Published online Feb 7, 2009. doi: 10.3748/wjg.15.538
Revised: January 4, 2009
Accepted: January 11, 2009
Published online: February 7, 2009
By modulating hepcidin production, an organism controls intestinal iron absorption, iron uptake and mobilization from stores to meet body iron need. In recent years there has been important advancement in our knowledge of hepcidin regulation that also has implications for understanding the physiopathology of some human disorders. Since the discovery of hepcidin and the demonstration of its pivotal role in iron homeostasis, there has been a substantial interest in developing a reliable assay of the hormone in biological fluids. Measurement of hepcidin in biological fluids can improve our understanding of iron diseases and be a useful tool for diagnosis and clinical management of these disorders. We reviewed the literature and our own research on hepcidin to give an updated status of the situation in this rapidly evolving field.
- Citation: Piperno A, Mariani R, Trombini P, Girelli D. Hepcidin modulation in human diseases: From research to clinic. World J Gastroenterol 2009; 15(5): 538-551
- URL: https://www.wjgnet.com/1007-9327/full/v15/i5/538.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.538
Hepcidin is a circulating peptide hormone that regulates the entry of iron into plasma. It is primarily, but not exclusively, secreted by hepatocytes and is highly conserved among different species. The mature bioactive form of hepcidin is a peptide of 25 amino acids that derives from a precursor (pre-prohepcidin) of 84 amino acids that undergoes two enzymatic cleavages. Other isoforms of 20 and 22 amino acids are detectable in human serum and urine, although the biological significance of these isoforms is uncertain[1–3]. There is evidence that hepcidin is also expressed in the heart, kidney, adipose tissue, pancreas and haematopoietic cells, although the biological relevance of extra-hepatic hepcidin is not well defined yet (see below).
Targeted deletion of the HAMP gene in mice or mutations in the human HAMP gene result in the most severe forms of iron-overload disease. Conversely, increased expression of hepcidin leads to decreased iron absorption and iron deficient anaemia. Hepcidin, therefore, is a negative regulator of iron transport into plasma. It acts by binding to ferroportin, the only known cellular iron exporter, causing ferroportin to be phosphorylated, internalized, ubiquitylated, sorted through the multivesicular body pathway and degraded in lysosomes[45]. The interaction between hepcidin and ferroportin can explain the systemic regulation of iron metabolism. In fact, hepcidin is mainly targeted to duodenal enterocytes, where it regulates dietary iron absorption, and to macrophages, where it inhibits the release of iron derived from senescent erythrocytes. By modulating hepcidin production, an organism controls intestinal iron absorption, iron uptake and mobilization from stores to meet body iron need[67].
Hepcidin is modulated by different stimuli, which act as positive or negative regulators. There are four main active regulation pathways (erythroid, iron store, inflammatory and hypoxia-mediated regulation) that control hepcidin production through different signalling pathways (Table 1). These pathways must be closely coordinated to match iron supply to erythropoietic demand and, in turn, to maintain adequate plasma iron concentrations.
| Positive regulators | Signalling pathway | Human disease | Negative regulators | Signalling pathway | Human disease |
| Inflammation [IL-6, IL-1α] | STAT-3 | Anemia of chronic disease | Hypoxia [HIF1, sHJV, ROS] | HIF-1, BMPs/SMAD Oxygenases inhibition | -- |
| Iron stores [mHJV] | BMPs/ SMAD4 | Hemochromatosis | Iron stores [Matriptase2, s-HJV, GDF15] | BMPs/SMAD inhibition | IRIDA, iron deficiency |
| Iron stores [HFE, TfR2] | BMPs/ SMAD4 | Hemochromatosis | Erythropoiesis [GDF15, others?] | BMPs/SMAD inhibition others? | Thalassemia, CDA1 |
| Liver metabolic activities | C/EBPα | Erythropoietin? | C/EBPα block | ||
| Oxygenases | ? | Oxidative stress [ROS] | C/EBPα, STAT3 block; Oxygenases inhibition HDAC activation | Hepatitis C viral and alcoholic liver disease |
Under normal conditions, iron store and inflammatory regulation activate hepcidin transcription in the hepatocytes through the bone morphogenetic proteins (BMPs)/SMAD4 and signal transducer and activator of transcription-3 (STAT-3) pathways, respectively[6]. Binding of specific BMPs by hepatocyte BMP receptors results in phosphorylation of receptor SMADs (R-SMADs) within the cytosol. These phosphorylated R-SMADs complex with SMAD4, and the complex of phosphorylated R-SMAD and SMAD4 then activates transcription of HAMP[8]. Accordingly, in vivo and in vitro experimental studies showed that specific BMPs can activate HAMP transcription[9] and a liver-specific Smad4-knockout (KO) abrogates hepcidin gene transcription in mice leading to massive iron overload[10]. The hemochromatosis protein HFE, transferrin receptor 2 (TfR2) and the membrane isoform of hemojuvelin (mHJV) are all positive modulators of HAMP transcription and, when defective, lead to hemochromatosis (HH) in humans and HH-like phenotype in knock-out animal models[7].
HFE: Recent studies in vitro and in animal models clarified the crucial role of HFE as a hepatocyte iron sensor and upstream regulator of hepcidin[1112]. Several mechanisms have been proposed by which HFE regulates iron metabolism. HFE competes with transferrin for binding to transferrin receptor (TfR)-1, lowering iron uptake into cells[1314]. Alternately, there is more recent evidence supporting a role for HFE as an important component of a larger iron-sensing complex that involves interactions with diferric transferrin, TfR1 and TfR2 at the plasma membrane of hepatocytes[15–17]. Defective HFE protein prevents formation of a functional iron sensor and signal transduction effector complex leading to dysregulated hepcidin expression as observed in human hereditary hemochromatosis[18–20] and mouse models of this disease[21]. Recent findings also support other HFE-dependent mechanisms of regulation of iron homeostasis through post-transcriptional regulation of Zip14, a metal transporter that mediates non-transferrin-bound iron into cells[22].
TfR2: Homozygous mutations of TfR2 have been linked to type 3 HH[23]. Animal models of mutated or knock-down TfR2 confirmed human findings indicating that TfR2 plays an important role in iron metabolism[24–26]. It has been shown that diferric transferrin levels may increase the stability of TfR2 protein and it has been postulated that TfR2 is a sensor of serum iron levels and works to modulate iron absorption through the induction of the iron regulatory hormone hepcidin[2728]. Accordingly, mutated or absent TfR2 is associated with reduced hepcidin expression either in animal models or in humans[1824]. TfR2-induced regulation of hepcidin synthesis likely occurs through interaction with HFE to form an iron-sensing complex that modulates hepcidin transcription through the activation of a still unclear signalling pathway upstream or independent of SMAD4[729]. Interestingly, hepatic HJV mRNA level was lower in complete TfR2 knock-out[25] than in controls suggesting an interaction between TfR2 and HJV in the regulation of hepcidin expression. This finding has been recently confirmed to also occur in humans (Pelucchi et al, Haematologica in press).
HJV: As BMP co-receptor, HJV is strongly involved in the regulation of hepcidin transcription through the BMP/SMAD4 pathway[93031]. Accordingly, Hjv-mutant mice exhibit iron overload as well as a dramatic decrease in hepcidin expression[32]. HJV is synthesized as a membrane-GPI-linked protein by hepatocytes but is also abundant in muscle. HJV exists in 2 forms: a membrane-bound form (m-HJV) and a soluble one (s-HJV), which in vitro reciprocally regulate hepcidin expression in response to opposite iron changes[33].
IL-6 and cytokines: Under inflammatory conditions, IL-6 and other cytokines induce HAMP transcription by activating STAT3 signalling to the hepcidin gene promoter[23435]. The inflammatory regulatory pathway is thought to have a dominant effect on the other regulatory pathways as shown by the fact that inactivation of HFE does not affect the increase of hepcidin expression and the hypoferremia induced by inflammation. However, STAT3 activation requires the presence of SMAD4 because absence of SMAD4 prevents STAT3-mediated hepcidin gene expression[10].
Hypoxia, anemia, increased erythropoiesis and reduced iron stores all negatively regulate hepcidin expression. Hypoxia and anemia regulate the production of erythrocytes through synthesis of erythropoietin (EPO) and are the two main signals that increase iron absorption independently of iron stores to meet the need of iron for erythrocyte production[3637].
