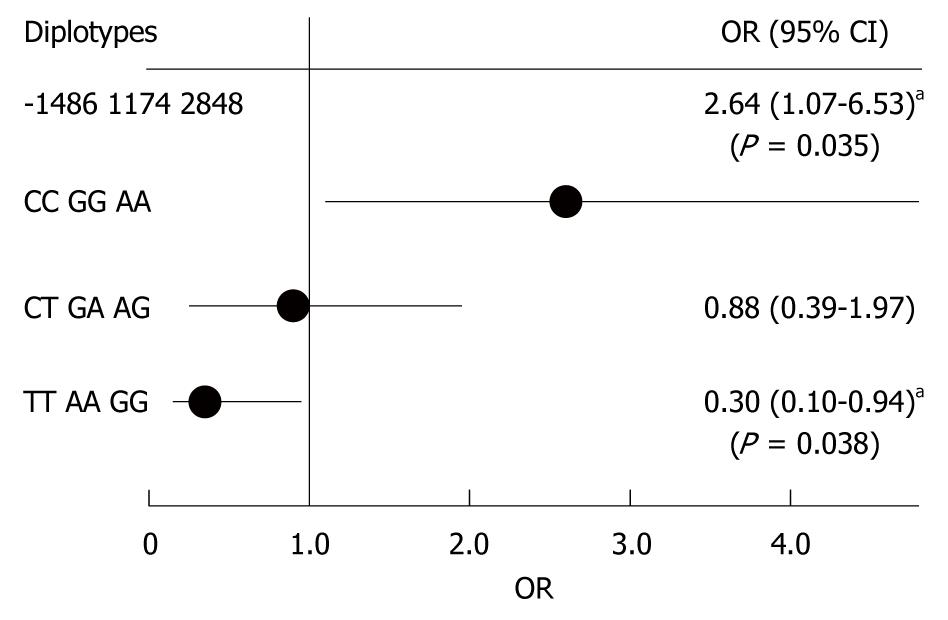
We next analyzed diplotype frequencies as pairs of haplotypes. Table 6 shows the distribution of three diplotype loci by disease status; an increased prevalence of CCGGAA diplotypes (Fisher’s exact test, P = 0.042) and a decreased prevalence of TTAAGG diplotypes (Fisher’s exact test, P = 0.039) was noted among UC cases, as compared with controls. Thus, we found an increased UC risk among CCGGAA diplotypes (OR 2.64, 95% CI: 1.73-6.53, P = 0.042) and a decreased UC risk among TTAAGG diplotypes (OR 0.30, 95% CI: 0.10-0.94, P = 0.039) (Figure 3). These data suggest that CCGGAA diplotypes are more susceptible to UC.
In order to further examine the contribution of genetic variations in TLR9 to UC phenotypes, we analyzed various clinical characteristics among UC patients: sex, age at diagnosis, disease location and disease severity. However, we did not observe any statistically significant correlations between disease status and diplotype, genotype or specific TLR9 allele.
In this study, we found that genetic variations of TLR9 are associated with an increased risk of UC in the Japanese population. TLR9, which is activated by unmethylated CpG DNA, triggers innate immune responses[19,20]. Although no variants have been found in Japanese patients, the NOD2/CARD15 gene was found to indicate CD susceptibility in Western populations[5,6], suggesting that abnormal innate immune responses toward luminal bacteria are involved in pathogenesis of IBD. With regard to TLRs, previous reports have indicated that TLR4 Asp299Gly polymorphism is associated with CD and UC in Caucasian populations[21]. Török et al[22] investigated possible associations between genetic variations in TLR9 and IBD in the German population, but did not detect any associations between TLR9 gene variations and UC susceptibility. Thus, as UC is a heterogeneous polygenic disease, association studies are expected to reveal various sets of susceptibility genes depending on the ethnic background of the study populations. Further genetic studies in different ethnic groups will resolve the role of the TLRs in UC susceptibility.
Associations between TLR9 and several autoimmune diseases or infectious diseases have been studied[23-26]. These reports have suggested that the -1237T/C SNP in TLR9 is associated with an increased risk of allergic asthma[27], while 2848G/A was associated with myocardial infarction, deep vein thrombosis and chronic obstructive pulmonary disease[27], and 1635A/G was associated with HIV phenotype[28]. In addition, there were no associations between TLR9 gene polymorphisms and susceptibility to systemic lupus erythematosus (SLE) and related phenotypes in Caucasian American subjects[23], but such an association was observed in a Japanese population[24]. These observations show that several candidate genes showing initial positive associations have generated negative findings in replication studies due to issues related to insufficient power or sample heterogeneity.
In this study, although relatively low numbers of subjects were enrolled, we observed statistically significant differences. According to the statistical methods used in this research, statistical differences in a category that has more than one subject indicate a reliable result. Moreover, we can expect to obtain the same results if more samples are added to the experiment. Therefore, we did not increase the power of this study, although more studies with large cohorts are necessary to characterize the role of TLR9 SNPs in the etiology of UC.
