Published online Jun 7, 2009. doi: 10.3748/wjg.15.2602
Revised: April 30, 2009
Accepted: May 7, 2009
Published online: June 7, 2009
AIM: To explore the effect of silencing of signal transducer and activator of transcription 3 (STAT3) expression by RNA interference (RNAi) on growth of human hepatocellular carcinoma (HCC) in tumor-bearing nude mice in vivo.
METHODS: To construct the recombinant plasmid of pSilencer 3.0-H1-STAT3-siRNA-GFP (pSH1-siRNA-STAT3) and establish the tumor-bearing nude mouse model of the HCC cell line SMMC7721, we used intratumoral injection together with electroblotting to transfect the recombinant plasmid pSH1-siRNA-STAT3 into the transplanted tumor. The weight of the nude mice and tumor volumes were recorded. STAT3 gene transcription was detected by semi-quantitative reverse transcription polymerase chain reaction (RT-PCR). Level of protein expression and location of STAT3 were determined by Western blotting and immunohistochemical staining. STAT3-related genes such as survivin, c-myc, VEGF, p53 and caspase3 mRNA and protein expression were detected in tumor tissues at the same time. The terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay was used to detect apoptosis of tumor cells.
RESULTS: The weight of the treated nude mice increased, and the tumor volume decreased markedly compared with those of the mock-treated and negative control groups (P < 0.01). The results of RT-PCR and Western blotting showed that mRNA and protein levels of STAT3 declined markedly in the treated group. The change in STAT3-related gene expression in tumor tissues at the mRNA and protein level also varied, the expression of survivin, VEGF and c-myc were obviously reduced, and expression of p53 and caspase3 increased (P < 0.01). Most of the tumor tissue cells in the treated group developed apoptosis that was detected by TUNEL assay.
CONCLUSION: Silencing of STAT3 expression by RNAi significantly inhibits expression of STAT3 mRNA and protein, and suppresses growth of human HCC in tumor-bearing nude mice. The mechanism may be related to down-regulation of survivin, VEGF and c-myc and up-regulation of p53 and caspase3 expression. Accordingly, the STAT3 gene may act as an important and effective target in gene therapy of HCC.
- Citation: Li J, Piao YF, Jiang Z, Chen L, Sun HB. Silencing of signal transducer and activator of transcription 3 expression by RNA interference suppresses growth of human hepatocellular carcinoma in tumor-bearing nude mice. World J Gastroenterol 2009; 15(21): 2602-2608
- URL: https://www.wjgnet.com/1007-9327/full/v15/i21/2602.htm
- DOI: https://dx.doi.org/10.3748/wjg.15.2602
RNA interference (RNAi) is a post-transcriptional gene-silencing mechanism, in which the homologous RNA sequences are introduced into the cells that inhibit the expression of a particular gene, through the introduction of short interfering RNAs[1]. There have been a large number of confirmed reports that RNAi targeting oncogenes may successfully inhibit the growth of tumor cells in vitro and in vivo. Hence, RNAi has been turned into a potent technology for tumor therapy.
Hepatocellular carcinoma (HCC) research has shown that RNAi targeting genes are related to initiation, development and metastasis of HCC[2–4]. Signal transducer and activator of transcription 3 (STAT3) is an important member of the family of STAT. Its signal pathway is closely associated with the proliferation, differentiation and apoptosis of cells, and constant activation of STAT3 can promote cell proliferation and carcinogenesis. At present, STAT3 is defined as an oncogene[5]. Studies of STAT3 have focused on a variety of tumor cell lines, including leukemia, multiple myeloma, breast cancer, prostate cancer, melanoma tumor, and lung cancer, while HCC has been rarely investigated. Persistently activated STAT3 is detected in many HCC cell lines and tissues.
Our preliminary research revealed that the recombination plasmid pSH1Si-STAT3 targeting the STAT3 gene could dramatically inhibit the expression of STAT3 mRNA and protein in the HCC cell line SMMC7721 and the proliferation of HCC cells, and induce apoptosis of HCC cells[6]. Therefore, we designed the present experiment to verify the therapeutic effect of recombination plasmid pSH1-siRNA-STAT3 in HCC in vivo.