Anemia could mediate hepcidin suppression through multiple mechanisms including increased EPO or erythropoietic activity, increased iron demand or liver hypoxia[38]. Pinto et al[39] found that EPO at high concentration is able to inhibit hepcidin expression in vitro, via signalling through EPO receptor and C/EBPα regulation. This finding contrasts with that previously reported by Pak et al[40] who showed that hepcidin regulation depended on erythropoietic activity and was not directly mediated by anemia, tissue hypoxia, or EPO in murine models and that secondary changes in plasma and tissue iron would also be expected to contribute to hepcidin regulation. The nature of the erythropoietic regulator of hepcidin is still uncharacterised, but may include one or more proteins released during active erythropoiesis. Recent observations in thalassemia patients has suggested that one of these regulators could be the cytokine growth differentiation factor-15 (GDF15)[41].
Hepcidin is suppressed in human cultured hepatoma cells exposed to hypoxia[42] but the physiological relevance and the mechanisms of hepcidin regulation by hypoxia are still uncertain and conflicting. Hypoxia-inducible factor (HIF)-1 and reactive oxygen species (ROS) have been both implicated in hepcidin regulation either directly or through modulation of 2-oxoglutarate-dependent oxygenases, respectively[42–44].
Studies in vitro and in vivo in animals[4546] have recently shown an alcohol-dependent decrease of hepcidin expression possibly through ROS-induced down-regulation of C/EBPα. Two other studies analysed the effect of 7 d alcohol exposure in HFE KO mice and results were discrepant, one showing an additive effect of alcohol on hepcidin down-regulation[47] and the other not[48]. There remains however a major discrepancy between these experimental findings and the absence of increased iron deposits in alcohol-fed animal models[4549], and clinical data in humans indicate a generally mild effect of chronic alcohol consumption on iron stores even in patients with hemochromatosis[50–53].
GDF15: GDF15 is a divergent member of the transforming growth factor-β superfamily that is secreted by erythroid precursors and other tissues. It has been identified as an oxygen-regulated transcript responding to hypoxia and as a molecule involved in hepcidin regulation[4154]. This is intriguing considering the strong interaction between iron and oxygen and indicates that some homeostatic systems for iron and oxygen are responsive to both stimuli. Tanno et al[41] showed that serum from thalassemia patients suppressed hepcidin mRNA expression in primary human hepatocytes and depletion of GDF15 reversed the hepcidin suppression. They suggested that GDF15 overexpression arising from an expanded erythroid compartment contributed iron overload in thalassemia syndromes by inhibiting hepcidin expression, possibly by antagonizing the BMP pathway. Very high levels of serum GDF15 were also observed in patients with congenital dyserythropoietic anemia type 1 (CDA I) suggesting that GDF15 contributes to the inappropriate suppression of hepcidin with subsequent secondary hemochromatosis in these patients[55]. Very recently Lakhal et al[54] partially elucidated some of the mechanisms regulating GDF15 and demonstrated robust and sensitive up-regulation of GDF15 mRNA and secreted protein in response to iron depletion in a range of human cell lines and in vivo in humans. They also demonstrated that this up-regulation was independent of HIF providing support for the involvement of a novel iron and oxygen-sensing pathway. Whether regulation of GDF15 by intracellular iron provides a mechanism by which intracellular iron might directly influence hepcidin production is unclear and requires further analysis.
HIF, ROS and Oxygenases: Hypoxia is primarily sensed in vertebrates by the HIF family of transcription factors. Under normal conditions, hydroxylation of HIF results in its recognition by the von-Hippel–Lindau (VHL) ubiquitin ligase, which targets it for degradation. In the absence of oxygen, HIF proteins are stabilized and function as transcription factors[37]. Mice with a liver-specific deletion of HIF1α, even if they are maintained on a low iron diet, show increased hepcidin expression because they have an impaired hypoxic response and cannot normally down-regulate hepcidin gene transcription[44]. In contrast, mice with a liver-specific deletion of the Vhl gene show extremely low levels of hepcidin expression[44]. The same authors showed that HIF1α can bind to the mouse and human HAMP promoter suggesting that HIF-family members can directly and negatively regulate hepcidin expression[644]. Thus, the VHL/HIF pathway could be an essential link between iron and oxygen homeostasis and hepcidin regulation in vivo that through coordinated down-regulation of hepcidin and up-regulation of erythropoietin and ferroportin, mobilizes iron to support erythrocyte production. Conflicting data were, however, obtained in hepatoma cell lines in which over-expression, knockout and chromatin immunoprecipitation (Chip) assays failed to demonstrate the involvement of HIF-1 in the regulation of hepcidin promoter[4243]. Choi et al[43] found that hypoxia-induced hepcidin suppression was related to ROS levels which prevented the binding of transcription factors, CCAAT/enhancer-binding protein alpha (C/EBPα) (a liver enriched transcription factor that accounts for developmental changes in liver metabolism after birth) and STAT-3, to the HAMP promoter. More recently, Braliou et al[42] showed that 2-oxoglutarate-dependent oxygenases (which include prolyl and asparagine hydroxylases, PHDs and FIH) were important to maintain high hepcidin mRNA expression in a HIF-1-independent manner These enzymes depend on molecular oxygen, 2-oxoglutarate and Fe2+ and act on their main substrate HIF-1α causing its inactivation and degradation by protein hydroxylation. Hypoxia and generation of ROS inhibit protein hydroxylation and cause stabilization and activation of HIF-α[37]. Braliou et al[42] demonstrated that hypoxia and chemical agents inhibiting the 2-oxoglutarate-dependent oxygenases repress hepcidin expression in hepatoma cell lines opening the way for further experimental studies to identify downstream factors mediating hepcidin regulation.
Soluble HJV: It has been shown that s-HJV can bind BMP and function as a competitive antagonist of m-HJV, leading to decreased hepcidin expression[33] and that chronic s-HJV injection in mice causes iron overload[56]. Silvestri et al[57] demonstrated that s-HJV is the product of a furin cleavage at the C-terminus of the protein that occurs mainly in the endoplasmic reticulum and that it is up-regulated in conditions of iron deficiency and hypoxia. They suggested that hypoxia and iron deficiency activate furin to release s-HJV and to rapidly reduce the amount of m-HJV, inhibiting hepcidin production[3357]. These events may occur in the same cells (hepatocytes) that produce hepcidin in order to suppress hepcidin up-regulation by an autocrine mechanism.
Transmembrane serine protease 6: Recently, Du et al[58] described mask, a recessive, chemically induced mutant mouse phenotype, characterized by progressive loss of body, but not facial, hair and microcytic anemia. The mask phenotype results from reduced absorption of dietary iron caused by high levels of hepcidin. High Hamp mRNA levels were in fact observed in the liver of mask homozygotes despite anemia, a finding consistent with insensitivity to low iron stores and failure to suppress hepcidin synthesis. A splicing defect was found in the transmembrane serine protease 6 gene Tmprss6 in the mask mutant. This gene is expressed in a limited number of tissues, but the major site of expression is the liver in both mice and humans. Du et al[58] demonstrated that TMPRSS6 cotransfection strongly inhibited Hamp reporter gene activation by each stimulus (IL-1α, IL-6, BMP2-4-9) in HepG2 cells, whereas the mask mutant version of TMPRSS6 showed a modest inhibitory effect. This indicates that TMPRSS6-dependent pathway predominates over all known Hamp-activating pathways and that TMPRSS6-mediated Hamp suppression is determinant for acquiring adequate iron uptake from dietary sources. They hypothesized that TMPRSS6, also known as matriptase-2, participates in a transmembrane signalling pathway triggered by iron deficiency and independent to the known Hamp activation pathways and demonstrated that the proteolytic activity of matriptase-2 is determinant to hepcidin suppression activity. Recent experiments provided evidence that this serine protease inhibits hepcidin by cleaving HJV from the plasma membrane and has no cleavage activity on s-HJV[59]. The tissue-restricted, strong liver expression of matriptase-2 is logical if we consider that its activity responds to iron deficiency, in order to suppress the mHJV-BMPs pathway of hepcidin activation. The authors suggest that the pathway may still be modulated by s-HJV, further increasing the inhibition of hepcidin transcription induced by matriptase-2 alone.
Since the discovery of hepcidin and the demonstration of its pivotal role in iron homeostasis, there has been a substantial interest in developing a reliable assay of the hormone in biological fluids. Measurement of hepcidin in biological fluids can improve our understanding of iron diseases and be a useful tool for diagnosis and clinical management of these disorders. However, this has proven to be a very challenging task.
Inherent problems: The development of traditional immunochemical methods based on the production of specific anti-hepcidin antibodies has been hampered by several factors, including: (1) the small size (25 amino acids) and the compact structure of the peptide, with few antigenic epitopes; (2) the high degree of conservation between animal species[60], with ensuing difficulties in elicitation of an appropriate immune response in host animals; (3) the limited availability of the antigen. Indeed, the production of synthetic hepcidin in its hairpin native conformation determined by four disulfide bonds among the eight cysteine residues (near one fourth of the molecular weight)[61] is a complex procedure, as well as the isolation of hepcidin from urine[62]. A further problem is represented by hepcidin tendency to aggregate and to stick to laboratory plastic tubes, necessitating the need for careful handling and for standardized pre-analytical procedures.
