Published online Nov 14, 2010. doi: 10.3748/wjg.v16.i42.5359
Revised: August 21, 2010
Accepted: August 28, 2010
Published online: November 14, 2010
AIM: To investigate p53 mutations in esophageal cancer in a high-risk population, and correlate them with smoking, alcohol consumption and betel chewing.
METHODS: One hundred and sixty-five tumor samples of esophageal squamous cell carcinoma (ESCC) obtained from a university hospital in Songkhla province, Southern Thailand were investigated for p53 mutations in exons 5-8, using polymerase chain reaction-single strand conformation polymorphism analysis, followed by direct sequencing. A polymerase chain reaction-restriction fragment length polymorphism (RFLP) assay was additionally used to confirm possible germline mutation in intron 6. A history of risk habits was obtained by interviews. The association between risk habits and mutation frequency was evaluated using the χ2 test.
RESULTS: The studied specimens were from 139 male and 26 female patients with ESCC, treated at Songklanagarind Hospital. Most of the patients were smokers (86.7%) and alcohol consumers (72.73%), and 38.3% were betel chewers. Forty-three mutations of the p53 gene were detected in 25.5% (42/165) of tumor samples. Mutations were most commonly found in exon 5 (25.6%) and exon 8 (25.6%). Mutations in the hot-spot codon 248 were found in four cases (9.3% of all mutations). G:C→C:G (30.23%), G:C→A:T (27.90%) and G:C→T:A (16.28%) were the prevalent spectra of mutations. Unexpectedly, among 10 intronic mutations, eight cases harbored a similar mutation: G→C substitution in intron 6 (nucleotide 12759, GenBank NC_000017). These were additionally confirmed by the RFLP technique. Similar mutations were also detected in their matched blood samples using RFLP and direct sequencing, which suggested germline mutations. There was no significant correlation between risk habits and p53 mutation frequency.
CONCLUSION: A proportion of Thai ESCC patients harbored specific intronic p53 mutations, which might be germline mutations. Further studies are needed to explore this novel finding.
-
Citation: Thongsuksai P, Boonyaphiphat P, Puttawibul P, Sudhikaran W. Specific intronic
p53 mutation in esophageal squamous cell carcinoma in Southern Thailand. World J Gastroenterol 2010; 16(42): 5359-5366 - URL: https://www.wjgnet.com/1007-9327/full/v16/i42/5359.htm
- DOI: https://dx.doi.org/10.3748/wjg.v16.i42.5359
Esophageal cancer is the eighth most common cancer worldwide, and there were 462 000 new cases in 2002[1]. It is a disease of high mortality, and ranks as the sixth most common cause of cancer death. There is a marked variation in incidence in different regions of the world; a 20-fold difference is observed between high-risk China and low-risk Western Africa. Other areas of moderately high risk are Southern and Eastern Africa, South-Central Asia, and Japan[1]. It seems that environmental carcinogens are responsible for these geographic differences and the different histological types. Tobacco and alcohol use are the main risk factors in Europe and North America[2,3], and other factors including betel chewing, hot beverages, fermented food, nutritional deficiencies or familial predisposition can be responsible for high rates in other high- or moderate-risk regions[4-7].
The incidence of esophageal cancer in Thailand is relatively low when one considers the country-wide estimates, with an age-standardized incidence rate (ASR) of 4.7 per 100 000 males in 1999[8]. However, the incidence is exceptionally high in Songkhla province in Southern Thailand, with an ASR of 8.1 per 100 000 males, which is close to worldwide incidence. In this region, oral cancer is event more prevalent, with the highest incidence (ASR 9.4 per 100 000 males) compared to other regions of the country. Most esophageal cancer cases in Thailand are squamous cell carcinomas. In our previous case-control study, alcohol consumption, cigarette smoking and betel quid chewing were found to be strong risk factors for esophageal squamous cell carcinoma (ESCC)[9].
The p53 tumor suppressor gene is an important gene in cell cycle regulation and apoptosis. Mutations in the p53 gene have been implicated as crucial events in the development of various cancers, including ESCC[10], and they have been identified as a vulnerable target for critical DNA damage. Analysis of p53 mutations in various human cancers has denoted a characteristic mutational pattern that is related to specific endogenous as well as exogenous carcinogen-related agents; a finding that has given rise to the term “mutagen fingerprints” in DNA[11].
p53 mutations in ESCC from Thailand have been reported by two groups in 1997 and 2000[12,13]. However, the numbers of cases were small and the relationship between mutations and risk habits were not explicitly evaluated. Here, we analyzed the p53 mutation profile of a larger sample set (165 cases) of ESCC using single-strand conformation polymorphism (SSCP) analysis and direct sequencing. In addition, the relationship between mutation frequency and risk habits, namely alcohol consumption, cigarette smoking and betel quid chewing, was examined.
Patients who were diagnosed with ESCC and treated at Songklanagarind Hospital during 1999-2005 were considered as candidates for the study. The study was approved by the Ethics Committee of the Faculty of Medicine, Prince of Songkla University, and informed consent was obtained from the patients. Data concerning detailed histories of tobacco use, alcohol consumption and betel chewing were obtained via face-to-face interviews using structured questionnaires. Only cases with available fresh-frozen tissue samples were included. Tissue samples were obtained from biopsy or surgical resection specimens, snapped frozen and stored at -80°C until DNA extraction. All of the cases were primary tumors that had not been treated with radiation or chemotherapy.
