INTRODUCTION
Colorectal adenomas and carcinomas are much more common than small bowel tumors, and colorectal cancer (CRC) is a leading cause of cancer deaths in Europe and the United States. The clinical symptoms of bleeding and/or obstruction found in the presence of large intestinal neoplasias are similar regardless of histologic type or etiology. It has been suggested that there is an association between colorectal carcinogenesis and pathways of fatty acid metabolism, with long-chain acyl-CoA thioesters as essential intermediates and cyclooxygenases (COXs) and acyl-CoA synthetases (ACSLs) as important enzymes. This observation is determined by molecular data indicating high functional diversity of activated lipid intermediates including formation of complex lipids (e.g. ceramide) and utilization in beta-oxidation as well as regulation of cellular signaling, transcriptional activity, cellular structure, and cellular behavior. Dietary lipids have been assumed as important additional variables in colorectal carcinogenesis.
In the following paragraphs the modifying role of lipids in colorectal carcinogenesis and the innovative technical approaches to elucidate the complex molecular network of lipids are addressed in detail.
EPIDEMIOLOGY AND DIVERSITY OF COLORECTAL CANCER
CRC continues to be one of the most common malignancies worldwide and the third leading cause of death among cancers[1]. In industrialized Western countries, the incidence and prevalence are higher than in Asia or developing countries. Although the molecular mechanism of CRC has become better understood in the past 2 decades, the prognosis of CRC, especially advanced cancer, has not significantly improved. Five-year survival rates have risen from 56.5% for patients diagnosed in the early 1980s to only 63.2% for those diagnosed in the early 1990s, and most recently to 64.9%[2]. The sparsely improving trend is most likely due to the fact that the prognosis for patients with CRC is highly dependent on disease stage at the point of diagnosis. Only 10% of CRCs are diagnosed early, and in most patients advanced disease is found. Therefore, early detection of CRC is essential in improvement of a patient’s prognosis.
CRCs comprise a heterogeneous group of malignancies including hereditary forms and so-called sporadic tumors[3,4]. At the current state of knowledge, the majority of CRCs are sporadic. However, experimental and epidemiological data give evidence that the class of sporadic CRCs comprises a heterogeneous group of malignancies with differences in pathogenesis and prognosis. It is suggested that the pathogenesis of some sporadic CRCs does not follow the traditional adenoma-carcinoma sequence. Recently, a new pathway in “sporadic” CRC development, the serrated pathway, has been discovered[5]. The molecular alterations seen in serrated tumors are different from those traditionally found in the adenoma-carcinoma sequence. In serrated lesions, loss of heterozygosity and mutations in APC or TP53 are rare, whereas hypermethylation of CpG islands and microsatellite instability are frequent. At present, 2 categories of serrated carcinogenesis are defined including 2 basic pathways, the sessile serrated pathway and the traditional serrated pathway. While in the sessile serrated pathway BRAF mutation and high-level microsatellite instability are molecular hallmarks in carcinogenesis, and tumor location is predominantly in the right colon, KRAS mutation and microsatellite stability have been characterized as important steps in the traditional serrated pathway, where cancers arise mainly in the left colon.
The diversity in colorectal carcinogenesis is accompanied by numerous genetic and epigenetic alterations. Epidemiological data indicate that the heterogeneity of such genomic lesions is determined by additive factors, called the modifiers of colorectal carcinogenesis[6]. Evidence is given that diet and nutrition are key factors acting as carcinogenesis modifiers. This activity is probably relevant in both steps of colorectal tumorigenesis, initiation and progression. Therefore, identification and characterization of nutritional components as promoters or silencers of intestinal cancer are of high interest in tumor preventive strategies, tumor epidemiology, and the health economy.
THE MODIFIER CONCEPT OF COLORECTAL CARCINOGENESIS
At present, the model of tumor genotype to phenotype/histomorphology correlation, best-known as the adenoma-carcinoma sequence, is well accepted. The genetic changes that occur in colorectal carcinogenesis are paralleled by the progression from small pre-malignant lesions to advanced carcinomas indicating that each successive genetic injury/modification confers an advantageous characteristic upon the expanding tumor. The MAPK-ERK (mitogen-activated protein kinase-extracellular signal-regulated kinase) pathway, which mediates cellular responses to many extracellular inputs, is involved in regulation of apoptosis, cell growth, secretion, and cellular differentiation. K-ras and B-raf are key players in this signaling cascade, because mutations result in continuous gene activation leading to autonomous cell proliferation and tumor growth. The KRAS gene belongs to the Ras oncogene family and encodes a 21 kDa protein with intrinsic GTPase activity. This protein is regulated by a cycle of de- and re-palmitoylation, which promotes its rapid exchange between the plasma membrane and the Golgi apparatus. While numerous different genes can become altered in carcinogenesis, it has been recently proposed that the number of deregulated signaling pathways is relatively small[7].