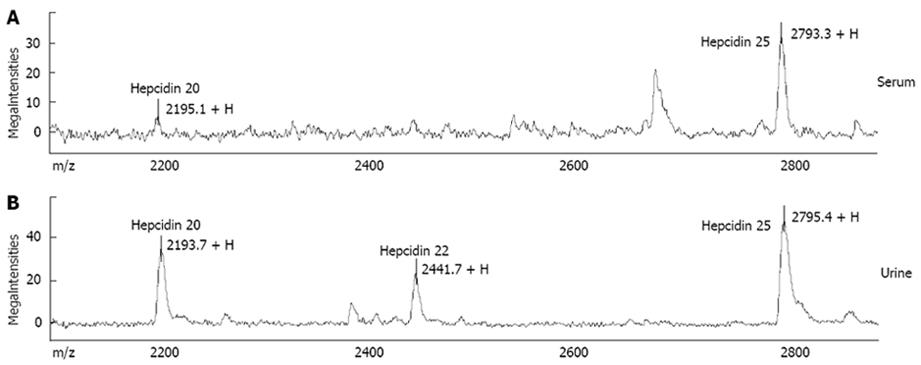
“First generation” methods: Early after hepcidin discovery, an ELISA kit able to detect serum pro-hepcidin became commercially available[63], based on an antibody recognizing an epitope outside the 25 amino acid sequence of the bioactive peptide. With few exceptions, this assay generally failed to give clinically useful information, since no correlation was reported with iron status and/or absorption[6465]. Most of the human studies published so far have relied on an immunodot assay for urinary hepcidin based on selective extraction of the peptide from urine by cation-exchange chromatography and its subsequent quantification by chemiluminescence using rabbit anti-human hepcidin primary antibodies[66]. Although this method provided very useful information on hepcidin regulation in human diseases[18–206768], it was quite laborious, and suitable only for relatively small series of patients. To circumvent inherent limitations of immunochemical methods, several research groups have focused on novel technologies, particularly on Mass Spectrometry (MS)-based methods, that rely on direct determination of the molecule of interest without the need for specific antibodies. Among these techniques, Surface-Enhanced Laser Desorption/Ionization Time-Of-Flight Mass Spectrometry (SELDI-TOF-MS) has emerged as a proteomic technique particularly promising for rapid direct detection of small sized biomarkers[69], like hepcidin. SELDI-TOF-MS combines matrix assisted laser-desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS) to surface chromatography. Depending on their chemical characteristics, peptides/proteins of interest are captured by specific interactions with protein surfaces (“chip-arrays”) that are used as a laser desorption ionization target. After the enriching step, proteins that do not bind to the surface are removed by a simple wash step, while bound proteins are analyzed by MS. Good preliminary results in hepcidin measurement were reported by several groups[70–73]. In particular, the SELDI-TOF-MS assay was able to discriminate different clinical conditions on the basis of the expected variations in hepcidin levels[7072], and correlated well with the immunodot assay in head-to-head comparison[70]. An important advantage of the SELDI-TOF-MS technique is the potential to evaluate not only the predominant 25 amino acid form, but also the forms with two peptides shorter at the amino terminus, i.e. hepcidin-22 and hepcidin-20 (Figure 1; also see below). Nevertheless, an inherent limitation of first generation SELDI-TOF-MS hepcidin assays (and in general, of all MS-based methods) is that peak heights in mass spectra do not always reflect absolute concentrations in clinical samples, because of competition during binding steps, and variations in ionisation efficiency. This implies that results can be expressed only in semi-quantitative arbitrary units, unless the use of a proper internal standard is implemented. But this again is a difficult task. Ideally, an internal standard is represented by a synthetic hepcidin-related peptide with no overlap with the endogenous peptide or other common peaks in serum/urine mass spectra, so that it could be spiked at known concentration to the clinical sample and co-analysed. Quantification of endogenous peptide is then obtained through the ratio internal standard:endogenous peptide. Murphy et al[74] first described a MS-based quantitative assay for either human or mouse hepcidin in serum using liquid chromatography tandem MS (LC-MS/MS) and calcitonin gene-related peptide (CGRP) as the internal standard. While a good accuracy was reported, the suitability of CGRP as non-hepcidin internal standard cannot be considered ideal because of its differences from the endogenous peptide in terms of mass, hydrophobicity, pI, and net charge[75].
Recent advances by “second generation” assays: This year has witnessed substantial advances in quantitative methods for hepcidin measurement in serum and/or urine. A variety of different approaches have been used by several research groups, summarized in Table 2. One of the most innovative was described by De Domenico et al[1]. They first identified the hepcidin binding domain (HBD) on the ferroportin molecule, and then synthesized a 19 amino acid corresponding peptide (spanning from amino acids 324 through 343 of the fourth extracellular loop of ferroportin). This peptide was then used in a competitive assay, i.e. by determining the ability of serum samples to compete with radioactive hepcidin for binding to the HBD peptide. In mice with targeted deletions of either HAMP or HFE gene, serum hepcidin measured by this method was found to vary as expected, as it did in a small group of healthy volunteers. This assay looks promising as an effective measure of true biologically active hepcidin, but needs further validation.
| Method | Reported normal range in serum (No. of controls) | Intra-assay precision (CV1) (%) | Internal standard | Note | LLOD2 | Reference |
| C-ELISA | 29-254 ng/mL (males) 17-286 ng/mL (females) (n = 114) | 5-19 | n.a. | Also used to quantify hepcidin in urine | 5 ng/mL | Ganz[76], 2008 |
| Quantitative SELDI-TOF-MS | 9-45 ng/mL (males) 7-15 ng/mL (females) (n = 23) | 5.7-11.7 | Des-Asp Hepcidin-24 (deletion of aspartate residue at position 25) | Also used to quantify hepcidin in urine | 3 ng/mL | Swinkels[77], 2008 |
| Quantitative SELDI-TOF-MS | 50 ng/mL (average in females) (n = 24) | 8-9 | Stable isotope labelled hepcidin (13C/15N phenylalanine at position 9) | 10 ng/mL | Ward[79], 2008 | |
| Micro-HPLC tandem tandem MS | 1-19.5 ng/mL (males) 1-6.5 ng/mL (females) (n = 10) | 4.8-21 | Stable isotope labelled hepcidin (13C/15N isoleucines at position 6-8) | 1 ng/mL | Kobold[78], 2008 | |
| Liquid Cromatography tandem MS | n.a. | 8.7-12.5 | Stable isotope labelled hepcidin (13C/15N glycines at position 12-20) | Only used in urine for the moment | n.a. | Bansal[75], 2008 |
| Hepcidin binding domain based test | 39-88 ng/mL (n = 40) | < 5 | n.a. | Potential for measuring true biologically active hepcidin (e.g. able to bind ferroportin) | n.r. | De Domenico[1], 2008 |
Ganz et al[76] were successful in obtaining sufficient amount of anti-hepcidin antibody to develop the first competitive enzyme-linked immunoassay (C-ELISA) for human hepcidin in a simple format (96-well plates) applicable to a relatively large series of patients. On the other hand, Swinkels et al[77] reported good quantification of hepcidin in both serum and urine updating the SELDI-TOF-MS protocol with the use of a proper internal standard clearly distinguishable from the endogenous peptide, i.e. a synthetic hepcidin-24 lacking the amino-terminal asparagine residue. Moreover, other groups working on MS-based methods reported reliable hepcidin quantification using stable isotope labelled 25 amino acid peptides with masses different from endogenous hepcidin because of the introduction of 13C/15N at different amino acid positions[757879]. Interestingly, the C-ELISA and the updated SELDI-TOF-MS protocol were used to measure hepcidin in series of serum/urine pair samples from healthy donors[7677], yielding quite similar results in terms of fractional excretion of the peptide (3%-5%). A preliminary direct head-to-head comparison of these two methods in split serum samples from the same individuals showed a high degree of correlation (Spearman’s rho up to 0.94; (Girelli D, unpublished observation)). Moreover, similar comparative analyses in rigorously selected healthy subjects without any potential confounder of iron status (e.g. subclinical inflammation or iron depletion) showed that the two methods were almost equally effective in reflecting the expected subtle physiologic variations of serum hepcidin as a function of established indices of body iron stores, with Spearman’s rho ranging from 0.7 to 0.8 (Girelli D, unpublished observation). In perspective, the two methods may be of complementary implementation in clinical and/or research setting. SELDI-TOF-MS has the disadvantage of relying on relatively expensive equipment and the need of dedicated skilled personnel. On the other hand, it has the positive advantage of being able to detect the two isoforms truncated at the N-terminus, i.e. hepcidin-22 and hepcidin-20. At present, the actual biological significance of these post-translational modifications is uncertain. Studies by Nemeth et al[3] have demonstrated that the five N-terminal amino acids are essential for the interaction with ferroportin, and thus for modulation of iron homeostasis. On the other hand, the 20 amino acid isoform has been demonstrated to possess greater antimicrobial activity than hepcidin-25[162]. Thus, the smaller forms may not simply reflect the catabolism of the “iron active” hepcidin-25 as postulated by some authors[80], but rather the generation of more potent defensins with antimicrobial peptides[1]. Further studies are needed in this direction, as it appears that we are just starting to know the complex role of hepcidin at the crossroads between iron homeostasis and infectious/inflammatory conditions.