We demonstrated that a C allele at -1486 (located in promoter region), a G allele at 1174 (located in intron 1) and an A allele at 2848 are associated with an increased risk of UC. SNPs in promoter regions potentially affect gene expression levels by altering the binding of gene transcription factors and SNPs in introns, thereby affecting mRNA splicing and/or enhancement of gene transcription. We were unable to identify down-regulation in TLR9 expression in UC patients. Evaluating the relationship between the TLR9 polymorphism and predefined clinical characteristics or biological markers also failed to demonstrate any impact on a particular UC phenotype. Taken together, these results indicate that TLR9 polymorphism is associated with the development of disease itself, which is probably based on functional impairment of TLR9.
A recent study suggested that TLR9-triggered type-1 IFN protects mice from experimental colitis[15]. It also underscores the important protective role of type-1 IFN in intestinal homeostasis, suggesting that type-1 IFN produced by TLR9 signaling affects the pathogenesis of IBD. Although further experiments are needed in order to identify the functional roles of TLR9 polymorphism, either downstream molecules of the TLR9 signaling pathway or cytokine production participating in the development of IBD is expected to be affected by these gene polymorphisms.
In conclusion, we identified an association between TLR9 polymorphisms and UC in Japanese patients. Our findings indicate the importance of TLR9 in the genetic control of reactions to intestinal microbes in UC. Studying SNPs among the molecules involved in bacterial recognition will be essential to understanding the individual responses to bacterial components and to define the genetic background associated with risk of IBD.
The pathogenesis of inflammatory bowel diseases (IBDs), which include ulcerative colitis (UC) and Crohn’s disease (CD), is multi-factorial, involving susceptibility genes as well as immunological and environmental factors. Recent studies on IBD have provided some evidence that commensal bacteria play a key role in the pathogenesis of the disease, and that the Toll-like receptor (TLR) signaling pathway activated by commensal bacteria plays an important role in maintaining colonic homeostasis. The signaling pathway of TLR9, which recognizes unmethylated CpG DNA in bacteria and viruses, protects mice from experimental colitis. As the relationships between UC and TLR9 gene variation have not been reported to date, in this report we focused on TLR9 gene mutations or polymorphisms in UC.
Among several candidate IBD-related chromosomal regions and genes, the caspase recruitment domain 15 (CARD15) gene coding for the nucleotide oligomerization domain 2 (NOD2) gene was identified as having the strongest linkage for CD susceptibility. Similarly to NOD2, TLRs are essential components of innate immunity that recognize microbial compounds from bacteria, fungi and viruses. While TLR activation leads to transcription of inflammatory and immunoregulatory genes, recent studies have demonstrated that TLR signaling in intestinal sites can inhibit inflammatory responses and maintain colonic homeostasis. A gene variation in NOD2/CARD15 has been reported in CD patients in Western countries, but this variation has not been identified in Japanese CD patients.
As the relationships between UC and TLR9 gene variation have not been reported to date, in this report we focused on TLR9 gene mutations or polymorphisms in UC. Three single nucleotide polymorphisms (SNPs) in TLR9 were identified in healthy controls, and were assessed in 48 UC patients. The authors found that TLR9 -1486CC, 1174GG and 2848AA increase the risk of UC, and TLR9 -1486TT, 1174AA and 2848GG decrease the risk of UC.
The authors’ findings indicate the importance of TLR9 in the genetic control of reactions to intestinal microbes in UC. Studying SNPs among the molecules involved in bacterial recognition will be essential to understanding the individual responses to bacterial components and to define the genetic background associated with risk of IBD.
TLR9 is a mammalian Toll-like receptor homologue that appears to function as an innate immune pattern recognition protein for motifs that are far more common in bacterial than in mammalian DNA.
It is interesting that the authors identified an association between TLR9 polymorphisms and UC in Japanese patients. My main concern is the sample size (not too large) and the fact that the P values obtained are not extremely significant (almost marginally significant on some occasions). This may limit the validity of the present work.