The human HCC cell line SMMC7721 was kept in the Department of Pathophysiology of the School of Basic Medicine in Jilin University, and cultured in Iscove’s Modified Dulbecco’s Medium (Gibco, BRL, USA) supplemented with 10% fetal bovine serum (Gibco, BRL), in an incubator containing 5% CO2 at 37°C, and then digested and passaged with 0.25% trypsin.
Male BALB/c nude mice were purchased from the Institute of Zoology, Chinese Academy of Sciences. They were 5-wk-old, SPF grade and weighed 17.76 ± 1.83 g, and were fed in an aseptic laminar flow room at 25°C and 60%-70% humidity, with a standard rodent diet and water.
The plasmids pSilencer3.0-H1-STAT3-siRNA and pSilencer1.0-U6-STAT3-siRNA-GFP were kindly provided by the Research Center of Prostate Diseases in Jilin University. The sequences of STAT3-siRNA were: 5'-GCAGCAGCTGAACAACATGTTCAAGAGACATGTTGTTCAGCTGCTGCTTTTT-3′ (forward), and 5'-AATTAAAAAGCAGCAGCTGAACAACATGTCTCTTGAACATGTTGTTCAGCTGCTGCTGCGGCC-3' (reverse). The two plasmids were linearized with EcoRI and HindII, and then pSilencer3.0-H1 and STAT3-siRNA-GFP were combined to form pSilencer3.0-H1-STAT3-siRNA-GFP (pSH1Si-STAT3), which was cloned and sequenced. A negative control pSH1Si-Scramble, which has no significant homology with human gene sequences, was constructed by the same method. The extraction of plasmids followed the routine Fastfiler Endo-free Plasmid Maxiprep protocol (BioDev, Beijing, China).
The cells in logarithmic growth phase were digested with 0.25% trypsin and centrifuged. The cells were collected and washed three times with PBS, and the living cells were counted under a microscope, and adjusted to a final concentration of 2.5 × 107 cells, with PBS.
The nude mice were fed on the super-clean biological laminar flow shelf for 1 wk. When the weight of the mice increased to 21.02 ± 1.81 g, tumor cells were injected subcutaneously into their hindquarters. Each mouse was seeded with 200 &mgr;L that contained 5 × 106 cells. Then, the mice were observed daily for mental state, diet and stools, and were monitored every 4 d for tumor size and body weight. The model was established successfully, when there were nodules with the size of a grain of rice after 1 wk, and a volume of 50-70 mm3 after 2 wk. We measured the largest (a) and smallest (b) superficial diameter with Vernier calipers and calculated the tumor volume (V) = a × b2× 0.5236.
The nude mice were divided into mock-treated, negative control and treated groups of five mice each. PBS was used for transfection in the mock-treated group, pSH1Si-Scramble was used for transfection in the negative control group, and pSH1Si-STAT3 was used for transfection in the treated group. The treatment volume was 40 &mgr;L and the plasmid concentration was 0.5 &mgr;g/&mgr;L, which was injected intratumorally at several points. At 30 s after injection, an electrical impulse was applied twice at intervals of > 1 min. Electrodes were placed at both ends of the tumor long and short axes; the electric field strength was 200 V/cm, electric pulse duration was 50 ms, and pulse frequency was 1 Hz. This process was repeated on days 10 and 17. Therefore, tumor volume and mouse body weight were measured every 4 d. On day 20, the tumors were removed and their weight and volume were measured, and then each tumor was divided into two parts, one was used for immunohistochemical analysis, and the other was used for screening mRNA expression of STAT3 and related genes and protein expression.