DNA was extracted from frozen tissues by standard methods. The tissue was digested overnight at 37°C in lysis buffer that contained 10% SDS, 10 mmol/L Tris, pH 8.0, 10 mmol/L NaCl, 10 mmol/L EDTA and 20 μL 10 mg/mL proteinase K. DNA extraction was performed using the phenol-chloroform method and it was precipitated by 1/10 volume of 4.0 mol/L NaCl and two volumes of cold absolute ethanol.
Exons 5-8 of the p53 gene were polymerase chain reaction (PCR)-amplified from tumor DNA, and mutations were detected by SSCP analysis. Samples that showed band shift were subjected to direct sequencing. Four sets of primer used were as follows: exon 5: TCTTCCTACAGTACTCCCCT sense, AGCTGCTCACCATCGCTATC antisense; exon 6: GATTGCTCTTAGGTCTGGCC sense, GCAAACCAGACCTCAGGCGG antisense; exon 7: TTATCTCCTAGGTTGGCTCT sense, GCTCCTGACCTGGAGTCTTC antisense; exon 8: TCCTGAGTAGTGGTAATCTA sense, GCTTGCTTACCTCGCTTAGT antisense.
PCR reactions were performed in a 50-μL volume reaction mixture that contained 0.5 μg genomic DNA, 10 pmol each primer, 100 mmol/L Tris, pH 8.3, 500 mmol/L KCl, 1.5 mmol/L MgCl2, 200 μmol/L dNTPs, and 1.25 U AmpliTag Gold (Perkin-Elmer, Foster City, CA, USA). Amplification was carried out in a Perkin-Elmer 480 DNA Thermal Cycler. The PCR conditions were 95°C for 10 min, followed by 35 cycles of 94°C denaturation for 1 min, 58°C annealing for 1 min, and 72°C extension for 1 min. The final extension was conducted at 72°C for 10 min.
For SSCP analysis, 2 μL PCR product was mixed with 5 μL 95% deionized solution that contained 0.1% bromophenol blue. The mixture was heat-denatured at 100°C for 5 min and rapidly placed on ice. Four microliters of each denatured product of exons 5 and 8 were loaded on to the 12% polyacrylamide gel (10 cm × 8 cm × 0.75 cm) with 5.26% crosslinking (19:1 acrylamide/bisacrylamide), supplemented with 5% glycerol. For exons 6 and 7, a ratio of 49:1 acrylamide/bisacrylamide (2.04% crosslinking) was used. Electrophoresis was performed in an ice box (12°C) at 2 W and constant 10 mA for 5 h for exons 5 and 8 and 1 h for exons 6 and 7. Positive controls, which consisted of samples that had been confirmed by direct sequencing to contain the p53 mutation, were run with each SSCP gel that was stained with silver nitrate. All positive cases were confirmed at least once by a separated PCR reaction and SSCP run.
The PCR products that showed band shift on the SSCP gel were purified using QIAquick PCR Purification Kit (Qiagen, Hilden, Germany) and then directly sequenced using the Ready Reaction Dye Terminator Cycle Sequencing kit (Perkin-Elmer). The primers used in sequencing were the same as those used in the PCR. Sequencing was performed on an automated sequencer (ABI-Prism 310, Applied Biosystems, Foster City, CA, USA). All mutations were confirmed by sequencing both DNA strands.
For intron 6 mutation at nucleotide 12759, other primers, not overlapped to the mutation point, were used (forward 5'-GCCTCTGATTCCTCACTGAT-3'; reverse 5'-TAAGCAGCAGGAGAAAGCCCC-3'). This experiment was also performed on four available matched blood samples and the sequencing was performed on an automated sequencer (ABI-Prism 3130).
PCR-restriction fragment length polymorphism analysis to detect intronic G→C at nucleotide 12759
As a significant number of cases showed G→C substitution in intron 6 at the 18th base after the end of exon 6 (corresponding to nucleotide 12759 based on GenBank NC_000017), we additionally confirmed this mutation through restriction fragment length polymorphism (RFLP) analysis. This analysis was also performed on matched blood samples of these cases to investigate whether they were germline mutations.
The 181-bp PCR product was amplified using primers, forward 5'-GCCTCTGATTCCTCACTGAT-3'; and reverse 5'-TTAACCCCTCCTCCCAGAGA-3'. The PCR was performed with 100 ng genomic DNA that contained 20 mmol/L Tris-HCl (pH 8.4), 50 mmol/L KCl, 2.0 mmol/L MgCl2, 37.5 μmol/L each nucleotide, and 1.25 U Taq polymerase. The cycling conditions were 95°C for 5 min, followed by 35 cycles of 95°C denaturation for 1 min, 60°C annealing for 1 min, and 72°C extension for 1 min, with a final extension of 72°C for 10 min. A 10-μL aliquot of each successful reaction was digested with 10 U BsaHI restriction enzyme (New England Biolabs, Beverly, MA, USA) in 2.5 μL 10 × NEB4 buffer with 12.5 μL water at 37°C for 2 h. The BsaHI-digested fragments were separated on 10% polyacrylamide gel. A complete digestion (denoted mutation) gave 158-bp and 23-bp DNA fragments.