Over the last few decades it has been shown that aberrant activity of some cellular pathways is crucial in colorectal carcinogenesis. Alterations in the coding genes, e.g. APC or TP53 mutations and loss of heterozygosity, were elucidated as key mechanisms for misdirected signaling[8]. Important insights into such principles of cancer-relevant molecular signaling were given by the characterization of hereditary colorectal carcinoma syndromes, i.e. hereditary non-polyposis colon cancer, familial adenomatous polyposis, and MUTYH-associated polyposis. Genotype to phenotype correlations in affected patients revealed that the type of variation in gene sequence and epigenetics is essential for onset and progress in cancer development. However, some inter-individual discrepancies were found in this correlation analysis, indicating additional molecular mechanisms that are likely to modify pathway execution[9-11]. These observations were substantiated by several experiments including studies in APC mice revealing examples of modifying genetic loci and non-genetic co-factors, e.g. nutritional components[12]. In mice, modifying genetic loci were addressed with the acronym Mom (modifiers of min) and demonstrated the genetic-based principle of modulation in gene expression with relevance to disease severity. One interesting candidate gene, firstly identified in the APC mice, is Pla2g2a coding secretory phospholipase A2 (PLA2)[13]. Activity of this enzyme (prototype group I) is associated with diverse functions including digestion, smooth muscle contraction, and cell proliferation. In addition, PLA2s of prototype group II are involved in inflammatory conditions and are upregulated by pro-inflammatory cytokines such as tumor necrosis factor and interleukin-1B. Recently, the PLA2-receptor (PLA2R) has been identified as a potential new tumor suppressor gene crucial in the induction of cellular senescence through activation of the p53 pathway[14].
In addition to genes acting as defined modifiers in colorectal carcinogenesis, matrix composition has been identified as an important factor to modify epithelial cell behavior and intestinal tumorigenesis[15]. The heterogeneity in matrix composition is marked and is a result of the complex mixture of non-epithelial cells embedded in a well-adapted set of extracellular stored lamellar and non-lamellar molecules. Additionally, different types of secreted intestinal products, such as mucins and other glycoproteins which are non-cellular matrix-components have been identified as important modifiers in intestinal pathogenesis[16]. Muc2, expressed by goblet cells, is the most abundant secreted intestinal mucin and important in intestinal homeostasis. The impaired expression and/or secretion of Muc2 have been correlated with the development of inflammatory intestinal conditions and colorectal carcinogenesis[17-20]. Recently, the molecular basis for this phenomenon was identified, when an interaction of Muc2 and Apc resulting in modification of Wnt signalling was determined[21,22].
In addition to Muc2, other molecules such as cytokines are suggested as important mediators in chronic intestinal inflammation and modifiers of intestinal tumor development. In the intestine, the main sources of cytokines and inflammation-triggered molecules are lymphocytes. The pathophysiological association of chronic inflammatory activity and intestinal tumorigenesis has been substantiated by epidemiological data demonstrating a high incidence of CRC in patients with chronic inflammatory bowel disease, a condition additionally associated with disturbances in composition of non-cellular matrix components.
Over the last decade, an additional modifier of colorectal carcinogenesis and pathogenesis of non-neoplastic intestinal diseases has been identified and characterized, namely the microbiome. The term intestinal microbiome describes the taxonomically complex and micro-ecological highly dynamic community of microorganisms on the gut mucosal surface[23]. The interaction of surface lining mucosal cells and the microbiome has an impact on the metabolome and lipidome, the maturation and proliferation of intestinal cells along the crypt-plateau axis, tissue homeostasis, and can be a factor in various diseases, such as inflammatory bowel disease and obesity[24,25]. The successful use of designed bacterial strains in the treatment of inflammatory intestinal conditions, such as necrotizing enterocolitis in preterm infants, underline that the microbiome modificatory activity is fundamental in intestinal homeostasis[26].