Tumor RNA was extracted using Trizol reagent (Gibco Life Technologies, Inc., Langley, OK, USA) following its manufacturer’s instructions. The concentration of the total RNA was detected by UV spectrophotometry. RT-PCR was performed by the two-step method. Synthesis of cDNA was performed using the cDNA Synthesis Kit (Genomy, Beijing, China). The conditions were: 94°C for 5 min, 94°C for 30 s, 56°C for 30 s, 72°C for 45 s, and 72°C for 7 min, for 30 cycles; the total volume was 25 &mgr;L. For quantitative analysis of c-myc, p53, survivin, VEGF and caspase3 mRNA, the expression of the housekeeping gene β-actin was used as an internal standard. The primer sequences were: STAT3: sense 5'-TTGCCAGTTGTGGTGATC-3', antisense 5'-AGACCCAGAAGGAGCCGC-3'; β-actin: sense 5'-AAGTACTCCGTGTGGATCGG-3', antisense 5'-ATGCTATCACCTCCCCTGTG-3'. The primers were synthesized by the Shanghai Sangon Biological Engineering Technology & Services Co. Ltd. The PCR fragments were separated and visualized in 20 g/L agarose gels stained with ethidium bromide. Semi-quantitative analysis was performed with the Gis gel analysis software (Shanghai, China). All experiments were done in triplicate. The ratio of the photo-density of the RT-PCR product of the target gene and β-actin was used to identify the expression intensity of the target gene.
The tumor tissues were ground and sonicated with supersonic lytic buffer that contained 50 mmol/L NaH2PO4, 100 mmol/L Tris-HCl, 250 mmol/L NaCl, 100 mg/L PMSF, 1 mg/L Aprotinin, pH 8.0, and then centrifuged at 12 000 ×g for 40 min. The Bio-Rad standard curve was used to determine the concentration of protein in each lysate. Loading buffer was added to each lysate, which was subsequently boiled for 5 min and then electrophoresed by SDS-PAGE. The proteins were mixed with 2 × loading buffer with the same volume before electrophoresis. After transferring onto nitrocellulose, proteins were incubated with antibodies (anti-p-STAT3 and β-actin, purchased from Santa Cruz Biotechnology, Santa Cruz, CA, USA) and then with peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology). Detection was performed with an enhanced chemiluminescence agent. The analysis was performed with the Bandscan analysis software (Sterling, VA, USA). All experiments were done in triplicate. The ratio of the proteins of STAT3 and β-actin was used to identify the expression intensity.
The tumor tissues were embedded in paraffin and then sliced. The slices were stained with hematoxylin and eosin (HE) and the expression of p-STAT3 protein (Santa Cruz Biotechnology) was determined using the streptavidin biotin complex immunohistochemical staining kit (Boster, Wuhan, China). A positive outcome was brown staining in the cytoplasm or nucleus, and the ratio of positive to total cells was > 15%.
The tumor tissues were fixed with 10% formalin for 4 h and then embedded in paraffin. The slices were deparaffinized in water and placed in 3% H2O2 for 10 min at room temperature. The TUNEL assay was carried out according to the manufacturer’s instructions (KGI Biotechnology Company, Nanjing, China). A positive result was brown staining in the nucleus.
Statistical analysis was performed using SPSS 12.0 software (Chicago, IL, USA). The results were expressed as mean ± SD. The data were treated by Student’s t test to determine statistical significance. P < 0.05 was considered statistically significant.