The study included 165 tumor samples from 139 male and 26 female patients with ESCC diagnosed during 1999-2005. The mean age of patients was 63.4 years with a range of 37-91 years (Table 1). Most patients were habitual current smokers (86.7%) and drinkers (72.73%), with most of them (71.5%) reporting both habits. Habitual betel chewing was reported in 17 out of 26 females (65.4%) and in 45 out of 136 males (33.1%).
| Variable | Category | No. of subjects | Frequency (%) |
| Sex | Male | 139 | 84.2 |
| Female | 26 | 15.8 | |
| Age (yr) | Mean, range | 63.4 (37-91) | |
| Smoking | Never | 19 | 11.6 |
| Habitual | 143 | 86.7 | |
| Occasional | 3 | 1.8 | |
| Drinking | Never | 33 | 20.0 |
| Habitual | 120 | 72.7 | |
| Occasional | 12 | 7.3 | |
| Betel chewing | Never | 78 | 48.1 |
| Habitual | 62 | 38.3 | |
| Occasional | 22 | 13.6 |
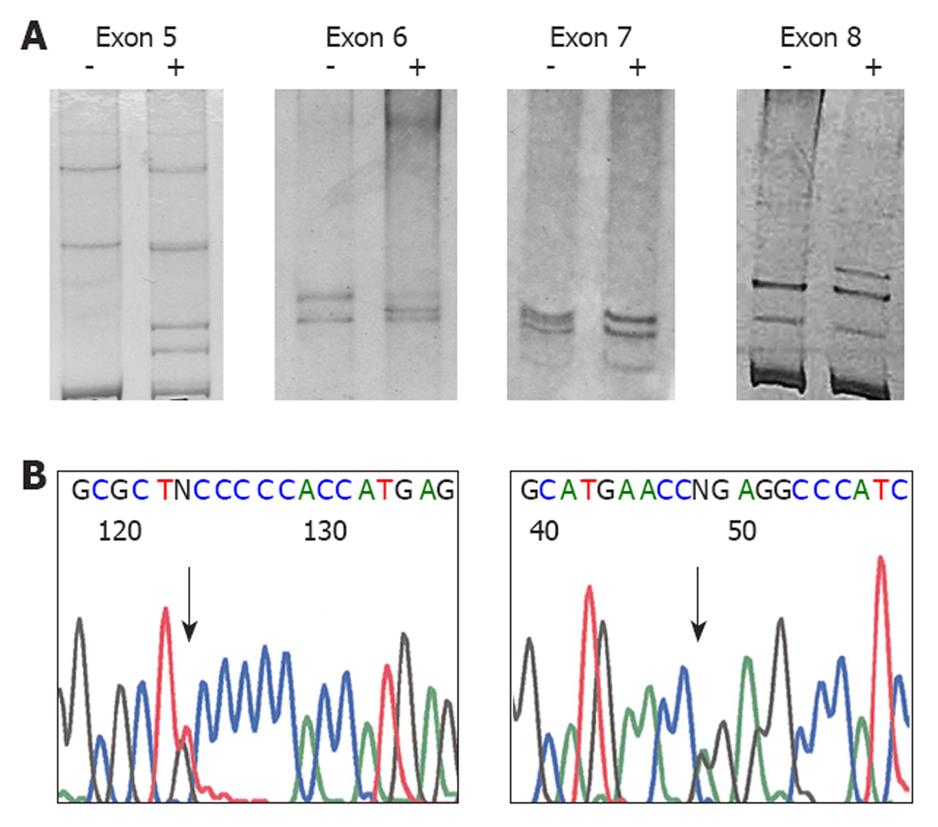
A total of 43 mutations were found in 42 tumors of the 165 samples (25.45%). The representative SSCP gels and sequencing chromatograms are shown in Figure 1.
Twenty-five mutations were missense mutations; one was nonsense, four were frameshift deletions, three were stop codons, and 10 were single base substitutions in the intron region. Mutations in coding sequences were most commonly found in exon 5 (25.58%) and exon 8 (25.58%) (Table 2). Among the 10 intronic mutations found, eight were intron 6 mutations.
| Location | n (%) |
| Exon 5 | 11 (25.58) |
| Exon 6 | 1 (2.33) |
| Exon 7 | 9 (20.93) |
| Exon 8 | 11 (25.58) |
| Intron 5 | 1 (2.33) |
| Intron 6 | 8 (18.60) |
| Intron 8 | 1 (2.33) |
| Exon-intron 6 | 1 (2.33) |
| Type of mutations | |
| Transitions | |
| G:C -> A:T | 8 (18.60) |
| G:C -> A:T at CpG | 4 (9.30) |
| A:T -> G:C | 2 (4.65) |
| Transversions | |
| G:C -> C:G | 13 (30.23) |
| G:C -> T:A | 7 (16.28) |
| A:T -> T:A | 4 (9.30) |
| Tandem | |
| GT -> TA | 1 (2.33) |
| Deletion | 4 (9.30) |
Of the five major mutation hot spots of the p53 gene (codon 175, 245, 248, 273 and 282), mutations at codon 248 were observed in four cases (accounting for 9.3% of all mutations), whereas mutations at other codons were not found.
The types of mutations are shown in Table 2. The most common type was G:C→C:G (30.23%), followed by G:C→A:T (27.90%) and G:C→T:A (16.28%). Surprisingly, eight out of 10 intronic mutations were found at the same location, that is, a G→C substitution at the 18th base after the last codon of exon 6 (nucleotide 12759, GenBank NC_000017). The details of the mutations of all the cases are shown in Table 3.