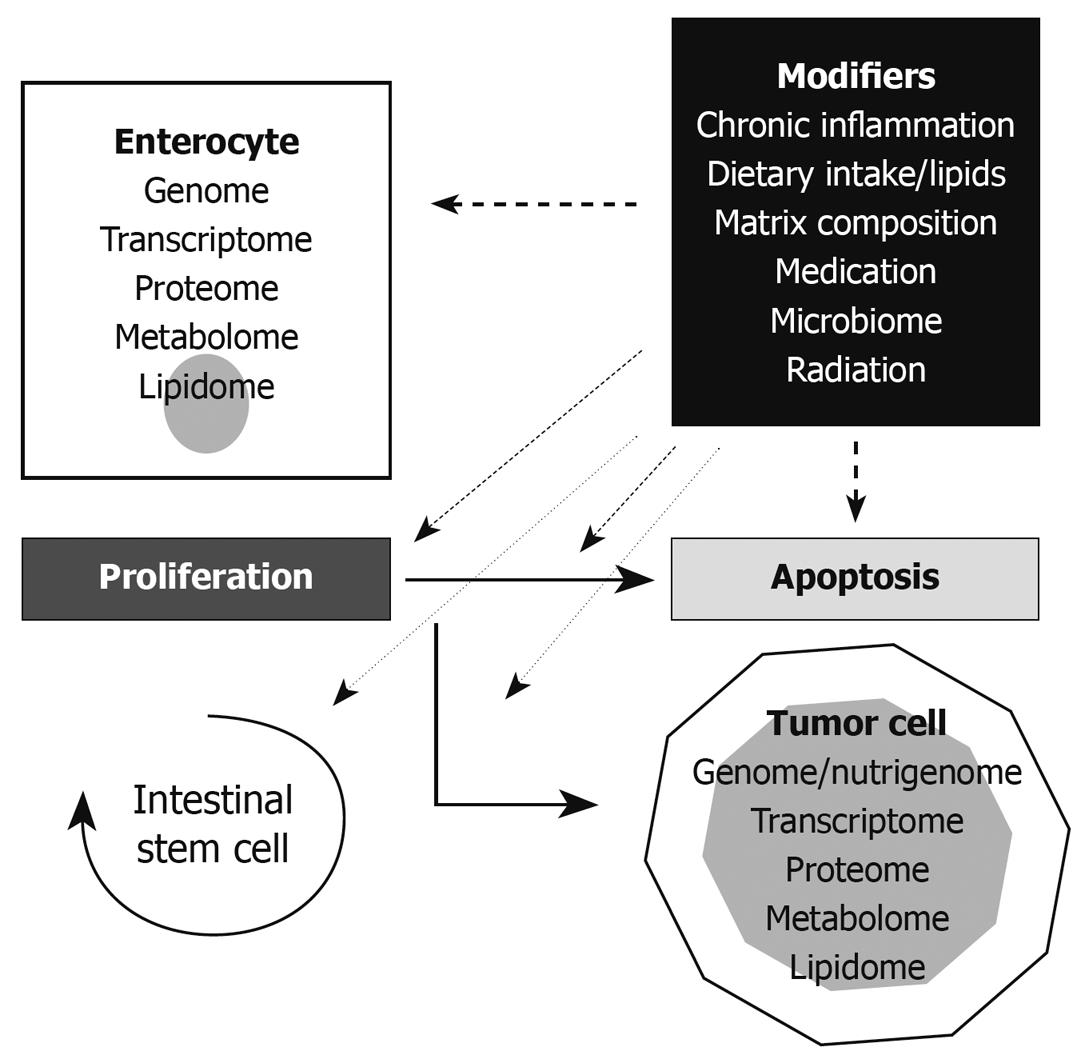
In summary, colorectal carcinogenesis is based on a sequence of molecular events in enterocytes including gene mutations, epigenetic modifications, and aberrant signaling in basic cellular pathways. However, proliferation, cellular activities, and cell death of enterocytes and the execution of so-called modifier genes of CRC-relevant pathways strongly depend on additional variables including matrix composition, the microbiome, chronic inflammatory activity, environmental effects, and dietary components. In particular, nutrition and diet, as stated below, are suggested as fundamental in modifying the initiation and progression of CRCs[6]. Key aspects in the current view of the CRC carcinogenesis modifier concept are summarized in Figure 1.