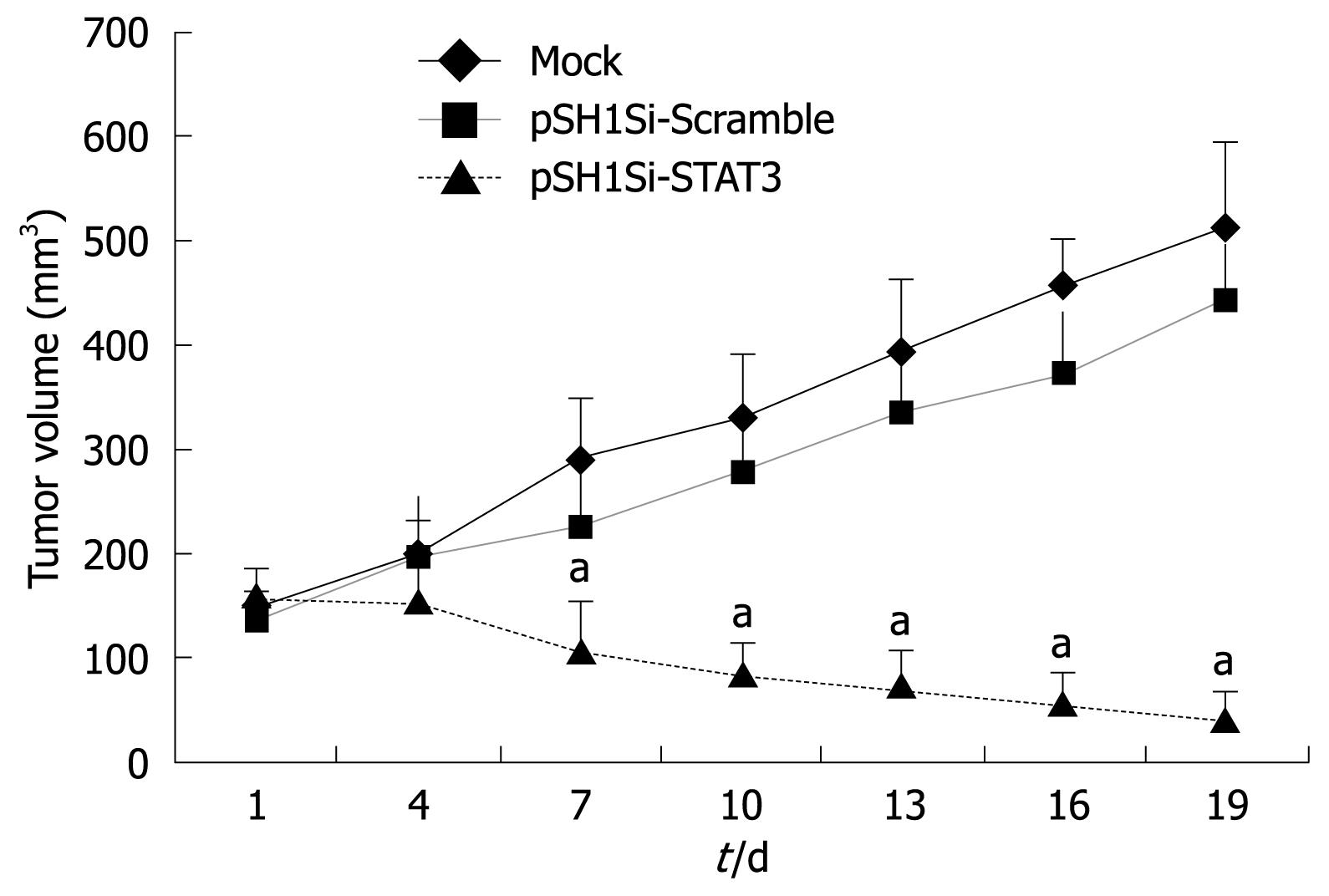
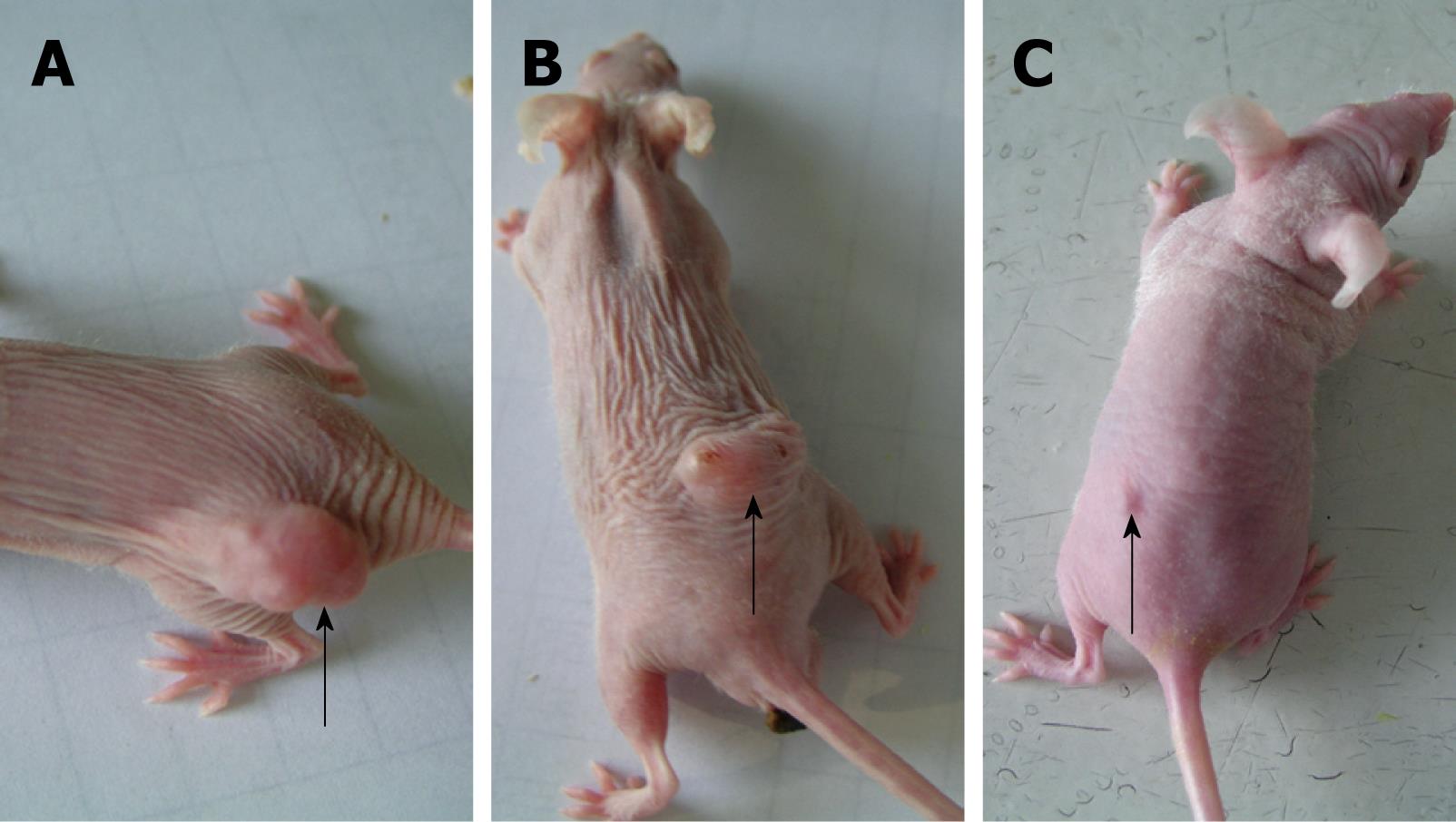
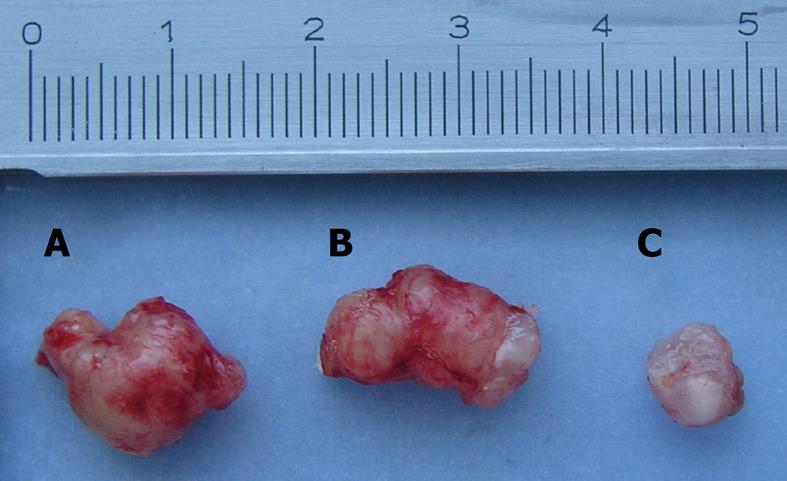
The body weight of tumor-bearing nude mice decreased gradually and tumor volume increased in the mock-treated and pSH1Si-Scramble groups (Figure 1). However, the tumor volume in the pSH1Si-STAT3 group was markedly diminished from day 7, and body weight clearly increased from day 10 (Figure 2). Compared with the mock-treated and pSH1Si-Scramble groups, there were marked differences in the pSH1Si-STAT3 group (P < 0.01). Although the changes in the pSH1Si-Scramble group were less than those in the mock-treated group, there was no statistical significance (P > 0.05). During excision of subcutaneous tumor, we found that tumor volumes were larger and blood supplies were richer in the mock-treated and pSH1Si-scramble groups than those in the pSH1Si-STAT3 group (Figure 3), and the tumor weight of the three groups was 0.67 ± 0.07, 0.6 ± 0.07 and 0.18 ± 0.09, respectively. There were great differences between the pSH1Si-STAT3 and the mock-treated and pSH1Si-Scramble groups (P < 0.01), but differences between the mock-treated and pSH1Si-Scramble groups were not significant (P > 0.05).