| Case ID | Age (yr)/sex | Exon | Codon | Nucleotide change | AA change | Exposure | ||
| Smoke | Alcohol | Betel | ||||||
| E111 | 60/M | 5 | 130-131 | 3 bp deletion | Frameshift | Yes | Yes | Yes |
| E303 | 73/M | 5 | 134-137 | 10 bp deletion | Frameshift | - | - | - |
| E006 | 63/M | 5 | 156 | CGC -> CCC | Arg -> Pro | Yes | Yes | No |
| E271 | 57/M | 5 | 158 | CGC -> CTC | Arg -> Leu | Yes | Yes | No |
| E072 | 58/F | 5 | 159 | GCC -> CCC | Ala -> Pro | Yes | Yes | Yes |
| E200 | 60/F | 5 | 161 | GCC -> ACC | Ala -> Thr | Yes | Yes | Yes |
| E397 | 56/M | 5 | 167 | CAG -> CGG | Gln -> Arg | Yes | Yes | No |
| E379 | 66/M | 5 | 168 | CAC -> CTC | His -> Leu | Yes | Yes | No |
| E355 | 61/M | 5 | 176 | TGC -> TTC | Cys -> Phe | Yes | No | Yes |
| E014 | 77/M | 5 | 176 | TGC -> TAC | Cys -> Tyr | No | Yes | Yes |
| E022 | 46/M | 5 | 184 | GAT -> AAT | Asp -> Asn | Yes | Yes | Yes |
| E464 | 68/M | 6 | 190 | CCT -> CTT | Pro -> Leu | Yes | Yes | No |
| E259 | 60/M | 7 | 228-232 | 21 bp deletion | Frameshift | Yes | Yes | No |
| E320 | 72/M | 7 | 234 | TAC -> TGC | Tyr -> Cys | No | Yes | Yes |
| E264 | 52/M | 7 | 238 | TGT -> TAT | Cys -> Tyr | Yes | Yes | No |
| E446 | 58/M | 7 | 245 | GGC -> CGC | Gly -> Arg | Yes | Yes | No |
| E106 | 56/M | 7 | 248 | CGG -> CAG (CpG site) | Arg -> Gln | Yes | Yes | No |
| E298 | 79/F | 7 | 248 | CGG -> TGG (CpG site) | Arg -> Trp | Yes | No | Yes |
| E294 | 52/M | 7 | 248 | CGG -> TGG (CpG site) | Arg -> Trp | Yes | Yes | No |
| E419 | 54/M | 7 | 248 | CGG -> TGG (CpG site) | Arg -> Trp | Yes | Yes | No |
| E022 | 53/M | 7 | 249 | AGG -> ATG | Arg -> Met | Yes | Yes | No |
| E012 | 74/M | 8 | 266 | GGA -> TGA | Gly -> Ter (end) | Yes | Yes | No |
| E231 | 54/M | 8 | 272 | GTG -> TAG | Val -> Ter (end) | Yes | Yes | Yes |
| E455 | 46/M | 8 | 272 | GTG -> ATG | Val -> Met | No | Yes | Yes |
| E449 | 47/M | 8 | 278 | CCT -> TCT | Pro -> Ser | No | Yes | Yes |
| E002 | 53/M | 8 | 279 | GGG -> GAG | Gly -> Glu | Yes | Yes | - |
| E027 | 54/M | 8 | 280 | AGA -> AGT | Arg -> Ser | Yes | Yes | No |
| E444 | 79/M | 8 | 283 | CGC -> CCC | Arg -> Pro | No | Yes | No |
| E387 | 74/F | 8 | 286 | GAA -> CAA | Glu -> Gln | Yes | Yes | No |
| E146 | 62/M | 8 | 287 | GAG -> TAG | Glu -> Ter (end) | Yes | Yes | No |
| E181 | 58/F | 8 | 287 | GAG -> TAG | Glu -> Ter (end) | No | No | No |
| E408 | 58/F | 8 | 296 | CAC -> CTC | His -> Leu | No | No | No |
| E462 | 74/M | Exon-intron 6 | 21 bp deletion | Affect splice site | Yes | Yes | No | |
| E023 | 51/F | Intron 5 | TGAGC -> TCTGC | - | No | No | Yes | |
| E158 | 61/M | Intron 6 | GGGG -> GGCG | - | Yes | Yes | Occ | |
| E169 | 53/M | Intron 6 | GGGG -> GGCG | - | Yes | Yes | No | |
| E189 | 60/M | Intron 6 | GGGG -> GGCG | - | Yes | Yes | Yes | |
| E199 | 48/M | Intron 6 | GGGG -> GGCG | - | Yes | Yes | Yes | |
| E240 | 61/M | Intron 6 | GGGG -> GGCG | - | Yes | Yes | - | |
| E302 | 41/M | Intron 6 | GGGG -> GGCG | - | Yes | Yes | Occ | |
| E329 | 63/M | Intron 6 | GGGG -> GGCG | - | Yes | Yes | Yes | |
| E435 | 45/M | Intron 6 | GGGG -> GGCG | - | Yes | Yes | No | |
| E409 | 56/M | Intron 8 | ACGAG -> ACTAG | - | Yes | Yes | Yes | |
Intronic G→C substitution at nucleotide 12759
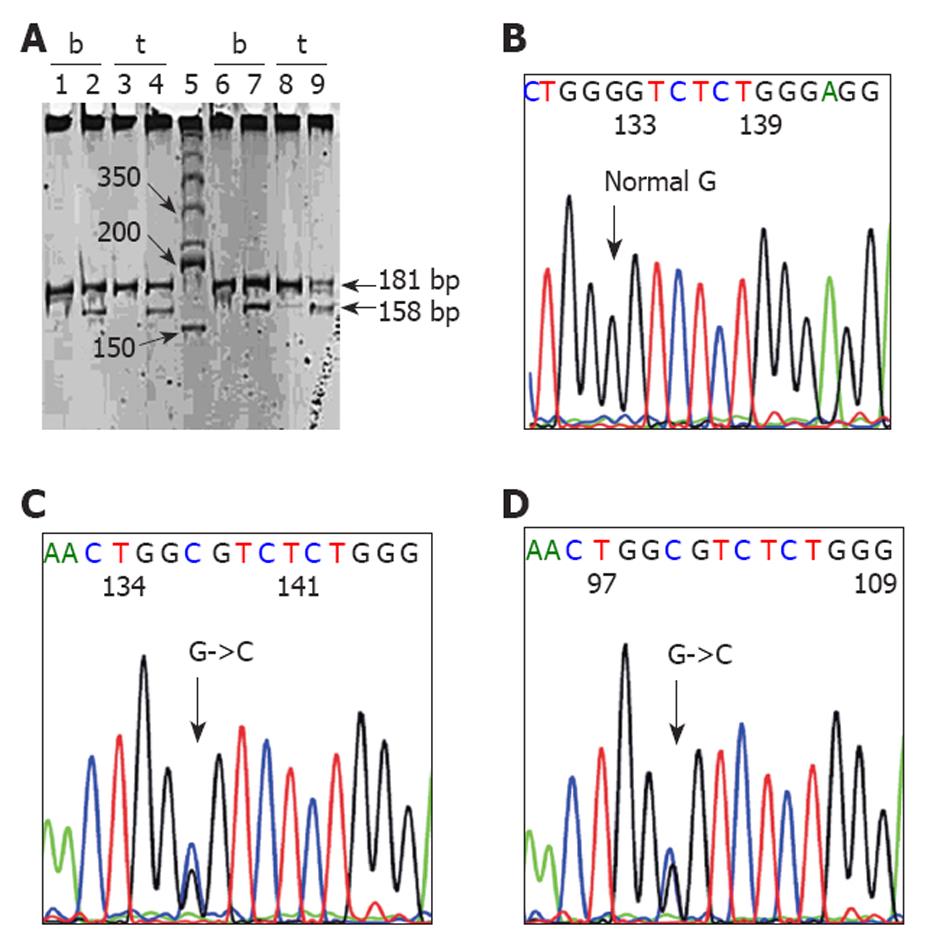