Figure 1 The modifier concept of colorectal carcinogenesis.
The important key points in enterocytic life span, proliferation and apoptosis, are connected by basal molecular pathways. Such pathways, as well as cell behavior determining molecular entities such as the genome, transcriptome, proteome, metabolome, and lipidome, are exposed to cellular stress and act as modifiers. In addition to enterocytes, the intestinal stem cells are affected by this evolutionary principle. The aberrant modification of pathways is suggested as an important variable in colorectal carcinogenesis. The term nutrigenome of tumor cells reflects the genome modification of malignant cells by dietary components with expression of so-called modifier genes as demonstrated in APC mice.
NUTRITION AND CRC - DIETARY FAT AND INTESTINAL LIPIDS
Among the many aspects of industrialization, such as obesity, exposure to electromagnetic fields, radiation, medications, and a reduced physical activity, diet seems to be a plausible variable in colorectal carcinogenesis because of its direct contact with the intestinal mucosa, i.e. the surface lining epithelial cells[27]. The literature is marked by epidemiological studies aiming to elucidate the constituents of a CRC-preventive diet, the optimal amount of dietary fiber, and to define biologic markers for nutritional epidemiology of CRC[28]. A reason for the controversial outcome of such studies could be that the entire diet, with its balance of animal and vegetable products, and grains that have been processed to varying degrees, is highly variable and not a single entity. However, some dietary components are described as putative candidates in modifying colorectal carcinogenesis. An increased risk for cancer development is found in individuals consuming diets high in red and processed meat[29]. In addition, alcohol in combination with a diet low in folate and methionine appears to increase cancer risk[30-32]. Vitamin D and calcium are suggested to have a protective effect in CRC carcinogenesis, but the molecular mechanism behind this has not been elucidated. Recently, it has been shown that deep-fried/oxidized fats such as hydroxyl- and hydroperoxy fatty acids, are able to influence lipid metabolism by activation of the transcription factor, peroxisome proliferator-activated receptor α (PPARα)[33]. Oxidized fats are established in diets containing large quantities of polyunsaturated fatty acids (PUFAs) by heating. Expression of PPARα-related genes is involved in degradation of fatty acids, affects the synthesis of cholesterol/triacylglyceride, and is associated with cellular behavior. These data indicate that lipid metabolism activity and cellular lipid-dependent signaling could be modified by a diet enriched in deep-fried fats.
In the last few decades, the highly diverse class of lipids has become the focus of intensive research activities, because epidemiological data have indicated a link between the intake of dietary lipids and development of (sporadic) CRCs[34,35]. In this concept, the composition of the different types of fatty acids and the expression/activity of lipid-metabolizing enzymes such as COXs and ACSLs are of high importance[36]. A growing number of reports support the findings that bioactive dietary components containing long-chain PUFAs modulate important determinants that link inflammation to cancer development and tumor progression[37], whereas short-chain fatty acids, e.g. acetate, propionate, and especially butyrate, mainly produced by the microbiome using fermentable dietary polysaccharides, are suggested to be cancer-preventive[38,39]. Propionate and acetate are utilized preferentially by extra-intestinal tissues, whereas butyrate is suggested to play an important role in intestinal mucosa homeostasis as an energy source for enterocytes. It has been shown that cancer-preventive intestinal butyrate levels correlate with increased epithelial cell differentiation, cell cycle arrest, and apoptosis of enterocytes. Additional aspects of butyrate activity, namely inhibition of the enzyme histone deacetylase, and a decrease in the transformation of primary to secondary bile acids as a result of colonic acidification, could be of relevance in colorectal carcinogenesis[40]. Recently, multiple free fatty acid receptors (FFAR) have been identified in the intestine[41]. The receptor activity highly depends on the length of the free fatty acids. Short-chain fatty acid signaling is mediated by FFAR2 and FFAR3, whereas FFAR1 and GPR120 can be activated by medium- and long-chain free fatty acids. It is suggested that defects in FFAR3 are associated with reduced extraction of energy from short-chain free fatty acids. The first experimental evidence was that at least FFAR2 could be associated with cellular transformation and colorectal carcinogenesis. In this experimental setting the putative mitogenic action of FFAR2 was aggravated by short-chain free fatty acids.