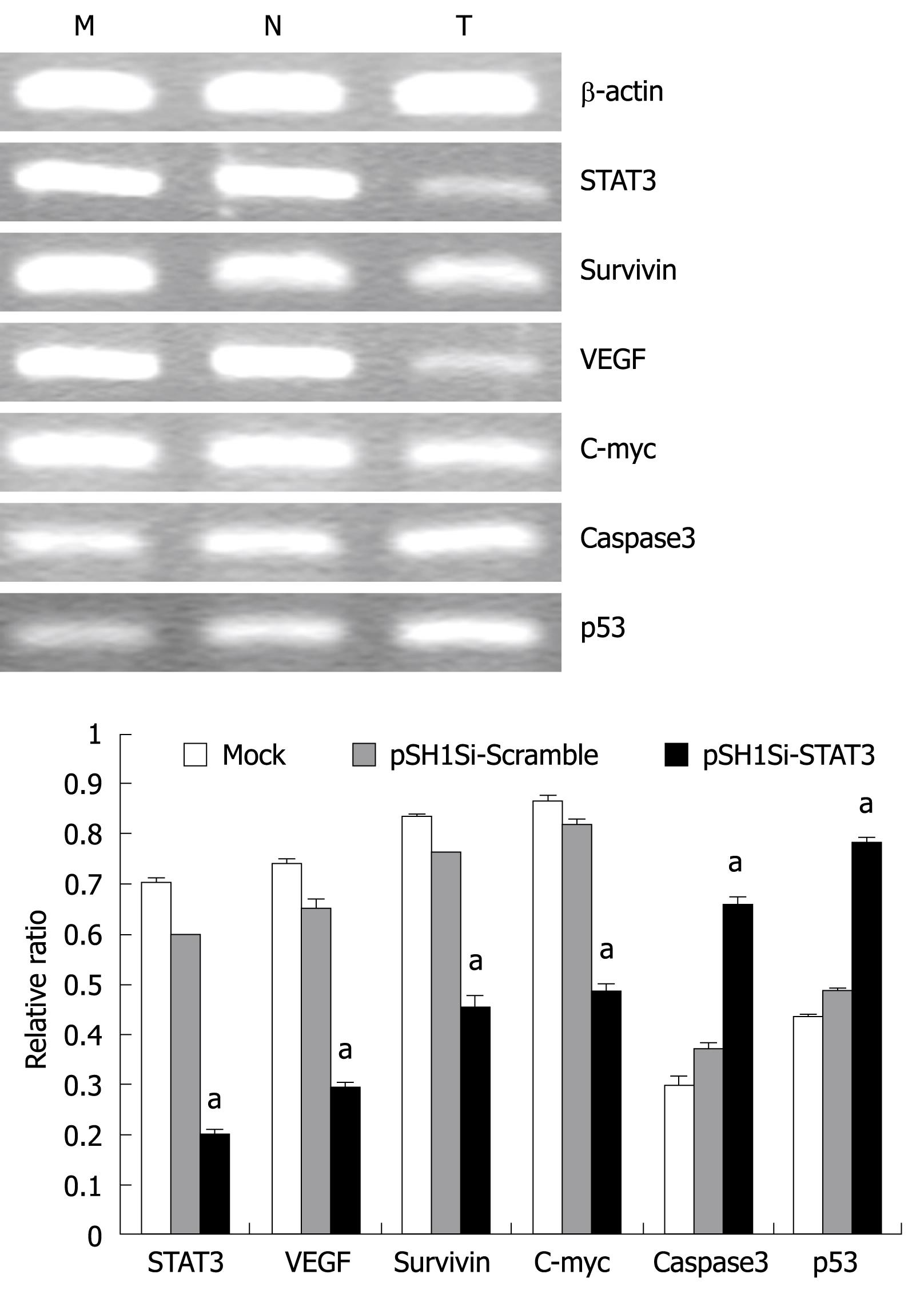
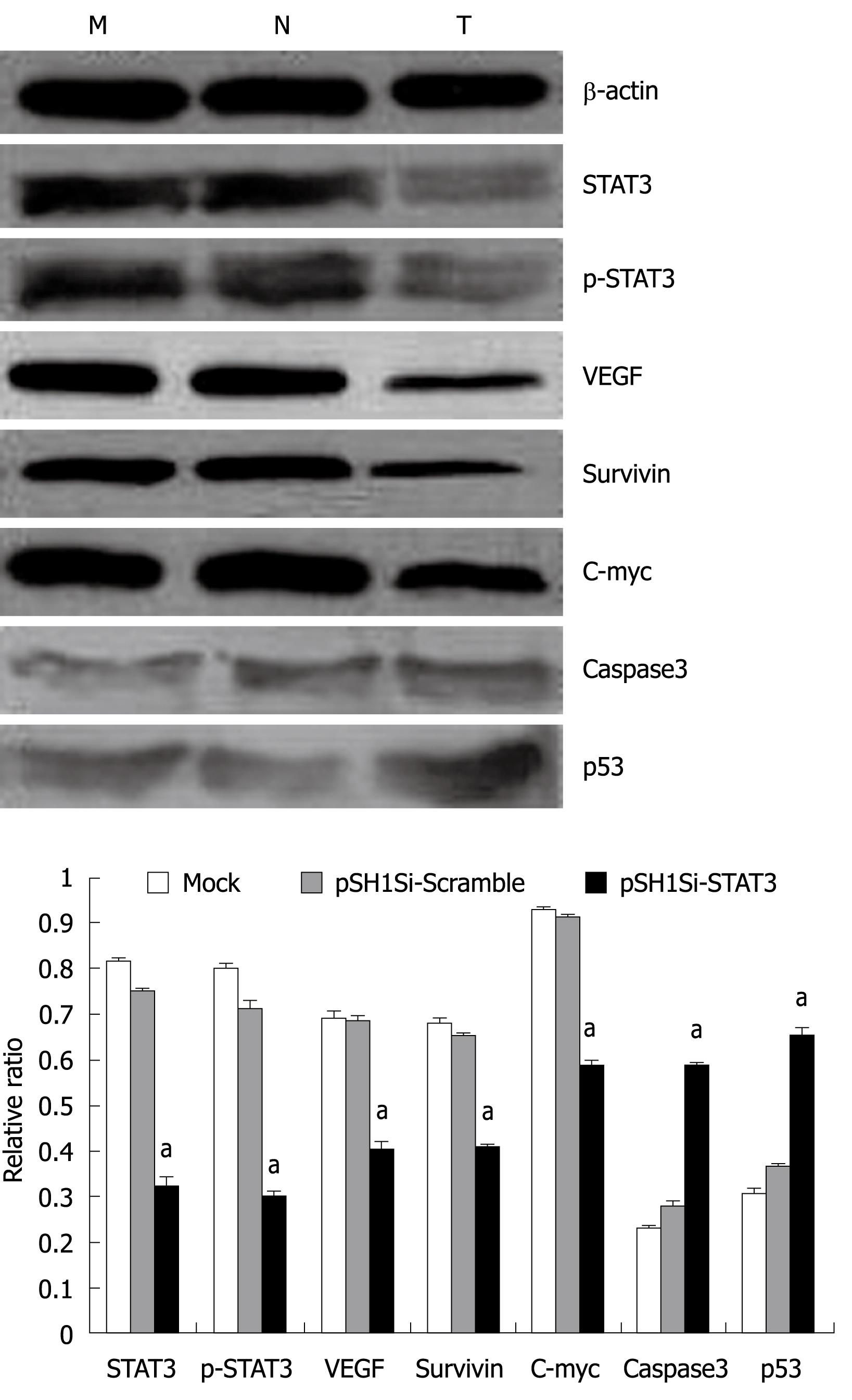
The mRNA expression of the target genes was analyzed by RT-PCR. The mRNA levels of STAT3, VEGF, survivin and c-myc genes declined obviously, but mRNA for the caspase3 and p53 genes in the pSH1Si-STAT3 group increased. The ratio of the photo-density of the RT-PCR product of the target genes and β-actin varied among the three groups, the differences between the pSH1Si-STAT3 and the mock-treated