As a result of the high frequency of intron 6 G→C substitution at 12759 (eight cases), we confirmed these mutations by the RFLP method and the results were similar. We further investigated whether these were germline mutations by examining their matched blood samples through RFLP and direct sequencing. Seven blood samples were available for RFLP and the results denoted a mutation in all of the cases, which suggested germline mutations (Figure 2). The DNA of only four blood samples was available for further direct sequencing and the mutations were confirmed in three out of four samples examined (Figure 2).
We also evaluated functional changes in the p53 proteins of these cases using an immunohistochemistry method (p53 antibody DO-7 clone; DakoCytomation, Carpinteria, CA, USA). Only five cases had adequate tissue for evaluation and the results showed diffuse strong expression in four cases and negative expression in one.
The frequency of mutations in relation to clinicopathological variables is shown in Table 4. Patients younger than 60 years had a significantly higher frequency of p53 mutations than older patients (38.7% vs 17.5%, P = 0.002). The mutation frequency was equal in both sexes. In relation to lifestyle habits, the frequency of mutations was slightly higher in non-smokers (36.4%) than smokers (23.8%), and in non-drinkers (33.3%) than drinkers (22.5%). The mutation frequency among betel and non-betel chewers was equal (24.2% and 25.0%). However, there were no statistically significant differences in mutation frequency between exposed and non-exposed patients to all of the three habits.
| Variables | Mutant p53 | Wild-type p53 | P value |
| Sex | |||
| Male | 36 (25.9) | 103 (74.1) | |
| Female | 6 (23.1) | 20 (76.9) | 0.762 |
| Age (yr) | |||
| ≤ 60 | 24 (38.7) | 38 (61.3) | |
| > 60 | 18 (17.5) | 85 (82.5) | 0.002 |
| Family history | |||
| Yes | 6 (37.5) | 10 (62.5) | |
| No | 33 (23.7) | 106 (76.3) | 0.230 |
| Smoking | |||
| Yes | 34 (23.8) | 109 (76.2) | |
| No/occasional | 8 (36.4) | 14 (63.6) | 0.207 |
| Drinking | |||
| Yes | 27 (22.5) | 93 (77.5) | |
| No/occasional | 15 (33.3) | 30 (66.7) | 0.155 |
| Betel chewing | |||
| Yes | 15 (24.2) | 47 (75.8) | |
| No/occasional | 25 (25.0) | 75 (75.0) | 0.908 |
The present study is the third on p53 mutations in Thai ESCC patients. All of the samples in these three studies were from the same hospital, a university hospital in Songkhla province, Southern Thailand. These studies were conducted at different times, and the present study confirmed the findings of the previous studies and found an additional unique mutation profile.
The present study demonstrated a 25.45% (42/165) frequency of p53 mutations. This frequency is relatively low compared to those of previous studies; however, wide variations in p53 mutation frequencies, ranging from 17% to 80%, have been reported. These variations might be related to several factors including the sensitivity of technique used in the detection of the mutations, the length of the examined regions, and the number of cases. However, the most notable factor responsible for the frequency variation could be the difference in mutagens in different populations. A high frequency of p53 mutation (> 50%) is usually reported in countries with a high incidence of ESCC, such as China and France[14,15], whereas lower frequency of mutation is found in low- or moderate-incidence countries[16-18]. Thailand is a moderate-risk area for ESCC; therefore, the frequency can be expected to be relatively low.