The important role of lipids, especially fatty acids, in the modifier concept of CRCs is underlined by the plethora of intestinal long-chain fatty acid (LCFA) activities. LCFAs were suggested as important cell cycle modifiers, particularly in enterocytes. In ACSL5-overexpressing enterocytes, an increase in apoptosis susceptibility was found, probably as a result of an aberrant and locally restricted increase in LCFA derivatives[42]. These investigations underline the importance of ACSL-mediated metabolic channeling of fatty acids in the regulation of cell behavior[43]. In addition, lipidation of proteins is suggested as a basic mechanism in the translation of LCFA modifier activities in several signaling cascades and receptor structures[44]. This mechanism was recently demonstrated for the CD95 death-inducing signaling complex (DISC)[45]. In the earliest events in DISC formation, palmitoylation of CD95 on cysteine 199 enhances receptor aggregation to a high molecular complex. Palmitoylation, as an important mechanism in fatty acid-based protein modification via thio-ester linkage to cysteine (S-palmitoylation), has been recently classified into 5 general classes[46]. The first class comprises transmembrane proteins S-acylated on cysteines adjacent to or in the transmembrane domain. In the second class, proteins are typified by Ras family molecules, where S-palmitoylation is dependent on prior prenylation within the C-terminal “CAAX” box. In the third class, palmitoylated proteins include molecules with S-palmitoylation at one or more cysteine residues near the N- or C-terminus. Dual acylation is found in the fourth class proteins such as members of the Src and G protein families. N-myristoylation is suggested to improve accessibility of the protein to a membrane-bound palmitoyl acyl transferase and subsequent protein palmitoylation. The hallmark of class 5 proteins is covalent binding of palmitate via amide-linkage to an N-terminal cysteine residue.
In summary, dietary fatty acids are suggested as important substrates for protein modification via direct molecular interaction. A complex metabolic conversion of absorbed lipids into cell-synthesized LCFA is not mandatory. However, synthesis of acyl-CoA derivatives from dietary fatty acids seems an important step for the modifying activity and bioavailability of lipids. Palmitoylation has been shown as a basic molecular mechanism in lipid-dependent protein modification. As detailed below, lipidomics is a powerful technical algorithm to elucidate key aspects in the interaction of dietary lipids and cellular lipid metabolism.
LIPIDOMICS - A PROMISING TOOL IN CRC-RELATED LIPID ANALYSIS
In the last 2 decades, laboratory technology to investigate carcinogenesis has primarily focused on genomic and proteomic analyses[47]. Many research-based studies on CRC combined genetic, translation-related and/or biochemical approaches to discover clinically useful DNA- or protein-based biomarkers for CRC screening, diagnosis and response to treatment. Since the crucial roles of lipids in numerous signaling pathways including carcinogenesis, apoptosis and proliferation, has been identified, the necessity to understand and analyze the impact of lipids in colorectal carcinogenesis, diagnosis, prognosis and treatment becomes more and more important.
In recent years, lipidomics, a novelty in the “-omic” research, has emerged as a promising tool to investigate cellular lipid metabolism and pathogenic alterations in lipid pathways[48]. Lipidomics, defined as the large-scale study of the pathways and networks of cellular lipids, is a chromatography- and mass-spectrometry-based research area that includes the study of cellular and functional lipidomics as well as lipids in health and disease. The cellular lipidome comprises over 1000 different, highly diverse and complex lipids of which many are metabolically interconvertible and structurally similar[49]. Because of this complexity, comprehensive lipid analyses are required to elucidate the physiological properties and functionality of different lipid metabolites. The ability to analytically distinguish different lipid species is critical. Despite these experimental challenges and the fact that lipidomics has only emerged as a distinct field within the past few years, the number of compounds that can be analyzed is growing rapidly[50]; including fatty acids, sphingolipids, glycerolipids, glycerophospholipids, sterol lipids and prenol lipids, each containing distinct classes and subclasses of molecules[51-56]. The following section gives a short overview of classical and state-of-the-art analytical techniques that are commonly used by lipidologists to investigate lipids in health and disease.
Thin layer chromatography (TLC) has been used for the qualitative study of lipids since the early 1960s[57,58]. The technique uses a thin layer of a stationary phase such as silica or cellulose on a flat support, usually a glass or aluminum plate. Although the combination of a wide variety of silica-based solid-phase and organic mobile-phase protocols enables the separation of almost all lipid classes, TLC does not offer sufficient structural specificity and sensitivity as other methods of lipid detection. This has led to its use primarily as a screening tool prior to use of more sensitive and elaborated methods.