In contrast, the C-ELISA method has the potential for a more widespread diffusion into clinical/hospital settings, since it does not need any specific equipment. However, the specificity of the antibody used with respect to various hepcidin isoforms remains to be verified.
Until recently, most of the studies of hepcidin in humans have relied on a human urinary hepcidin assay and on comparisons between patients and small groups of normal subjects without distinction between gender and age, all factors that might influence hepcidin concentrations. Very recently, Ganz et al[76] reported data on 114 healthy subjects, showing that the medians of hepcidin concentrations, measured by C-ELISA, significantly differed between men and women (112 and 65 ng/mL, respectively), likely due to difference in iron stores. They also showed a trend for age-related increase in serum hepcidin in both genders needing further confirm, and that there was a direct correlation between serum hepcidin and ferritin concentrations within the range of normality for ferritin.
Iron overload: In most cases iron overload results from inadequate hepcidin production relative to body iron stores[6781]. However, the causes that lead to reduced hepcidin production are different and include: hereditary defects that disrupt one or another protein involved in the normal pathway of positive regulation of hepcidin transcription (HFE, TfR2, HJV) and hepcidin itself; hereditary defects of proteins involved in iron transport (hypo-transferrinemia); ineffective erythropoiesis leading to increased and endless iron need by the erythroid marrow independent of iron stores (iron loading anemias)[38]. Exceptions are the iron overload due to hereditary defects of ferroportin, in which urinary hepcidin was found to be variably increased[198283] and pure transfusional iron overload in which the increased iron stores stimulate hepatic hepcidin production and can reach into the thousand of ng/mg creatinine[84]. In both these conditions, however, the cases tested are few and not homogeneous, and results need confirmation. There are no available data on aceruloplasminemia.
Hemochromatosis: Based on the current knowledge of the pathophysiological mechanisms of hepcidin regulation and on hepcidin measurement at either protein or mRNA level, hemochromatosis (HH) can be divided into two groups: primary iron overload disorders associated with defective or suppressed HAMP expression and ferroportin diseases.
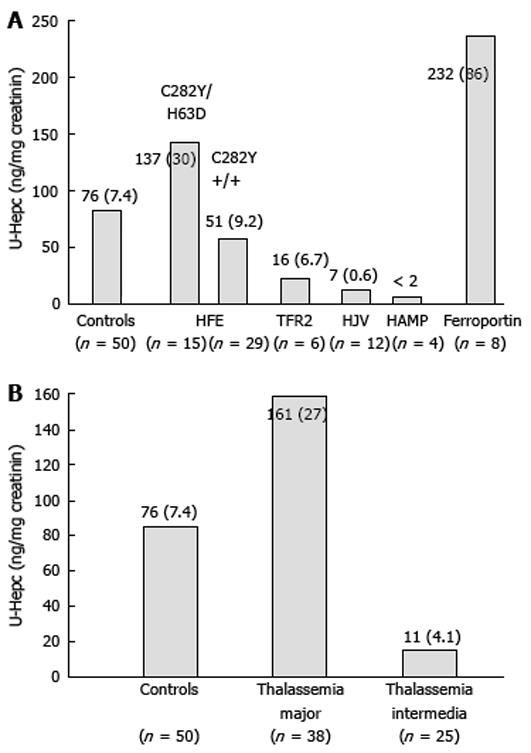
The first group includes HH type 1, 2 and 3, which are determined by defects in four different genes (HFE, TfR2, HJV and HAMP). Since all these proteins are involved in the same pathway, the physiopathological mechanism leading to iron overload in all these forms of HH depends on absent or defective synthesis, or the inability to up-regulate hepcidin production appropriately in response to increased iron stores[6711]. This in turn induces increased iron absorption in the plasma which exceeds the binding capacity of transferrin, causing the production of non-transferrin-bound iron and the accumulation of iron in parenchymal tissues. Thus, the differences among these forms of primary iron overload is quantitative (the amount of iron overload and the severity of iron-related damage), rather than qualitative (similar alterations of serum iron indices, similar iron distribution in the liver, same targets of iron deposition and damage). It can be hypothesised that the more important a defective protein is in the regulatory pathway of hepcidin transcription, the stronger is its suppression and the higher is the rate and amount of iron accumulation and, consequently, the severity of iron-related damage. In fact, in juvenile hemochomatosis (JH), hepcidin concentrations are lower than in adult forms of hemochromatosis, despite massive iron overload[19]. Figure 2 shows the concentration of urinary hepcidin measured by the same method in different forms of HH derived from personal and unpublished observations and on available data in the literature. For HFE-related HH patients we report only data collected at baseline. For the rare forms of HH, the available data on hepcidin are very limited in number and in most of these patients hepcidin measurement was not done at baseline. Thus, we took care, as far as possible, to select those still not fully iron depleted and sampled some time from phlebotomies, to limit the effect of phlebotomy-induced erythropoiesis and iron depletion on hepcidin synthesis.
Due to the higher prevalence of HFE-related HH, most of the data available on hepcidin measurements concern this form of HH. At diagnosis, when C282Y homozygous patients are iron overloaded, hepcidin values either at mRNA, urine or serum levels are only slightly lower than in controls[206785–87]. Interestingly, in only one study hepcidin was measured also in the C282Y/H63D genotype, showing that it was higher than in controls at diagnosis[20]. In both genotypes, however, the hepcidin/ferritin ratio (a derived parameter used to normalize hepcidin levels by the amount of iron overload) was significantly lower than in controls[20]. When analysed after iron depletion, patients of both genotypes, even at a time remote from the last phlebotomy, showed hepcidin concentrations significantly lower than that observed in iron overloaded patients and in controls. These findings allowed two considerations to be made: first, assuming that after normalization of iron stores HH patients return to their inborn status, van Dijk et al[87] concluded that hepcidin levels are innately low in HFE-HH; second, that patients with HFE-HH can modulate, although inappropriately, the chronic hepcidin increase in response to iron stores[20]. This impairment is less evident when the defect is mild as observed in patients with the C282Y/H63D genotype[20]. Based on the evidence that genetic background influences iron overload in HFE knockout mice and on recent findings in humans showing that some polymorphisms in the BMPs/SMAD4 signalling cascade can influence the iron phenotype in C282Y homozygotes[88], it can be hypothesised that phenotype variability in HFE-HH depends on the whole effect of major disease locus (HFE) together with genetic polymorphisms and modifiers on hepcidin production that can protect or aggravate the effect of the defective protein. Acquired factors may contribute to phenotype variability through modulation of hepcidin production in HFE-HH[89], but no data are available yet.
Recently, we demonstrated that in contrast to healthy subjects, hepcidin response after acute oral iron challenge was blunted in most patients with HFE-HH at diagnosis and after iron depletion[20]. This supports a role of HFE in the iron-sensing pathways regulating hepcidin synthesis. Interestingly, we also found a blunted response to acute oral iron challenge in a severe iron overloaded patient affected by type 3 HH due to a novel splicing defect of TFR2 (Pelucchi et al, Haematologica in press). Although this single observation needs caution, it provides the first evidence in vivo, in humans, that TFR2 may act as a sensor of serum iron levels to modulate hepcidin production[2728].
A handful of data is available for ferroportin disease (type 4 hemochromatosis) and these data are reported in Figure 2. The high concentration of urinary hepcidin has been ascribed to response to iron overload, but also to “hepcidin resistance” that may vary according to different mutations of ferroportin. However, there are not enough data to define how hepcidin is regulated in the two different forms of ferroportin disease (types A and B) and if hepcidin measurement can be of some help in distinguishing one type from the other.
Iron loading anemias: The erythroid regulator exerts a negative effect on hepcidin production that overwhelms the store regulator as shown by the marked iron overload which develops in hypotransferrinemia and iron loading anemias[38].
