Published online Feb 21, 2009. doi: 10.3748/wjg.15.865
Revised: January 10, 2009
Accepted: January 17, 2009
Published online: February 21, 2009
AIM: To explore the method for early diagnosis of gastric cancer by screening the expression spectrum of saliva protein in gastric cancer patients using mass spectrometry for proteomics.
METHODS: Proportional peptide mass fingerprints were obtained by analysis based on proteomics matrix-assisted laser desorption ionization time-of-flight/mass spectrometry. A diagnosis model was established using weak cation exchange magnetic beads to test saliva specimens from gastric cancer patients and healthy subjects.
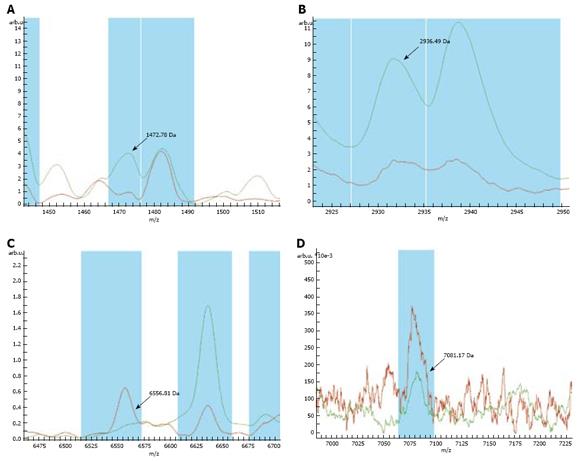
RESULTS: Significant differences were observed in the mass to charge ratio (m/z) peaks of four proteins (1472.78 Da, 2936.49 Da, 6556.81 Da and 7081.17 Da) between gastric cancer patients and healthy subjects.
CONCLUSION: The finger print mass spectrum of saliva protein in patients with gastric cancer can be established using gastric cancer proteomics. A diagnostic model for distinguishing protein expression mass spectra of gastric cancer from non-gastric-cancer saliva can be established according to the different expression of proteins 1472.78 Da, 2936.49 Da, 6556.81 Da and 7081.17 Da. The method for early diagnosis of gastric cancer is of certain value for screening special biological markers.
- Citation: Wu ZZ, Wang JG, Zhang XL. Diagnostic model of saliva protein finger print analysis of patients with gastric cancer. World J Gastroenterol 2009; 15(7): 865-870
- URL: https://www.wjgnet.com/1007-9327/full/v15/i7/865.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.865
Gastric cancer is one of the most common cancers. Even though its mortality rate has decreased in recent years, its morbidity still ranks first among all kinds of malignant tumor. Moreover, gastric cancer obviously occurs in juvenescence, and is diagnosed at its advanced stage. About 150-200 thousand patients die of gastric cancer every year in our country, which accounts for almost a quarter of all deaths from malignant tumors. About 200000 new gastric cancer patients are diagnosed each year. At present, available tumor markers have a relatively high false-positive rate for the diagnosis of gastric cancer, and thus cannot predict early gastric cancer[12]. Therefore, it has become important to find a special way to predict early stage gastric cancer.
Saliva can be used in diagnosing gastric cancer, and it has been verified for many years that detection of salivary components is a valuable tool to diagnose a variety of diseases[3]. Salivaas, a diagnostic specimen has received attention. Along with the wide application of proteomics and its related techniques, proteomics has been used more widely in the study of saliva[45]. In this study, we employed matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF-MS) technique to analyze the protein mass peak of saliva from gastric cancer patients and healthy subjects, and to screen for the special biological markers for predicting early gastric cancer.
Saliva samples were collected from gastric cancer patients in the Medical Department of Nanfang Hospital, Southern Medical University. Control samples were collected from healthy volunteers. Saliva was collected and put into a 15-mL centrifuge tube, and centrifuged at 10000 r/min for 5 min at 4°C. Fifty microliters of each saliva sample was put into in a 1-mL EP tube and stored at -80°C. When experiments were conducted, the samples were taken out from the refrigerator at -80°C. All samples were defrosted at room temperature. The healthy subjects at the age of 23-68 years served as a normal group (n = 18, 12 males, 6 females) and the patients at the age of 25-78 years served as a gastric cancer group (n = 23, 15 males, 8 females).