High pressure liquid chromatography (HPLC), a more sophisticated chromatographic technique, is routinely employed in lipidomic analysis for separation and identification of lipid species. HPLC provides resolution of lipids based on headgroup/class (normal phase) or fatty acid residues (reverse phase). For example, fatty acids and acyl-CoA are almost always separated on a reverse-phase column where the separation occurs on the basis of polarity, effective chain length, and degree of unsaturation[51], whereas separation of phospholipids can be achieved by either normal-phase HPLC[59] or reverse-phase HPLC[60]. However, HPLC alone can not guarantee the accurate quantification of lipids since this often depends on definite resolution and structural elucidation.
Mass spectrometry based techniques are currently the analytical tool of choice in lipidomic methodology. Two powerful approaches that provide sensitivity and structural specificity are electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) mass spectrometry. ESI allows lipids in solution to be continuously infused directly into the ion source of a mass spectrometer, typically a triple quadrupole mass analyzer. By applying a high voltage to the infusion capillary, an electric field is created along which the charged droplets travel. As they travel, the solvent rapidly evaporates and droplets divide into individual charged ions to enter the mass spectrometer. The combination of classical separation techniques such as HPLC with state of the art ESI-mass spectrometry provides the necessary quantitative accuracy, sensitivity and structural specificity to detect and quantify a wide range of lipid species. Thus, liquid chromatography-mass spectrometry (LC-MS) and tandem mass spectrometry (LC-MS/MS)-based methods are currently one of the most popular technologies in lipid research[52,61-63].
In contrast to the LC-based lipid analysis, the recently developed direct infusion-based shotgun lipidomics approach, an analytical platform without direct coupling of any chromatography for lipid separation, allows the absolute quantification of hundreds of lipid species in small quantities with high throughput. This powerful strategy provides a promising future application in clinical studies and medical approaches to investigate pathogenic changes in lipid metabolism and lipid-mediated disorders[64,65].
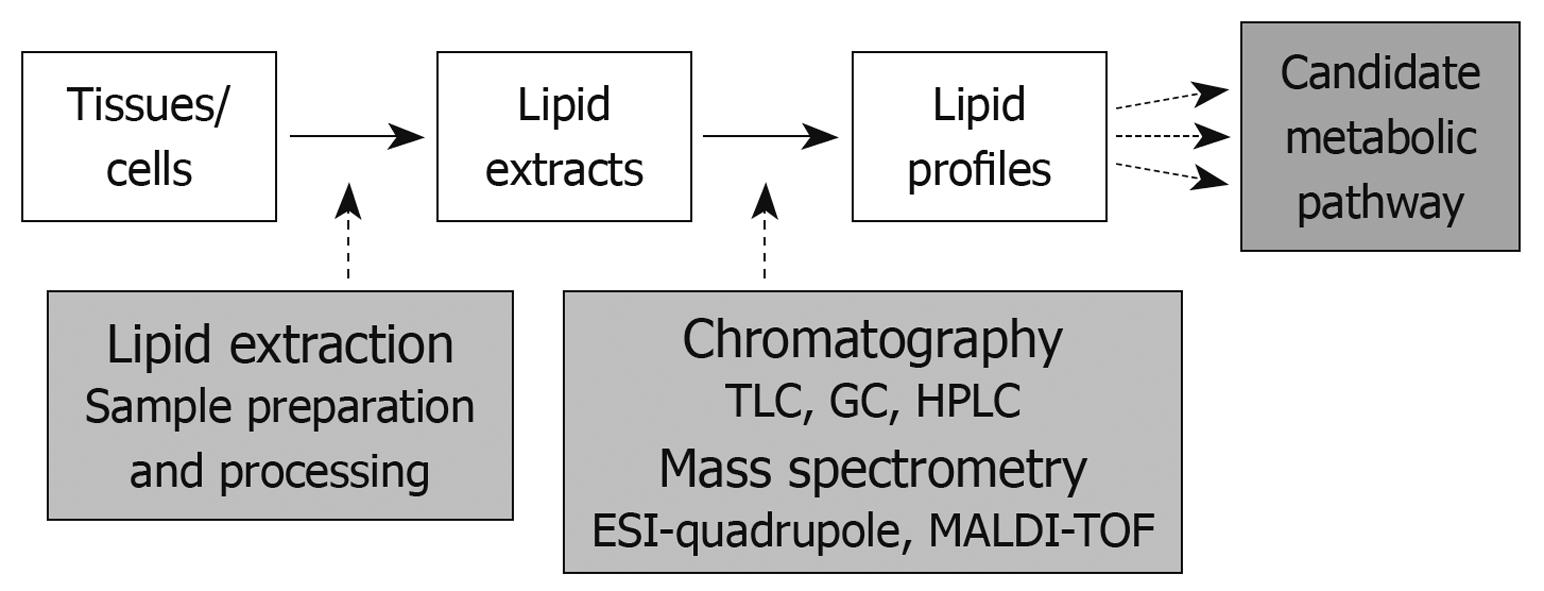
In addition to ESI mass spectrometry, MALDI mass spectrometry, typically used in conjunction with time-of flight mass analyzers, is a laser-based soft ionization method that has been successfully used for large lipid species. Here, the lipid sample is mixed with an energy absorbing matrix and irritated with a laser to ionize the lipid molecules which then enter the mass spectrometer. Although MALDI is most often used for protein analyses and only used to a small extent for lipid analyses, the power of this technology lies in its application for tissue-imaging mass spectrometry. Direct tissue analysis of lipid species provides the opportunity to identify not only quantities but also the distribution and subcellular localization of lipids within the tissue[66,67]. MALDI tissue imaging (MSI), thus, greatly enhances the application spectrum of lipidomic analysis in disease-related alterations in lipid metabolism. A typical lipidomic work flow, including sample preparation and analysis is summarized in Figure 2.