Thalassemia intermedia and thalassemia major are the best studied human models of hepcidin modulation by ineffective erythropoiesis alone and the combined and opposite effect of both ineffective erythropoiesis and transfusion dependent iron overload, respectively. Regular transfusions, in fact, induce a huge tissue iron accumulation, but also inhibit erythropoietic drive. As shown in Figure 2, hepcidin levels are markedly reduced in thalassemia intermedia, due to the erythropoietic drive and despite systemic tissue iron overload[9091]. Similar results were observed in not transfused hereditary diserythropoietic and acquired sideroblastic anemias[19] that share common pathogenetic mechanisms of iron overload with thalassemia intermedia[3638]. In thalassemia major, hepcidin production is higher than in thalassemia intermedia although still inappropriate to the massive transfusional iron loading that partially counteracts the erythropoietic-dependent hepcidin down-regulation[90–93]. However, there is a large variability in thalassemia major probably dependent on several factors that might influence hepcidin production: time of transfusion[93], iron chelation therapy, amount of iron overload. Very few data are presently available for other iron loading anemias that share pathogenetic mechanisms of iron overload with thalassemic syndromes, such as sideroblastic and congenital dyserythropoietic anemias. As expected, hepcidin either at mRNA level in the liver or at protein level in the urine was found to be suppressed despite marked iron overload[19]. There are scanty and discrepant results in sickle cell disease where iron status and hepcidin regulation are influenced by various factors that may act in opposite directions: anemia and hypoxia that lead to suppression of hepcidin production, transfusional iron overload and inflammation that stimulate hepcidin synthesis by different pathways[9394].
Due to the rarity of congenital hypo-transferrinemia in humans, most data on hepcidin regulation comes from studies in hypotransferrinemic mice which recapitulate the human disorder, showing very low hepatic mRNA levels that support the notion of a dominant erythroid signal in hepcidin regulation and the importance of low hepcidin levels in the development of iron overload[93–95]. There is a single study in a child with congenital hypo-transferrinemia which describes the modulation of urinary hepcidin during plasma transfusions[96]. The study suggests that hepcidin production is regulated by the balance between iron requirements of the erythroid marrow and iron supply by transferrin, in agreement with the concept that iron supply to the erythron is the most important factor influencing iron absorption and iron release from stores[3696].
Transfusional iron overload: The majority of studies relate to thalassemic patients in which ineffective erythropoiesis and transfusions frequently cooperate to produce iron overload and induce opposite effects on hepcidin transcription as extensively described. In contrast, very few and sometimes contradictory data are available for patients with pure transfusional iron overload. Nemeth et al[84] showed that hepcidin can reach into the thousands of ng/mg creatinine in two polytransfused patients with myelodysplastic syndrome (MDS), suggesting that the ranges of variation for urinary hepcidin and serum ferritin induced by iron overload are actually very similar. A recent study in 20 patients with MDS and myelofibrosis reported different results, mostly suggesting suppression rather than induction of hepcidin synthesis[97]. However, the group was not homogeneous and included patients with different MDS subtypes and alterations of iron status such as iron deficiency, iron overload secondary to transfusions or ineffective erythropoiesis. Overall, these findings indicate that the causes of hepcidin down-regulation in patients with MDS and myelofibrosis are much more heterogeneous than in thalassemia and need to be defined in more selected series[98].
Other acquired iron overload disorders: Preliminary data are available for some common chronic liver diseases frequently associated with slight to moderate iron overload. Aoki et al[99] measured hepatic mRNA expression in patients with chronic hepatitis C showing that hepcidin expression did not correlate with markers of inflammation, but correlated with hepatic iron stores, suggesting that iron overload in chronic hepatitis C is not due to inappropriate hepcidin production. In contrast, Fujita et al[100] found that patients with chronic hepatitis C had serum hepcidin-to-ferritin ratios significantly lower than HCV negative controls and that this relative impairment of hepcidin production was fully reversible after successful HCV eradication by PEG-IFN plus ribavirin. The hypothesis that HCV might suppress hepcidin expression is also supported by recent experiments in HCV replicon cells and in HCV core-expressing Huh7 cells. Miura et al[101] found that hepcidin expression was inversely correlated with the amount of reactive oxygen species (ROS) production and that HCV-induced oxidative stress caused hypoacetylation of histones and inhibited binding of two positive regulators (C/EBPα and STAT3) of hepcidin transcription. Interestingly, anti-oxidants restored hepcidin expression in these cell lines and reduced HDAC activity in a dose-dependent manner. Hepatic hepcidin mRNA expression correlated with hepatic iron in advanced chronic liver disease and may also be affected by hepatic dysfunction[68]. A reduced hepcidin synthesis indeed might be one of the mechanisms leading to iron overload in advanced liver disease of any origin, but no further studies have clarified this issue.
Increased hepatic iron deposits have been frequently described in association with obesity and alterations of lipid or glucose metabolism, insulin resistance and non-alcoholic fatty liver (NAFLD) and variably named dysmetabolic or insulin-resistance hepatic iron overload syndrome (IR-HIO). Urinary hepcidin levels, although inappropriate for the iron overload, were indeed significantly higher in patients with dysmetabolic iron overload than in controls[67]. This finding has been also confirmed by others, as recently reviewed by Deugnier et al[89]. Hepcidin resistance or increased extra-hepatic production have been hypothesised[6789] to explain these findings. Another explanation is that IR-HIO might be the result of an association of a mild-moderate, maybe polygenic, defect of hepcidin production and insulin resistance or metabolic syndrome (MS). These patients may retain some ability to increase hepcidin production in response to iron load as observed in subjects carrying the low expressing C282Y/H63D HFE genotype[20]. In IR-HIO patients, NAFLD and MS might induce hepcidin production through cytokine-mediated pathways[102] leading to the typical phenotype[103]. Further studies are needed to clarify this issue and the role of dysmetabolism in dysregulating iron regulatory pathways.
Iron deficiency: Urinary hepcidin levels were undetectable or low in patients with iron deficiency anemia[7684], in agreement with the suppressive effect of deficient iron stores and iron deficient erythropoiesis on hepcidin production. Very recently, the TMPRSS6 gene has been identified as the hepcidin negative regulator required to sense low iron stores[104]. TMPRSS6 mutations cause the rare iron-refractory iron deficiency anemia (IRIDA)[105106] and, in accordance with the suppressive effect of TMPRSS6 protein on hepcidin production, IRIDA patients show inappropriately elevated urinary hepcidin levels. This may explain the failure to absorb dietary iron despite systemic iron deficiency as well as the partial failure to respond to parenteral iron, which must be processed and then released by macrophages before being driven to the erythroid marrow[105106].
Although the liver is the main site of hepcidin synthesis, recent studies demonstrated the presence of measurable amounts of hepcidin mRNA and protein in cells and tissues other than liver in humans and animals: heart, kidney, retina, monocytes and macrophages, splenocyte and alveolar cells, adipocytes and pancreatic β-cells[102107–113]. In all these tissues the basal expression rate of hepcidin is lower than in the liver, suggesting a local role for hepcidin regulating iron homeostasis in these organs and tissues in an autocrine and paracrine fashion. On the other hand, it has been hypothesised that pancreatic hepcidin may contribute to the systemic hepcidin pool since it is exclusively synthesized by β-cells that secrete their product into the blood[108]. Nonetheless, this seems unlikely since expression of hepcidin in the pancreas is lower than in the liver and β-cells represent only a small portion of the total pancreatic parenchyma, in contrast to hepatocytes.
A major limitation of these studies is that they analysed hepcidin expression in response to only one of the major hepcidin regulators (iron, inflammation or hypoxia). In addition, only few evaluated the coupled modifications of hepcidin and ferroportin expression in response to such stimuli. There is emerging evidence, in fact, that hepcidin and its molecular target ferroportin may be expressed in the same cells, suggesting that ferroportin may be regulated by hepcidin generated within these cells independent of hepcidin in the circulation[107108113114].
Most studies showed that hepcidin responds to an acute phase reaction caused by either LPS, turpentine, group A Streptococcus strain or Pseudomonas aeruginosa. They can induce a 20-80 fold increase of hepcidin expression in murine macrophages, splenocytes and retinal cells by a Toll-like receptor 4 (TLR-4) dependent pathway[107110112], and in human monocytes via IL-6 induction, which is TLR-4 independent and involves STAT-3 dependent activation[114]. It is suggested that formation of hepcidin may locally contribute to the development of iron retention as part of the innate defensive mechanism generally aimed at reducing the availability of the essential nutrient iron from pathogens. This may be important for tissues such as the retina and the brain, which are protected by blood barriers or at inflammatory sites with poor perfusion where circulating hepcidin is scanty. In particular, it has been proposed that hepcidin formation by activated monocytes/macrophages may result in biologically significant accumulation of this peptide in the inflammatory environment[114]. Hepcidin produced by monocytes targets membrane bound ferroportin primarily as a secreted peptide in an autocrine way, but hepcidin also affects, although to a much lesser extent, ferroportin expression within the cell, suggesting two pathways of hepcidin trafficking within macrophages.