A weak cation exchange (WCX) magnetic bead kit, alpha-cyano-4-hvdroxycinnamic acid (HCCA), and AutoFlex III MALDI-TOF mass spectrometer were purchased from Bruker Company. Mass concentration (0.3 g/L) and ethanol (chromatographic grade)/acetone (chromatographic grade) = 2/1 were freshly prepared.
The WCX magnetic bead kit was taken out from a refrigerator at 4°C, washed and eluted. Finally, the separated magnetic beads and eluted polypeptide samples were transferred into a 0.5-mL clean sample tube for further MS analysis.
One microliter polypeptide sample separated with the magnetic beads was applied. After the polypeptide sample was dried at room temperature, into which 1 &mgr;L HCCA substrate solution (3 g/L, dissolved in 50% acetonitrile and 2% trifluoroacetic acid) was added. Then, the prepared sample was applied and analyzed on MALDI-TOF-MS. A linear mode was used to collect peptides with a molecular weight of 1000-10000. Twenty percent of laser energy was used with 400 shots. Peptide mass finger prints were obtained by accumulating 50 single scanning of MS signals.
FlexAnalysis 3.0 and ClinProTools 2.1 (from Bruker Company) were used to analyze the grouping index and its dependability of data. P < 0.05 was considered statistically significant.
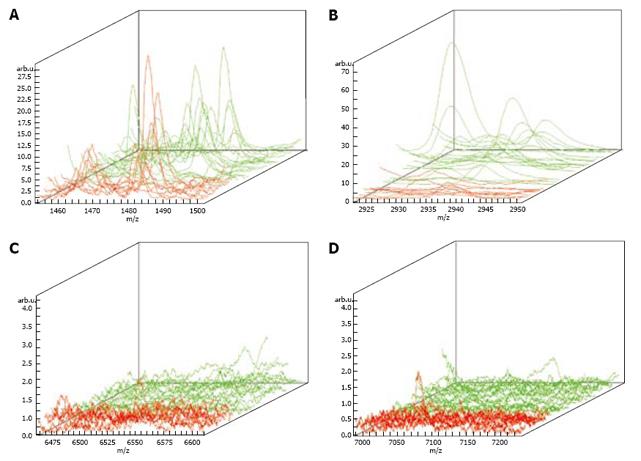
The mass spectrum of samples from 41 subjects was analyzed and compared. Seventy-four protein mass peaks were found. Mass peaks were proved to be significantly different in four proteins (1472.78 Da, 2936.49 Da, 6556.81 Da and 7081.17 Da) (Figures 1,2,3,4). The protein mass peak at 1472.78 Da was higher in the gastric cancer group than in the normal group. Based on the identification of these four protein mass peaks, 22 patients were accurately diagnosed with gastric cancer. All the 18 healthy subjects were confirmed to have no gastric cancer. The present method for the diagnosis of gastric cancer had a sensitivity of 95.65% (22/23), and a specificity of 100% (18/18). The results are shown in Table 1.
| Groups | Classification of proteome mass spectrum model | |||
| n | Gastric cancer | Non-gastric cancer | Ratio (%) | |
| Gastric cancer group | 23 | 22 | 1 | 95.65 |
| Normal group | 18 | 0 | 18 | 100 |
Saliva is an important and necessary body fluid. Blood constituents such as hormones, amino acids, electrolyte, immunoglobulin and creatinine, can enter saliva through the blood barriers of the capillary walls. As one of the fluids in the human body, variation in saliva constituents is influenced by various pathophysiological changes in the body. Saliva constituents are related to serum and the essential proteins in saliva are positively correlated with serum. For example, there is an extremely good dependability between blood plasma free F and salvia F, which is not influenced by the flow rate and stimulation of saliva. Many proteins in saliva, such as anti-HIV antibody, secretory type leukoprotease inhibitor (SLPI) and IgA, have important physiological functions. SLPI, a kind of single strand polypeptide, can be separated from human parotid gland saliva and only has anti-HIV effects in the oral cavity. Saliva can be atraumatically and conveniently taken, and easily observed at any time. The test results are stable and the sensitivity is rather high, and the tests can be repeated. Since biochemical microanalysis techniques have been significantly improved, saliva can be used as a diagnostic index instead of blood[36–9].