Figure 2 Summary of the lipidomics work flow - lipid extraction and analysis.
TLC: Thin layer chromatography; GC: Gas chromatography; HPLC: High pressure liquid chromatography; ESI: Electrospray ionization; MALDI-TOF: Matrix-assisted laser desorption ionization-time-of flight.
The potential and the feasibility of lipidomic technologies in studies of diabetes, obesity, atherosclerosis and Alzheimer’s disease have been described recently[68]. A lipidomic study of breast cancer pathogenesis demonstrated that the complete profile of adipose tissue lipids in patients with benign and malignant breast tumors, rather than a single lipid has the capability to quantify the dietary part of breast cancer risk and to identify dietary modification in order to reduce breast cancer occurrence[69]. As the relevance of lipids as potential biomarkers becomes apparent, an increasing number of studies have focused on the discovery of reliable lipid biomarkers for disease prognosis, prevention and therapeutic application. A recent study showed that the analysis of fatty acyl chains from ethanolamine-containing plasmalogens provided a suitable biomarker to identify malignancy and metastatic capacity of frequent human cancers, such as breast, lung, and prostate cancer[70]. Although there are currently not many studies focusing on lipidomics-based approaches in colorectal carcinogenesis, the implications of this strategy for the future of CRC treatment and prevention are vast. As the field of lipidomics advances, the role of the lipidome in cellular functions and pathologic states will become clearer, and the identification and establishment of preventive and therapeutic approaches will become more focused. The integration of lipidomic data with genetic, proteomic and metabolomic data will provide a powerful analytical platform for elucidating the mechanism of lipid-based disease, for biomarker screening, and for monitoring pharmacological therapy[71].
CONCLUSION
In current opinion, the basic molecular pathways determining colorectal carcinogenesis are few in number. However, the number of mechanisms that could act in modifying initiation and progression of intestinal neoplasia is constantly growing. Among dietary components, the modifying capacity of lipids, especially of LCFAs, has been demonstrated by experimental and epidemiological studies. Lipidomics, the large-scale study of the pathways and networks of lipids in cells or tissues, is established as a modern and powerful tool to further elucidate the colorectal carcinogenesis modifying activities of lipids.
Peer reviewers: Takayuki Yamamoto, MD, Inflammatory Bowel Disease Center, Yokkaichi Social Insurance Hospital, 10-8 Hazuyamacho, Yokkaichi 510-0016, Japan; Dr. Devinder Kumar Dhawan, Professor, Department of Biophysics & Coordinator, Nuclear Medicine, Panjab University, Chandigarh 160014, India
S- Editor Tian L L- Editor Cant MR E- Editor Lin YP