Further studies are needed to understand the function of hepcidin in extrahepatic tissues and to evaluate possible influences at the systemic level. In fact, it has been hypothesised that in massively obese patients hepcidin production by adipocytes might contribute to the development of iron deficiency anemia in some patients[102]. Other authors, based on the co-localization of hepcidin with insulin-storing secretory granules, suggested that regulation of iron and glucose metabolism are distinctly coupled at the pancreatic level by the co-release of insulin and hepcidin[108].
Despite these recent important advances, much work still needs to be done. For example, the presence in blood of pro-hepcidin at concentrations greater than the iron active peptide remains puzzling. Huang et al[115] recently suggested that pro-hepcidin conversion into hepcidin may occur in the circulation as part of an iron-dependent regulation at the post-translational level, but this needs to be confirmed in larger studies. Other issues to be clarified are fluctuations of hepcidin related to circadian rhythm and/or meals[7276], and if urinary hepcidin quantification can give information that might be complementary to single point serum measurement, similar to many other hormones. Since hepcidin is directly implicated in the regulation of iron homeostasis, its measurement might turn out to be a useful tool in the differential diagnosis of iron overload disorders and iron deficiency. The definition of the biological value of the hepcidin/ferritin ratio will be important to understand the appropriateness of hepcidin production in the clinical setting. Similarly the response of hepcidin to oral iron might prove to be a useful test to evaluate iron absorption in iron deficiency and the iron-sensing pathway in iron overload and in targeting phlebotomy treatment in patients with hemochromatosis[20]. Finally, of special importance are collaborative studies among various laboratories aimed at comparing the analytical performance of different methods, as well as for promoting standardization of hepcidin assay. In this regard, the results of the ongoing first international round robin for quantification of urinary and plasma hepcidin (Kroot et al, Blood Suppl. in press) are eagerly awaited.
| 1. | De Domenico I, Nemeth E, Nelson JM, Phillips JD, Ajioka RS, Kay MS, Kushner JP, Ganz T, Ward DM, Kaplan J. The hepcidin-binding site on ferroportin is evolutionarily conserved. Cell Metab. 2008;8:146-156. |
| 2. | Kemna EH, Kartikasari AE, van Tits LJ, Pickkers P, Tjalsma H, Swinkels DW. Regulation of hepcidin: insights from biochemical analyses on human serum samples. Blood Cells Mol Dis. 2008;40:339-346. |
| 3. | Nemeth E, Preza GC, Jung CL, Kaplan J, Waring AJ, Ganz T. The N-terminus of hepcidin is essential for its interaction with ferroportin: structure-function study. Blood. 2006;107:328-333. |
| 4. | Ganz T. Cellular iron: ferroportin is the only way out. Cell Metab. 2005;1:155-157. |
| 5. | Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090-2093. |
| 7. | De Domenico I, McVey Ward D, Kaplan J. Regulation of iron acquisition and storage: consequences for iron-linked disorders. Nat Rev Mol Cell Biol. 2008;9:72-81. |
| 8. | Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531-539. |
| 9. | Xia Y, Babitt JL, Sidis Y, Chung RT, Lin HY. Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood. 2008;111:5195-5204. |
| 10. | Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399-409. |
| 11. | Ganz T. Iron homeostasis: fitting the puzzle pieces together. Cell Metab. 2008;7:288-290. |
| 12. | Vujic Spasic M, Kiss J, Herrmann T, Kessler R, Stolte J, Galy B, Rathkolb B, Wolf E, Stremmel W, Hentze MW. Physiologic systemic iron metabolism in mice deficient for duodenal Hfe. Blood. 2007;109:4511-4517. |
| 13. | Giannetti AM, Björkman PJ. HFE and transferrin directly compete for transferrin receptor in solution and at the cell surface. J Biol Chem. 2004;279:25866-25875. |
| 14. | Lebrón JA, West AP Jr, Bjorkman PJ. The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. J Mol Biol. 1999;294:239-245. |
| 15. | Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281:28494-28498. |
| 16. | Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7:205-214. |
| 17. | Waheed A, Britton RS, Grubb JH, Sly WS, Fleming RE. HFE association with transferrin receptor 2 increases cellular uptake of transferrin-bound iron. Arch Biochem Biophys. 2008;474:193-197. |
| 18. | Nemeth E, Roetto A, Garozzo G, Ganz T, Camaschella C. Hepcidin is decreased in TFR2 hemochromatosis. Blood. 2005;105:1803-1806. |
| 19. | Papanikolaou G, Tzilianos M, Christakis JI, Bogdanos D, Tsimirika K, MacFarlane J, Goldberg YP, Sakellaropoulos N, Ganz T, Nemeth E. Hepcidin in iron overload disorders. Blood. 2005;105:4103-4105. |
| 20. | Piperno A, Girelli D, Nemeth E, Trombini P, Bozzini C, Poggiali E, Phung Y, Ganz T, Camaschella C. Blunted hepcidin response to oral iron challenge in HFE-related hemochromatosis. Blood. 2007;110:4096-4100. |
| 21. | Zhou XY, Tomatsu S, Fleming RE, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt EM, Ruddy DA, Prass CE. HFE gene knockout produces mouse model of hereditary hemochromatosis. Proc Natl Acad Sci USA. 1998;95:2492-2497. |
| 22. | Gao J, Zhao N, Knutson MD, Enns CA. The hereditary hemochromatosis protein, HFE, inhibits iron uptake via down-regulation of Zip14 in HepG2 cells. J Biol Chem. 2008;283:21462-21468. |
| 23. | Roetto A, Totaro A, Piperno A, Piga A, Longo F, Garozzo G, Calì A, De Gobbi M, Gasparini P, Camaschella C. New mutations inactivating transferrin receptor 2 in hemochromatosis type 3. Blood. 2001;97:2555-2560. |
| 24. | Kawabata H, Fleming RE, Gui D, Moon SY, Saitoh T, O'Kelly J, Umehara Y, Wano Y, Said JW, Koeffler HP. Expression of hepcidin is down-regulated in TfR2 mutant mice manifesting a phenotype of hereditary hemochromatosis. Blood. 2005;105:376-381. |
| 25. | Wallace DF, Summerville L, Lusby PE, Subramaniam VN. First phenotypic description of transferrin receptor 2 knockout mouse, and the role of hepcidin. Gut. 2005;54:980-986. |
| 26. | Wallace DF, Summerville L, Subramaniam VN. Targeted disruption of the hepatic transferrin receptor 2 gene in mice leads to iron overload. Gastroenterology. 2007;132:301-310. |
| 27. | Johnson MB, Enns CA. Diferric transferrin regulates transferrin receptor 2 protein stability. Blood. 2004;104:4287-4293. |
| 28. | Robb A, Wessling-Resnick M. Regulation of transferrin receptor 2 protein levels by transferrin. Blood. 2004;104:4294-4299. |
| 29. | Calzolari A, Raggi C, Deaglio S, Sposi NM, Stafsnes M, Fecchi K, Parolini I, Malavasi F, Peschle C, Sargiacomo M. TfR2 localizes in lipid raft domains and is released in exosomes to activate signal transduction along the MAPK pathway. J Cell Sci. 2006;119:4486-4498. |
| 30. | De Domenico I, Ward DM, Kaplan J. Hepcidin regulation: ironing out the details. J Clin Invest. 2007;117:1755-1758. |
| 31. | Verga Falzacappa MV, Casanovas G, Hentze MW, Muckenthaler MU. A bone morphogenetic protein (BMP)-responsive element in the hepcidin promoter controls HFE2-mediated hepatic hepcidin expression and its response to IL-6 in cultured cells. J Mol Med. 2008;86:531-540. |
| 32. | Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115:2180-2186. |
| 33. | Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106:2884-2889. |
| 34. | Fleming RE. Iron and inflammation: cross-talk between pathways regulating hepcidin. J Mol Med. 2008;86:491-494. |
| 35. | Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353-358. |