Studies on saliva proteomics have received extensive attention worldwide[45]. Till now, 309 kinds of protein have been identified in full salivary fluid of healthy subjects, using proteomics techniques, among which, the most important acidic proteins are catheptic enzyme L and hyaluronan-conjugated protein; the most important basic proteins are saliva rich pyrrolidinecarboxylic acid, glycoprotein PRB2, and an unknown protein with a level of 12.8; while the smallest proteins are T cell receptor 8 catenin fragment and hylaxin HNP-3, and the protein with a maximal relative molecular mass is mucoprotein 5B. Of the 309 saliva proteins identified according to their functions, 28.7% are uncertain functional proteins, 21% are associated with immunity, 1.6% are associated with protein replication and reparation, 4.8% are associated with cell mobility and secretion, 2.3% are associated with transcription and ribosomes, 4.2% are associated cell multiplication and the cell cycle, 9.7% are associated with signal transduction, 5.2% are associated with metabolism, and 7.1% are associated with the cytoskeleton and endomembrane, respectively. Saliva is an important body fluid with complicated constituents and multiple biological functions. Its distinctive and abundant protein constituents undoubtedly can be used as biological markers of cancer and other diseases. The oral saliva protein flux analysis and precise determination have been restricted by old techniques, since the biological functions of most saliva proteins are unknown, and the value of saliva diagnosis and prognosis remains unclear. With the application of high-flux and high-precision proteomics, it has become possible that saliva proteins can be used as biological markers in prevention and bioprotein-targeted therapy, and in predicting the prognosis of diseases[3–10]. At present, no study on saliva proteomics is available in China, and synchronic detection and comparative study of saliva and serum proteins have not been reported worldwide.
Early stage markers of gastric cancer are a hot spot of investigation around the world. How to find effective and better methods and techniques that can be applied to the treatment of gastric cancer has become the focus of research. There are more than 1000 proteins in saliva, and saliva possesses atraumatic and convenient features, and can be used as a simple and abundant resource. In addition, recent studies have demonstrated that 20% of saliva proteins are similar in the blood, and certain saliva proteins are matched with blood proteins that influence senile dementia, breast cancer and diabetes[10–12]. MS technique is one of the most important techniques in proteomics studies. The most widely used MS technique in proteomics studies is MALDI-TOF MS. Application of MALDI-TOF MS, in study of biological markers of disease, is undoubtedly at the leading edge of molecular diagnosis. Proteomics has become a powerful tool for the discovery of new biological markers. Combining MS techniques and proteomics also provides good prospects for research of biological markers[1314]. Investigation of proteomics provides not only a material basis for vital movement rules, but also a theoretical basis for overcoming diseases. Certain diseases are related to special protein molecules. Proteins may be the molecular targets of new drugs or molecular markers for the early diagnosis of certain diseases. MS techniques, are important in studies of proteomics, and can be used in the study of protein/peptide spectra, biological marker spectra, or single biological markers for complicated diseases, such as cardiovascular and cerebrovascular disease, tumors, stroke and neuro-degenerative disease. Furthermore, they are also expected to open new avenues for research into pathogenesis, diagnosis and treatment of complicated diseases[15–17].