Nevertheless, the two previous studies from Thailand have demonstrated higher frequency of p53 mutations compared to the present study[12,13]. The first study by Suwiwat et al [12] has reported 10 mutations in eight out of 16 (50%) cases. The second study by Tanière et al [13] has reported 25 mutations in 23 out of 56 cases (41%). The lower frequency might represent underestimated data, whereas the higher frequency might represent overestimated data. The low frequency of mutations in the present study might have been due to various factors, among which was the fact that we used tumor samples that could have contained both tumor and non-tumor cells, in contrast to the microdissected tumor cells used in the study of Tanière et al[13]. With regard to the screening method used, both SSCP and denaturing gradient gel electrophoresis (Tanière study) have been reported to have comparable sensitivity[19]. However, the small number of cases examined could result in over-figured data due to sampling bias.
The present study demonstrated heterogeneous mutation types, which predominantly involved the G:C base pair. This is similar to previous Thai reports except for a relatively higher proportion of G:G to C:G transversion (30% vs 23%) and a lower proportion of G:C to A:T transition at CpG (9.3% vs 17.14%). The patterns of predominant G:A to A:T transition and G:C to T:A transversion have also been reported in high-risk areas such as China[14,20] and moderate-risk countries such as Japan and India[18,21]. This is different, however, from the high-risk area of Western Europe where a relatively higher proportion of mutations at the A:T base pair has been reported[22].
The G:C to A:T transition accounted for 28% of all mutations in the present study. One-third of these (4/12 mutations) were G:C to A:T transition at the CpG site, and all were found at the hot spot codon 248. A G:C to A:T transition at the CpG site was thought to have resulted from spontaneous deamination of 5-methylcytosine to form thymine[23], which preferentially occurred at codons 175, 245, 248, 273 and 282 in the p53 gene. The previous Thai studies have reported transition at the CpG site of codon 175 (one case), 273 (one case) and 248 (three cases)[12,13]. These findings suggest that codon 248 is the most common hot spot codon in Thai ESCC cases.
In reference to the G:C to A:T transition at a non-CpG site, laboratory studies have found that it is the most common mutation caused by alkylating agents, consistent with O6-methylguanine mispairing with thymine[24]. Mutagenic alkylating N-nitrosamines in tobacco smoke might be responsible for this mutation. In China and India, dietary N-nitrosamines might also contribute to this mutation type[20,25]. Our previous study has demonstrated that betel chewing also is a strong risk factor for ESCC in Thailand[9]. Nitroso derivatives from areca alkaloids have been proven to be oncogenic in animal models[26]. They have been found probably to account for the predominant G:A to A:T transition in betel-chewing-related oral cancers[27]. Most of the patients in the current study had a history of drinking and smoking, as well as betel chewing, therefore, smoking and betel chewing might both contribute to the G:C to A:T transition in Thai ESCC patients. However, it is difficult to identify a specific type of mutation with a specific risk factor because the mutation patterns are considerably heterogeneous and most patients have multiple risk habits.
In the present study, we unexpectedly found a high frequency of G to C substitution at the 18th base after the end of exon 6 (nucleotide 12759, GenBank NC_000017). We additionally found that these were germline mutations because similar mutations were also found in their blood samples. We validated these results by a second method, the RFLP.
From the total of 26597 somatic mutation records in the IARC TP53 database, version R14[28], intronic mutations have been found in 699 records, which represents 2.63% of the total mutations. G to C substitution at nucleotide 12759, similar to the present study, has been found in three cases; two were gastric lymphomas from Hong-Kong[29] and one was small-cell lung carcinoma from Russia[30]. There was a case of ESCC reported to have intron 6 G to C at nucleotide 12758. Surprisingly, this was a case from the study of Tanière et al[13], which was the previous study from our hospital. Looking at the details of the mutation in this published article, we found it to be GGGG→GGCG (case 9, Table II)[13], which represents a change at the 18th base after the end of exon 6 or nucleotide 12759, based on the GenBank NC_000017 reference sequence, rather than at nucleotide 12758. Surprisingly, in this case, a similar mutation was also found in the adjacent uninvolved tissue and gastric mucosa, which suggests a germline mutation. These findings suggest that intronic G to C substitution at nucleotide 12759, which might be a germline mutation, is prevalent in Thai ESCC. It should be noted that the cases included in the Tanière study would not have been included in the present study because the periods of sample collection did not overlap (1990-1998 vs 1999-2005).
The role of intronic base changes on the function of genes has been questioned. However, some studies have demonstrated alterations in introns or splice donors that affect the expression or function of the p53 gene[31,32]. In particular, Lehman et al[32] have demonstrated functional change of the immortalized lymphoblastoid cell lines derived from familial breast cancer patients who had germline G to C substitution in intron 6 at nucleotide 13964 (or nucleotide 13274 based on the GenBank NC_000017). In addition, immunohistochemical analysis of breast tumors from these patients also has revealed high levels of mutant p53 protein, which suggests a functional mutation. Our results were consistent with this study, which confirms that cases with suspected germline G to C substitution at 12759 have a high level of p53 expression. All this evidence indicates that germline intronic G to C substitution at 12759 is prevalent and associated with inherited risk of ESCC in Songkhla, Thailand. In a recent IARC TP53 database, version R14[28], this intronic base change has not been reported as any polymorphic sequence variation (polymorphism) or germline mutation. However, as this mutation was not investigated in healthy controls in the current study, any conclusion on the role of this mutation is still limited. Further studies to detect this mutation in healthy controls as well as in familial members of affected patients should be performed.