| 38. | Nemeth E. Iron regulation and erythropoiesis. Curr Opin Hematol. 2008;15:169-175. |
| 39. | Pinto JP, Ribeiro S, Pontes H, Thowfeequ S, Tosh D, Carvalho F, Porto G. Erythropoietin mediates hepcidin expression in hepatocytes through EPOR signaling and regulation of C/EBPalpha. Blood. 2008;111:5727-5733. |
| 40. | Pak M, Lopez MA, Gabayan V, Ganz T, Rivera S. Suppression of hepcidin during anemia requires erythropoietic activity. Blood. 2006;108:3730-3735. |
| 41. | Tanno T, Bhanu NV, Oneal PA, Goh SH, Staker P, Lee YT, Moroney JW, Reed CH, Luban NL, Wang RH. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med. 2007;13:1096-1101. |
| 42. | Braliou GG, Verga Falzacappa MV, Chachami G, Casanovas G, Muckenthaler MU, Simos G. 2-Oxoglutarate-dependent oxygenases control hepcidin gene expression. J Hepatol. 2008;48:801-810. |
| 43. | Choi SO, Cho YS, Kim HL, Park JW. ROS mediate the hypoxic repression of the hepcidin gene by inhibiting C/EBPalpha and STAT-3. Biochem Biophys Res Commun. 2007;356:312-317. |
| 44. | Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, Nizet V, Johnson RS. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs). J Clin Invest. 2007;117:1926-1932. |
| 45. | Bridle K, Cheung TK, Murphy T, Walters M, Anderson G, Crawford DG, Fletcher LM. Hepcidin is down-regulated in alcoholic liver injury: implications for the pathogenesis of alcoholic liver disease. Alcohol Clin Exp Res. 2006;30:106-112. |
| 46. | Harrison-Findik DD. Role of alcohol in the regulation of iron metabolism. World J Gastroenterol. 2007;13:4925-4930. |
| 47. | Harrison-Findik DD, Klein E, Crist C, Evans J, Timchenko N, Gollan J. Iron-mediated regulation of liver hepcidin expression in rats and mice is abolished by alcohol. Hepatology. 2007;46:1979-1985. |
| 48. | Flanagan JM, Peng H, Beutler E. Effects of alcohol consumption on iron metabolism in mice with hemochromatosis mutations. Alcohol Clin Exp Res. 2007;31:138-143. |
| 49. | Olynyk J, Hall P, Reed W, Williams P, Kerr R, Mackinnon M. A long-term study of the interaction between iron and alcohol in an animal model of iron overload. J Hepatol. 1995;22:671-676. |
| 50. | Adams PC, Agnew S. Alcoholism in hereditary hemochromatosis revisited: prevalence and clinical consequences among homozygous siblings. Hepatology. 1996;23:724-727. |
| 51. | Brissot P, Bourel M, Herry D, Verger JP, Messner M, Beaumont C, Regnouard F, Ferrand B, Simon M. Assessment of liver iron content in 271 patients: a reevaluation of direct and indirect methods. Gastroenterology. 1981;80:557-565. |
| 52. | Chapman RW, Morgan MY, Laulicht M, Hoffbrand AV, Sherlock S. Hepatic iron stores and markers of iron overload in alcoholics and patients with idiopathic hemochromatosis. Dig Dis Sci. 1982;27:909-916. |
| 53. | LeSage GD, Baldus WP, Fairbanks VF, Baggenstoss AH, McCall JT, Moore SB, Taswell HF, Gordon H. Hemochromatosis: genetic or alcohol-induced? Gastroenterology. 1983;84:1471-1477. |
| 54. | Lakhal S, Talbot NP, Crosby A, Stoepker C, Townsend AR, Robbins PA, Pugh CW, Ratcliffe PJ, Mole DR. Regulation of growth differentiation factor 15 expression by intracellular iron. Blood. 2008;84. |
| 55. | Tamary H, Shalev H, Perez-Avraham G, Zoldan M, Levi I, Swinkels DW, Tanno T, Miller JL. Elevated growth differentiation factor 15 expression in patients with congenital dyserythropoietic anemia type I. Blood. 2008;112:5241-5244. |
| 56. | Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117:1933-1939. |
| 57. | Silvestri L, Pagani A, Camaschella C. Furin-mediated release of soluble hemojuvelin: a new link between hypoxia and iron homeostasis. Blood. 2008;111:924-931. |
| 58. | Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EM. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320:1088-1092. |
| 59. | Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8:502-511. |
| 60. | Ganz T, Nemeth E. Iron imports. IV. Hepcidin and regulation of body iron metabolism. Am J Physiol Gastrointest Liver Physiol. 2006;290:G199-G203. |
| 61. | Hunter HN, Fulton DB, Ganz T, Vogel HJ. The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J Biol Chem. 2002;277:37597-37603. |
| 62. | Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806-7810. |
| 63. | Kulaksiz H, Gehrke SG, Janetzko A, Rost D, Bruckner T, Kallinowski B, Stremmel W. Pro-hepcidin: expression and cell specific localisation in the liver and its regulation in hereditary haemochromatosis, chronic renal insufficiency, and renal anaemia. Gut. 2004;53:735-743. |
| 64. | Hadley KB, Johnson LK, Hunt JR. Iron absorption by healthy women is not associated with either serum or urinary prohepcidin. Am J Clin Nutr. 2006;84:150-155. |
| 65. | Roe MA, Spinks C, Heath AL, Harvey LJ, Foxall R, Wimperis J, Wolf C, Fairweather-Tait SJ. Serum prohepcidin concentration: no association with iron absorption in healthy men; and no relationship with iron status in men carrying HFE mutations, hereditary haemochromatosis patients undergoing phlebotomy treatment, or pregnant women. Br J Nutr. 2007;97:544-549. |
| 66. | Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, Ganz T. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271-1276. |
| 67. | Barisani D, Pelucchi S, Mariani R, Galimberti S, Trombini P, Fumagalli D, Meneveri R, Nemeth E, Ganz T, Piperno A. Hepcidin and iron-related gene expression in subjects with Dysmetabolic Hepatic Iron Overload. J Hepatol. 2008;49:123-133. |
| 68. | Détivaud L, Nemeth E, Boudjema K, Turlin B, Troadec MB, Leroyer P, Ropert M, Jacquelinet S, Courselaud B, Ganz T. Hepcidin levels in humans are correlated with hepatic iron stores, hemoglobin levels, and hepatic function. Blood. 2005;106:746-748. |
| 69. | Cristea IM, Gaskell SJ, Whetton AD. Proteomics techniques and their application to hematology. Blood. 2004;103:3624-3634. |
| 70. | Bozzini C, Campostrini N, Trombini P, Nemeth E, Castagna A, Tenuti I, Corrocher R, Camaschella C, Ganz T, Olivieri O. Measurement of urinary hepcidin levels by SELDI-TOF-MS in HFE-hemochromatosis. Blood Cells Mol Dis. 2008;40:347-352. |
| 71. | Kemna E, Tjalsma H, Laarakkers C, Nemeth E, Willems H, Swinkels D. Novel urine hepcidin assay by mass spectrometry. Blood. 2005;106:3268-3270. |
| 72. | Kemna EH, Tjalsma H, Podust VN, Swinkels DW. Mass spectrometry-based hepcidin measurements in serum and urine: analytical aspects and clinical implications. Clin Chem. 2007;53:620-628. |
| 73. | Tomosugi N, Kawabata H, Wakatabe R, Higuchi M, Yamaya H, Umehara H, Ishikawa I. Detection of serum hepcidin in renal failure and inflammation by using ProteinChip System. Blood. 2006;108:1381-1387. |
| 74. | Murphy AT, Witcher DR, Luan P, Wroblewski VJ. Quantitation of hepcidin from human and mouse serum using liquid chromatography tandem mass spectrometry. Blood. 2007;110:1048-1054. |
| 75. | Bansal SS, Halket JM, Bomford A, Simpson RJ, Vasavda N, Thein SL, Hider RC. Quantitation of hepcidin in human urine by liquid chromatography-mass spectrometry. Anal Biochem. 2009;384:245-253. |
| 76. | Ganz T, Olbina G, Girelli D, Nemeth E, Westerman M. Immunoassay for human serum hepcidin. Blood. 2008;112:4292-4297. |
| 77. | Swinkels DW, Girelli D, Laarakkers C, Kroot J, Campostrini N, Kemna EH, Tjalsma H. Advances in quantitative hepcidin measurements by time-of-flight mass spectrometry. PLoS ONE. 2008;3:e2706. |
| 78. | Kobold U, Dülffer T, Dangl M, Escherich A, Kubbies M, Röddiger R, Wright JA. Quantification of hepcidin-25 in human serum by isotope dilution micro-HPLC-tandem mass spectrometry. Clin Chem. 2008;54:1584-1586. |