Gastric cancer usually has a long latent period. Early gastric cancer does not present with overt symptoms. Gastric cancer is at an advanced stage when it is diagnosed and has characteristics such as easy metastasis and poor prognosis. Therefore, early diagnosis is extremely important for the control and treatment of gastric cancer. Even though advanced diagnostic methods, such as X-ray barium meal examination, dual-phase helical computed tomography (CT), virtual CT, and gastroscopy, can improve gastric cancer diagnosis, their sensitivity and specificity are low for early discovery of gastric cancer. Under such circumstances, gastric cancer can be found only after cancer cells have metastasized to their surrounding tissues, or whole-body aggravation occurs. Furthermore, cancer biological markers have a rather low positive rate for the early diagnosis of gastric cancer. At present, the sensitivity of cancer biological markers is only about 18%-40%. Combined examination of multiple cancer biological markers can achieve a sensitivity of 60%-80%. However, since the false positive rate is rather high, it is difficult to employ it as a biological index for the early diagnosis of gastric cancer[18–21]. We used the MALDI technique and WCX magnetic beads to examine gastric cancer samples, which can accurately identify gastric cancer. A difference in mass spectra was found between the normal subjects and gastric cancer patients, indicating that the present method can greatly improve the diagnosis of gastric cancer and might be used in its treatment. Twenty-three saliva samples from gastric cancer patients and 18 saliva samples from healthy volunteers were examined using MALDI-TOF-MS (AutoFlex III, from German Bruker Company) and WCX magnetic beads. The mass peaks of four proteins (1472.78 Da, 2936.49 Da, 6556.81 Da and 7081.17 Da) were significantly different in gastric cancer and normal groups. The mass peak of protein 1472.78 Da was significantly higher in the gastric cancer group than in the normal group, indicating that the protein of 1472.78 Da may play an important role in the occurrence and development of gastric cancer. Among the four peaks, only the mass peak of protein 6556.81 Da in samples from gastric cancer patients was lower than that in samples from normal subjects, suggesting that the protein expression mass spectrum diagnosis model for classification of gastric cancer and non-gastric cancer can be developed by comparative analysis of mass peaks of proteins 1472.78 Da, 2936.49 Da, 6556.81 Da and 7081.17 Da, thus providing a new tool for predicting early gastric cancer. The positive identification rate for the mass peaks of these four proteins was significantly higher than that for the common serum cancer biological markers. Since the number of gastric cancer patients and normal controls was small, a larger sample size is needed to verify the mass peaks of the four proteins identified in this study.
In conclusion, the saliva proteome technique is an attractive prospect in the early clinical diagnosis of gastric cancer. Gastric cancer can be identified at an early stage by screening the high-risk group using MALDI technique and further studies on these proteins are needed to reveal their clinical value. Earlier detection and treatment of gastric cancer are important in decreasing the death rate. However, the saliva proteome technique needs to be further studied, because its cost is rather high and some other problems remain to be solved.
Gastric cancer is one of the most common cancers and threatens human health. At present, available tumor markers have a low sensitivity and a relatively high false-positive rate for the diagnosis of gastric cancer, thus cannot predict early gastric cancer. Therefore, it is necessary to find new methods to predict early stage gastric cancer.
Saliva is an important and necessary body fluid. Studies on saliva proteomics have received extensive attention worldwide. At present, markers of early stage gastric cancer are a hot topic of investigation around the world. How to find effective and better methods that can be applied in clinical practice has become the target of many studies. Recent studies reveal that there are more than 1000 proteins in saliva, and saliva possesses atraumatic and convenient features, and can be used as a simple and abundant resource.
A diagnosis model was developed using weak cation exchange (WCX) magnetic beads to test saliva specimens from gastric cancer patients and healthy subjects. Significant differences in the mass to charge ratio peaks of four proteins (1472.78 Da, 2936.49 Da, 6556.81 Da and 7081.17 Da) were observed between gastric cancer patients and healthy subjects.
The saliva proteomic model for distinguishing gastric and non-gastric cancer can be applied to the early diagnosis of gastric cancer.
Saliva: An important and necessary body fluid. Blood constituents such as hormones, amino acids, electrolytes, immunoglobulin and creatinine etc, can enter saliva through the blood barriers of the capillary walls.
The authors studied the differences in saliva protein spectra between normal subjects and gastric cancer patients. The study is valuable in aiding early diagnosis of gastric cancer. The method for screening patients with the risk of gastric cancer is simple, fast, and easy to use. Therefore, this study is innovative and practical, and the method developed by the authors can be used in the early diagnosis of gastric cancer.