It is believed that p53 mutations result from specific carcinogens[11]. In some cancers, such as those of the lung or urinary bladder, the link between risk factors, in particular smoking, and p53 mutation frequency and/or pattern have been consistently demonstrated[30,33]. However, such data on esophageal carcinoma are limited and inconsistent[20,34]. Consistent with some of these reports, the present study did not find any association between p53 mutation frequency and smoking, alcohol consumption or betel chewing. Various reasons could account for the lack of association. ESCC might be associated with many risk factors. This hypothesis is supported by studies from India that have found a significant correlation of p53 mutation frequency in ESCC with diets rich in nitrosamines[25,35]. In addition, risk of cancer development might be different among exposed individuals due to genetic polymorphism of carcinogen-metabolizing enzymes, which determine individual capacity to detoxify carcinogens. This could modify the relationship between the exposure and gene mutation. Finally, the sample size in the current study could have been too small to detect any significant association between exposure and p53 gene mutation.
In conclusion, our results have demonstrated that the Thai population, which is in a moderate-risk area for ESCC, has p53 mutational spectra that are likely related to specific endogenous and exogenous carcinogens. However, a statistically significant relation between the mutation frequency among exposure groups was not demonstrated. We unexpectedly found a high frequency of G to C mutation at intron 6, which might be germline mutations. Further studies are needed to explore the questions arising from the results observed.
Cancer of the esophagus is prevalent in some regions of the world including Thailand. It is a dreadful disease that patients may die shortly after diagnosis. Environmental factors as well as familial predisposition have been shown to be associated with the development of this cancer, possibly via an alteration of the p53 tumor-suppressor gene.
Mutations in the p53 gene have been implicated to be critical events in the development of various cancers. Significant association between specific exposures and the p53 mutations has been evident in some cancers, but the data in esophageal squamous cell carcinoma are limited.
The mutation profiles identified are consistent with most previous reports. The mutation types, G:C to C:G (30.2%), G:C to A:T (27.9%) and G:C to T:A (16.3%) were prevalent and likely to be associated with combination of exposures. Exceptionally, a unusually high frequency (8 from 42 cases) of intron 6 mutation (G to C substitution) at nucleotide 12759 was found and they were proofed to be germline mutations.
The results indicated that a proportion of esophageal cancer in this region is heritable. Further study is to be conducted to identify this specific germline mutation in healthy population and in familial members of the patients. The information would be valuable for designing diagnosis and preventive intervention in high-risk population.
An intron is a region within a gene that is not translated into protein. It is transcribed to pre-mRNA and subsequently removed by a process called splicing. A germline mutation is a heritable variation in the lineage of germ cells. Mutations in these cells are transmitted to offspring while those in somatic cells are not. Germline mutations play a key role in genetic diseases and also in certain types of cancer.
The authors found intronic p53 mutation in esophageal squamous cell carcinoma in Southern Thailand, which was considered as a germline mutation. This is a novel finding and interesting.
Peer reviewer: Mitsuyoshi Urashima, MD, PhD, MPH, Division of Molecular Epidemiology, Jikei University School of Medicine, 3-25-8 Nishi-shimbashi, Minato-ku, Tokyo 105-8461, Japan
S- Editor Cheng JX L- Editor Kerr C E- Editor Lin YP
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. |
| 2. | Hashibe M, Boffetta P, Janout V, Zaridze D, Shangina O, Mates D, Szeszenia-Dabrowska N, Bencko V, Brennan P. Esophageal cancer in Central and Eastern Europe: tobacco and alcohol. Int J Cancer. 2007;120:1518-1522. |
| 3. | Xu XC. Risk factors and gene expression in esophageal cancer. Methods Mol Biol. 2009;471:335-360. |
| 4. | Hu N, Dawsey SM, Wu M, Bonney GE, He LJ, Han XY, Fu M, Taylor PR. Familial aggregation of oesophageal cancer in Yangcheng County, Shanxi Province, China. Int J Epidemiol. 1992;21:877-882. |
| 5. | Castellsagué X, Muñoz N, De Stefani E, Victora CG, Castelletto R, Rolón PA. Influence of mate drinking, hot beverages and diet on esophageal cancer risk in South America. Int J Cancer. 2000;88:658-664. |
| 6. | Kamangar F, Chow WH, Abnet CC, Dawsey SM. Environmental causes of esophageal cancer. Gastroenterol Clin North Am. 2009;38:27-57, vii. |
| 7. | Islami F, Malekshah AF, Kimiagar M, Pourshams A, Wakefield J, Goglani G, Rakhshani N, Nasrollahzadeh D, Salahi R, Semnani S. Patterns of food and nutrient consumption in northern Iran, a high-risk area for esophageal cancer. Nutr Cancer. 2009;61:475-483. |
| 8. | Khuhaprema T, Srivatanakul P, Sriplung H, Wiangnon S, Sumitsawan Y, Attasara P. Cancer in Thailand. Vol IV, 1998-2000. Bangkok: Ministry of Public Health 2007; . |
| 9. | Boonyaphiphat P, Thongsuksai P, Sriplung H, Puttawibul P. Lifestyle habits and genetic susceptibility and the risk of esophageal cancer in the Thai population. Cancer Lett. 2002;186:193-199. |