| 79. | Ward DG, Roberts K, Stonelake P, Goon P, Zampronio CG, Martin A, Johnson PJ, Iqbal T, Tselepis C. SELDI-TOF-MS determination of hepcidin in clinical samples using stable isotope labelled hepcidin as an internal standard. Proteome Sci. 2008;6:28. |
| 80. | Kemna EH, Tjalsma H, Willems HL, Swinkels DW. Hepcidin: from discovery to differential diagnosis. Haematologica. 2008;93:90-97. |
| 81. | Pietrangelo A. Hemochromatosis: an endocrine liver disease. Hepatology. 2007;46:1291-1301. |
| 82. | Cremonesi L, Forni GL, Soriani N, Lamagna M, Fermo I, Daraio F, Galli A, Pietra D, Malcovati L, Ferrari M. Genetic and clinical heterogeneity of ferroportin disease. Br J Haematol. 2005;131:663-670. |
| 83. | Zoller H, McFarlane I, Theurl I, Stadlmann S, Nemeth E, Oxley D, Ganz T, Halsall DJ, Cox TM, Vogel W. Primary iron overload with inappropriate hepcidin expression in V162del ferroportin disease. Hepatology. 2005;42:466-472. |
| 84. | Nemeth E, Valore EV, Territo M, Schiller G, Lichtenstein A, Ganz T. Hepcidin, a putative mediator of anemia of inflammation, is a type II acute-phase protein. Blood. 2003;101:2461-2463. |
| 85. | Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, Subramaniam VN, Powell LW, Anderson GJ, Ramm GA. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361:669-673. |
| 86. | Gehrke SG, Kulaksiz H, Herrmann T, Riedel HD, Bents K, Veltkamp C, Stremmel W. Expression of hepcidin in hereditary hemochromatosis: evidence for a regulation in response to the serum transferrin saturation and to non-transferrin-bound iron. Blood. 2003;102:371-376. |
| 87. | van Dijk BA, Laarakkers CM, Klaver SM, Jacobs EM, van Tits LJ, Janssen MC, Swinkels DW. Serum hepcidin levels are innately low in HFE-related haemochromatosis but differ between C282Y-homozygotes with elevated and normal ferritin levels. Br J Haematol. 2008;142:979-985. |
| 88. | Milet J, Dehais V, Bourgain C, Jouanolle AM, Mosser A, Perrin M, Morcet J, Brissot P, David V, Deugnier Y. Common variants in the BMP2, BMP4, and HJV genes of the hepcidin regulation pathway modulate HFE hemochromatosis penetrance. Am J Hum Genet. 2007;81:799-807. |
| 89. | Deugnier Y, Brissot P, Loréal O. Iron and the liver: update 2008. J Hepatol. 2008;48 Suppl 1:S113-S123. |
| 90. | Kattamis A, Papassotiriou I, Palaiologou D, Apostolakou F, Galani A, Ladis V, Sakellaropoulos N, Papanikolaou G. The effects of erythropoetic activity and iron burden on hepcidin expression in patients with thalassemia major. Haematologica. 2006;91:809-812. |
| 91. | Origa R, Galanello R, Ganz T, Giagu N, Maccioni L, Faa G, Nemeth E. Liver iron concentrations and urinary hepcidin in beta-thalassemia. Haematologica. 2007;92:583-588. |
| 92. | Camberlein E, Zanninelli G, Détivaud L, Lizzi AR, Sorrentino F, Vacquer S, Troadec MB, Angelucci E, Abgueguen E, Loréal O. Anemia in beta-thalassemia patients targets hepatic hepcidin transcript levels independently of iron metabolism genes controlling hepcidin expression. Haematologica. 2008;93:111-115. |
| 93. | Kearney SL, Nemeth E, Neufeld EJ, Thapa D, Ganz T, Weinstein DA, Cunningham MJ. Urinary hepcidin in congenital chronic anemias. Pediatr Blood Cancer. 2007;48:57-63. |
| 95. | Nemeth E, Ganz T. Hepcidin and iron-loading anemias. Haematologica. 2006;91:727-732. |
| 96. | Trombini P, Coliva T, Nemeth E, Mariani R, Ganz T, Biondi A, Piperno A. Effects of plasma transfusion on hepcidin production in human congenital hypotransferrinemia. Haematologica. 2007;92:1407-1410. |
| 97. | Winder A, Lefkowitz R, Ghoti H, Leiba M, Ganz T, Nemeth E, Rachmilewitz EA. Urinary hepcidin excretion in patients with myelodysplastic syndrome and myelofibrosis. Br J Haematol. 2008;142:669-671. |
| 98. | Mariani R, Pelucchi S, Pozzi M, Paolini V, Piperno A. Reduced expression of hepcidin in patients with myelodysplastic syndrome and myelofibrosis: the causes might be more heterogeneous than in thalassaemia. Br J Haematol. 2008;143:746-747. |
| 99. | Aoki CA, Rossaro L, Ramsamooj R, Brandhagen D, Burritt MF, Bowlus CL. Liver hepcidin mRNA correlates with iron stores, but not inflammation, in patients with chronic hepatitis C. J Clin Gastroenterol. 2005;39:71-74. |
| 100. | Fujita N, Sugimoto R, Motonishi S, Tomosugi N, Tanaka H, Takeo M, Iwasa M, Kobayashi Y, Hayashi H, Kaito M. Patients with chronic hepatitis C achieving a sustained virological response to peginterferon and ribavirin therapy recover from impaired hepcidin secretion. J Hepatol. 2008;49:702-710. |
| 101. | Miura K, Taura K, Kodama Y, Schnabl B, Brenner DA. Hepatitis C virus-induced oxidative stress suppresses hepcidin expression through increased histone deacetylase activity. Hepatology. 2008;48:1420-1429. |
| 102. | Bekri S, Gual P, Anty R, Luciani N, Dahman M, Ramesh B, Iannelli A, Staccini-Myx A, Casanova D, Ben Amor I. Increased adipose tissue expression of hepcidin in severe obesity is independent from diabetes and NASH. Gastroenterology. 2006;131:788-796. |
| 103. | Riva A, Trombini P, Mariani R, Salvioni A, Coletti S, Bonfadini S, Paolini V, Pozzi M, Facchetti R, Bovo G. Revaluation of clinical and histological criteria for diagnosis of dysmetabolic iron overload syndrome. World J Gastroenterol. 2008;14:4745-4752. |
| 104. | Muckenthaler MU. Fine tuning of hepcidin expression by positive and negative regulators. Cell Metab. 2008;8:1-3. |
| 105. | Melis MA, Cau M, Congiu R, Sole G, Barella S, Cao A, Westerman M, Cazzola M, Galanello R. A mutation in the TMPRSS6 gene, encoding a transmembrane serine protease that suppresses hepcidin production, in familial iron deficiency anemia refractory to oral iron. Haematologica. 2008;93:1473-1479. |
| 106. | Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM, Strouse JJ, Markianos K. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet. 2008;40:569-571. |
| 107. | Gnana-Prakasam JP, Martin PM, Mysona BA, Roon P, Smith SB, Ganapathy V. Hepcidin expression in mouse retina and its regulation via lipopolysaccharide/Toll-like receptor-4 pathway independent of Hfe. Biochem J. 2008;411:79-88. |
| 108. | Kulaksiz H, Fein E, Redecker P, Stremmel W, Adler G, Cetin Y. Pancreatic beta-cells express hepcidin, an iron-uptake regulatory peptide. J Endocrinol. 2008;197:241-249. |
| 109. | Kulaksiz H, Theilig F, Bachmann S, Gehrke SG, Rost D, Janetzko A, Cetin Y, Stremmel W. The iron-regulatory peptide hormone hepcidin: expression and cellular localization in the mammalian kidney. J Endocrinol. 2005;184:361-370. |
| 110. | Liu XB, Nguyen NB, Marquess KD, Yang F, Haile DJ. Regulation of hepcidin and ferroportin expression by lipopolysaccharide in splenic macrophages. Blood Cells Mol Dis. 2005;35:47-56. |
| 111. | Merle U, Fein E, Gehrke SG, Stremmel W, Kulaksiz H. The iron regulatory peptide hepcidin is expressed in the heart and regulated by hypoxia and inflammation. Endocrinology. 2007;148:2663-2668. |
| 112. | Peyssonnaux C, Zinkernagel AS, Datta V, Lauth X, Johnson RS, Nizet V. TLR4-dependent hepcidin expression by myeloid cells in response to bacterial pathogens. Blood. 2006;107:3727-3732. |
| 113. | Theurl M, Theurl I, Hochegger K, Obrist P, Subramaniam N, van Rooijen N, Schuemann K, Weiss G. Kupffer cells modulate iron homeostasis in mice via regulation of hepcidin expression. J Mol Med. 2008;86:825-835. |
| 114. | Theurl I, Theurl M, Seifert M, Mair S, Nairz M, Rumpold H, Zoller H, Bellmann-Weiler R, Niederegger H, Talasz H. Autocrine formation of hepcidin induces iron retention in human monocytes. Blood. 2008;111:2392-2399. |
| 115. | Huang X, Fung ET, Yip C, Zeleniuch-Jacquotte A. Serum prohepcidin is associated with soluble transferrin receptor-1 but not ferritin in healthy post-menopausal women. Blood Cells Mol Dis. 2008;41:265-269. |