Peer reviewer: José Manuel Martin-Villa, Professor, PhD, Department of Immunología, Facultad de Medicina, Universidad Complutense de Madrid, Pabellón V. Planta 4ª, Madrid 28040, Spain
S- Editor Sun H L- Editor Logan S E- Editor Lin YP
| 1. | Sartor RB. Pathogenesis and immune mechanisms of chronic inflammatory bowel diseases. Am J Gastroenterol. 1997;92:5S-11S. [Cited in This Article: ] |
| 2. | Blumberg RS, Saubermann LJ, Strober W. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr Opin Immunol. 1999;11:648-656. [Cited in This Article: ] |
| 3. | Podolsky DK. Mucosal immunity and inflammation. V. Innate mechanisms of mucosal defense and repair: the best offense is a good defense. Am J Physiol. 1999;277:G495-G499. [Cited in This Article: ] |
| 4. | Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603-606. [Cited in This Article: ] |
| 5. | Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599-603. [Cited in This Article: ] |
| 6. | Inoue N, Tamura K, Kinouchi Y, Fukuda Y, Takahashi S, Ogura Y, Inohara N, Núñez G, Kishi Y, Koike Y. Lack of common NOD2 variants in Japanese patients with Crohn's disease. Gastroenterology. 2002;123:86-91. [Cited in This Article: ] |
| 7. | Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782-787. [Cited in This Article: ] |
| 8. | Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257-263. [Cited in This Article: ] |
| 9. | Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131-137. [Cited in This Article: ] |
| 10. | Rachmilewitz D, Katakura K, Karmeli F, Hayashi T, Reinus C, Rudensky B, Akira S, Takeda K, Lee J, Takabayashi K. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology. 2004;126:520-528. [Cited in This Article: ] |
| 11. | Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229-241. [Cited in This Article: ] |
| 12. | Lee J, Mo JH, Katakura K, Alkalay I, Rucker AN, Liu YT, Lee HK, Shen C, Cojocaru G, Shenouda S. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat Cell Biol. 2006;8:1327-1336. [Cited in This Article: ] |
| 13. | Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, Sanjo H, Matsumoto M, Hoshino K, Wagner H, Takeda K. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740-745. [Cited in This Article: ] |
| 14. | Lund J, Sato A, Akira S, Medzhitov R, Iwasaki A. Toll-like receptor 9-mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J Exp Med. 2003;198:513-520. [Cited in This Article: ] |
| 15. | Katakura K, Lee J, Rachmilewitz D, Li G, Eckmann L, Raz E. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J Clin Invest. 2005;115:695-702. [Cited in This Article: ] |
| 16. | Podolsky DK. Inflammatory bowel disease (2). N Engl J Med. 1991;325:1008-1016. [Cited in This Article: ] |
| 17. | Podolsky DK. Inflammatory bowel disease (1). N Engl J Med. 1991;325:928-937. [Cited in This Article: ] |
| 18. | Lennard-Jones JE. Classification of inflammatory bowel disease. Scand J Gastroenterol Suppl. 1989;170:2-6; discussion 16-19. [Cited in This Article: ] |
| 20. | Krieg AM. CpG motifs in bacterial DNA and their immune effects. Annu Rev Immunol. 2002;20:709-760. [Cited in This Article: ] |
| 21. | Franchimont D, Vermeire S, El Housni H, Pierik M, Van Steen K, Gustot T, Quertinmont E, Abramowicz M, Van Gossum A, Devière J. Deficient host-bacteria interactions in inflammatory bowel disease? The toll-like receptor (TLR)-4 Asp299gly polymorphism is associated with Crohn's disease and ulcerative colitis. Gut. 2004;53:987-992. [Cited in This Article: ] |
| 22. | Török HP, Glas J, Tonenchi L, Bruennler G, Folwaczny M, Folwaczny C. Crohn's disease is associated with a toll-like receptor-9 polymorphism. Gastroenterology. 2004;127:365-366. [Cited in This Article: ] |
| 23. | Demirci FY, Manzi S, Ramsey-Goldman R, Kenney M, Shaw PS, Dunlop-Thomas CM, Kao AH, Rhew EY, Bontempo F, Kammerer C. Association study of Toll-like receptor 5 (TLR5) and Toll-like receptor 9 (TLR9) polymorphisms in systemic lupus erythematosus. J Rheumatol. 2007;34:1708-1711. [Cited in This Article: ] |
| 24. | Tao K, Fujii M, Tsukumo S, Maekawa Y, Kishihara K, Kimoto Y, Horiuchi T, Hisaeda H, Akira S, Kagami S. Genetic variations of Toll-like receptor 9 predispose to systemic lupus erythematosus in Japanese population. Ann Rheum Dis. 2007;66:905-909. [Cited in This Article: ] |
| 25. | Kikuchi K, Lian ZX, Kimura Y, Selmi C, Yang GX, Gordon SC, Invernizzi P, Podda M, Coppel RL, Ansari AA. Genetic polymorphisms of toll-like receptor 9 influence the immune response to CpG and contribute to hyper-IgM in primary biliary cirrhosis. J Autoimmun. 2005;24:347-352. [Cited in This Article: ] |
| 26. | Carvalho A, Pasqualotto AC, Pitzurra L, Romani L, Denning DW, Rodrigues F. Polymorphisms in toll-like receptor genes and susceptibility to pulmonary aspergillosis. J Infect Dis. 2008;197:618-621. [Cited in This Article: ] |
| 27. | Lazarus R, Klimecki WT, Raby BA, Vercelli D, Palmer LJ, Kwiatkowski DJ, Silverman EK, Martinez F, Weiss ST. Single-nucleotide polymorphisms in the Toll-like receptor 9 gene (TLR9): frequencies, pairwise linkage disequilibrium, and haplotypes in three U.S. ethnic groups and exploratory case-control disease association studies. Genomics. 2003;81:85-91. [Cited in This Article: ] |