| 10. | Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49-53. |
| 11. | Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855-4878. |
| 12. | Suwiwat S, Oda H, Shimizu Y, Ishikawa T. Prevalence of p53 mutations and protein expression in esophageal cancers in southern Thailand. Int J Cancer. 1997;72:23-26. |
| 13. | Tanière P, Martel-Planche G, Puttawibul P, Casson A, Montesano R, Chanvitan A, Hainaut P. TP53 mutations and MDM2 gene amplification in squamous-cell carcinomas of the esophagus in south Thailand. Int J Cancer. 2000;88:223-227. |
| 14. | Hu N, Huang J, Emmert-Buck MR, Tang ZZ, Roth MJ, Wang C, Dawsey SM, Li G, Li WJ, Wang QH. Frequent inactivation of the TP53 gene in esophageal squamous cell carcinoma from a high-risk population in China. Clin Cancer Res. 2001;7:883-891. |
| 15. | Robert V, Michel P, Flaman JM, Chiron A, Martin C, Charbonnier F, Paillot B, Frebourg T. High frequency in esophageal cancers of p53 alterations inactivating the regulation of genes involved in cell cycle and apoptosis. Carcinogenesis. 2000;21:563-565. |
| 16. | Pütz A, Hartmann AA, Fontes PR, Alexandre CO, Silveira DA, Klug SJ, Rabes HM. TP53 mutation pattern of esophageal squamous cell carcinomas in a high risk area (Southern Brazil): role of life style factors. Int J Cancer. 2002;98:99-105. |
| 17. | Gamieldien W, Victor TC, Mugwanya D, Stepien A, Gelderblom WC, Marasas WF, Geiger DH, van Helden PD. p53 and p16/CDKN2 gene mutations in esophageal tumors from a high-incidence area in South Africa. Int J Cancer. 1998;78:544-549. |
| 18. | Egashira A, Morita M, Kakeji Y, Sadanaga N, Oki E, Honbo T, Ohta M, Maehara Y. p53 gene mutations in esophageal squamous cell carcinoma and their relevance to etiology and pathogenesis: results in Japan and comparisons with other countries. Cancer Sci. 2007;98:1152-1156. |
| 19. | Condie A, Eeles R, Borresen AL, Coles C, Cooper C, Prosser J. Detection of point mutations in the p53 gene: comparison of single-strand conformation polymorphism, constant denaturant gel electrophoresis, and hydroxylamine and osmium tetroxide techniques. Hum Mutat. 1993;2:58-66. |
| 20. | Bennett WP, von Brevern MC, Zhu SM, Bartsch H, Muehlbauer KR, Hollstein MC. p53 mutations in esophageal tumors from a high incidence area of China in relation to patient diet and smoking history. Cancer Epidemiol Biomarkers Prev. 1997;6:963-966. |
| 21. | Ralhan R, Arora S, Chattopadhyay TK, Shukla NK, Mathur M. Circulating p53 antibodies, p53 gene mutational profile and product accumulation in esophageal squamous-cell carcinoma in India. Int J Cancer. 2000;85:791-795. |
| 22. | Hollstein MC, Peri L, Mandard AM, Welsh JA, Montesano R, Metcalf RA, Bak M, Harris CC. Genetic analysis of human esophageal tumors from two high incidence geographic areas: frequent p53 base substitutions and absence of ras mutations. Cancer Res. 1991;51:4102-4106. |
| 24. | Horsfall MJ, Gordon AJ, Burns PA, Zielenska M, van der Vliet GM, Glickman BW. Mutational specificity of alkylating agents and the influence of DNA repair. Environ Mol Mutagen. 1990;15:107-122. |
| 25. | Murtaza I, Mushtaq D, Margoob MA, Dutt A, Wani NA, Ahmad I, Bhat ML. A study on p53 gene alterations in esophageal squamous cell carcinoma and their correlation to common dietary risk factors among population of the Kashmir valley. World J Gastroenterol. 2006;12:4033-4037. |
| 26. | Rivenson A, Hoffmann D, Prokopczyk B, Amin S, Hecht SS. Induction of lung and exocrine pancreas tumors in F344 rats by tobacco-specific and Areca-derived N-nitrosamines. Cancer Res. 1988;48:6912-6917. |
| 27. | Thongsuksai P, Boonyaphiphat P, Sriplung H, Sudhikaran W. p53 mutations in betel-associated oral cancer from Thailand. Cancer Lett. 2003;201:1-7. |
| 28. | Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622-629. |
| 29. | Chan WY, Chan EK, Chow JH. Epstein-Barr virus-associated gastric lymphomas are distinct from mucosa-associated lymphoid tissue-type lymphomas: genetic abnormalities of p53 gene. Diagn Mol Pathol. 2001;10:153-160. |
| 30. | Le Calvez F, Mukeria A, Hunt JD, Kelm O, Hung RJ, Tanière P, Brennan P, Boffetta P, Zaridze DG, Hainaut P. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former, and current smokers. Cancer Res. 2005;65:5076-5083. |
| 31. | Lozano G, Levine AJ. Tissue-specific expression of p53 in transgenic mice is regulated by intron sequences. Mol Carcinog. 1991;4:3-9. |
| 32. | Lehman TA, Haffty BG, Carbone CJ, Bishop LR, Gumbs AA, Krishnan S, Shields PG, Modali R, Turner BC. Elevated frequency and functional activity of a specific germ-line p53 intron mutation in familial breast cancer. Cancer Res. 2000;60:1062-1069. |
| 33. | Moore LE, Smith AH, Eng C, DeVries S, Kalman D, Bhargava V, Chew K, Ferreccio C, Rey OA, Hopenhayn C. P53 alterations in bladder tumors from arsenic and tobacco exposed patients. Carcinogenesis. 2003;24:1785-1791. |
| 34. | Saeki H, Ohno S, Araki K, Egashira A, Kawaguchi H, Ikeda Y, Morita M, Kitamura K, Sugimachi K. Alcohol consumption and cigarette smoking in relation to high frequency of p53 protein accumulation in oesophageal squamous cell carcinoma in the Japanese. Br J Cancer. 2000;82:1892-1894. |
| 35. | Gaur D, Arora S, Mathur M, Nath N, Chattopadhaya TK, Ralhan R. High prevalence of p53 gene alterations and protein overexpression in human esophageal cancer: correlation with dietary risk factors in India. Clin Cancer Res. 1997;3:2129-2136. |