| 1. | Vitzthum F, Behrens F, Anderson NL, Shaw JH. Proteomics: from basic research to diagnostic application. A review of requirements & needs. J Proteome Res. 2005;4:1086-1097. |
| 2. | Liao AJ, Su Q, Wang X, Zeng B, Shi W. Isolation and bioinformatics analysis of differentially methylated genomic fragments in human gastric cancer. World J Gastroenterol. 2008;14:1333-1338. |
| 4. | Hu S, Loo JA, Wong DT. Human saliva proteome analysis and disease biomarker discovery. Expert Rev Proteomics. 2007;4:531-538. |
| 5. | Papale M, Pedicillo MC, Di Paolo S, Thatcher BJ, Lo Muzio L, Bufo P, Rocchetti MT, Centra M, Ranieri E, Gesualdo L. Saliva analysis by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF/MS): from sample collection to data analysis. Clin Chem Lab Med. 2008;46:89-99. |
| 6. | Løvås K, Husebye ES. [Salivary cortisol in adrenal diseases]. Tidsskr Nor Laegeforen. 2007;127:730-732. |
| 7. | Flink H, Tegelberg A, Lagerlöf F. Influence of the time of measurement of unstimulated human whole saliva on the diagnosis of hyposalivation. Arch Oral Biol. 2005;50:553-559. |
| 8. | Christodoulides N, Mohanty S, Miller CS, Langub MC, Floriano PN, Dharshan P, Ali MF, Bernard B, Romanovicz D, Anslyn E. Application of microchip assay system for the measurement of C-reactive protein in human saliva. Lab Chip. 2005;5:261-269. |
| 9. | Castro M, Elias PC, Martinelli CE Jr, Antonini SR, Santiago L, Moreira AC. Salivary cortisol as a tool for physiological studies and diagnostic strategies. Braz J Med Biol Res. 2000;33:1171-1175. |
| 10. | Helmerhorst EJ, Oppenheim FG. Saliva: a dynamic proteome. J Dent Res. 2007;86:680-693. |
| 11. | Amado FM, Vitorino RM, Domingues PM, Lobo MJ, Duarte JA. Analysis of the human saliva proteome. Expert Rev Proteomics. 2005;2:521-539. |
| 12. | Denny P, Hagen FK, Hardt M, Liao L, Yan W, Arellanno M, Bassilian S, Bedi GS, Boontheung P, Cociorva D. The proteomes of human parotid and submandibular/sublingual gland salivas collected as the ductal secretions. J Proteome Res. 2008;7:1994-2006. |
| 13. | Engwegen JY, Gast MC, Schellens JH, Beijnen JH. Clinical proteomics: searching for better tumour markers with SELDI-TOF mass spectrometry. Trends Pharmacol Sci. 2006;27:251-259. |
| 14. | Johnson CJ, Zhukovsky N, Cass AE, Nagy JM. Proteomics, nanotechnology and molecular diagnostics. Proteomics. 2008;8:715-730. |
| 15. | Ma QW, Cheng XR. MALDI-TOF-MS Guides A New Era for The Molecular Diagnosis. Shengwu Jishu Shijie. 2006;5:66-68. |
| 16. | Xiao Z, Prieto D, Conrads TP, Veenstra TD, Issaq HJ. Proteomic patterns: their potential for disease diagnosis. Mol Cell Endocrinol. 2005;230:95-106. |
| 17. | Wouters BG. Proteomics: methodologies and applications in oncology. Semin Radiat Oncol. 2008;18:115-125. |
| 18. | Liang Y, Liu CB, Li JC. Application of Serum Proteomic Patterns for Detection of Gastric Adenocarcinoma. Zhongguo Zhongliu. 2006;15:127-130. |
| 19. | Wu B, Wilmouth RC. Proteomics analysis of immunoprecipitated proteins associated with the oncogenic kinase cot. Mol Cells. 2008;25:43-49. |
| 20. | Chu YQ, Ye ZY, Tao HQ, Wang YY, Zhao ZS. Relationship between cell adhesion molecules expression and the biological behavior of gastric carcinoma. World J Gastroenterol. 2008;14:1990-1996. |
| 21. | Abbaszadegan MR, Moaven O, Sima HR, Ghafarzadegan K, A'rabi A, Forghani MN, Raziee HR, Mashhadinejad A, Jafarzadeh M, Esmaili-Shandiz E. p16 promoter hypermethylation: a useful serum marker for early detection of gastric cancer. World J Gastroenterol. 2008;14:2055-2060. |