and pSH1Si-Scramble groups were significant (P < 0.01, Figure 4), but the difference between the mock-treated and pSH1Si-Scramble groups was not significant (P > 0.05).
The protein expression of the target genes was analyzed by Western blotting. The protein levels of STAT3, p-STAT3, VEGF, survivin and c-myc genes were down-regulated clearly, but those of active caspase3 and p53 genes were up-regulated in the pSH1Si-STAT3 group. The ratio of the proteins of the target genes and β-actin varied among the three groups. The differences in the pSH1Si-STAT3 group were significantly greater than those in the mock-treated and pSH1Si-Scramble groups (P < 0.01, Figure 5), however, there was no significant difference between the mock-treated and pSH1Si-Scramble groups (P > 0.05).
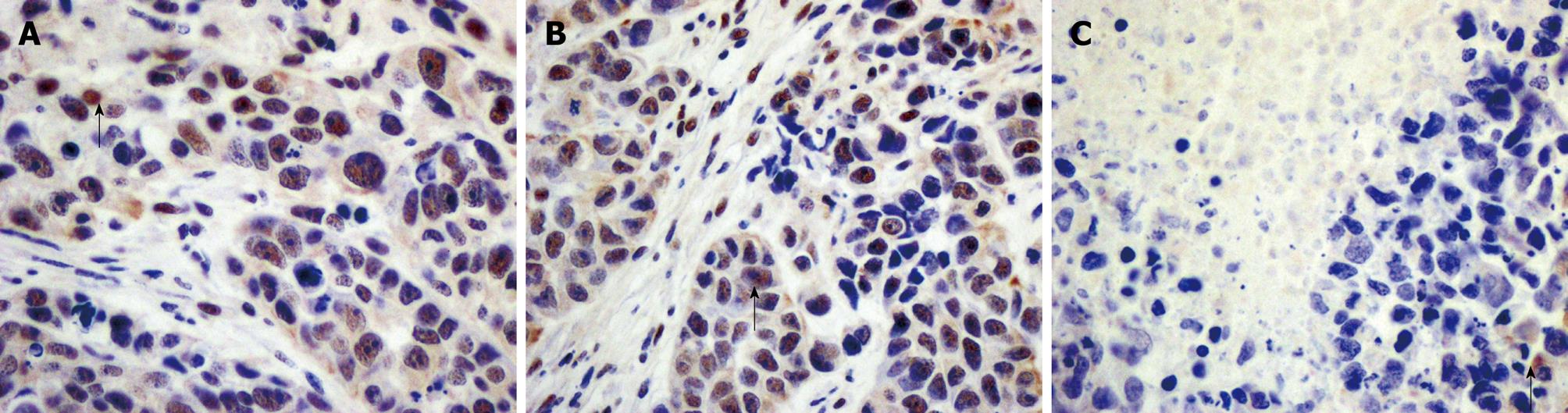
Immunohistochemistry showed that p-STAT3 was stained brown in the nuclei of many cells in the mock-treated and pSH1Si-Scramble groups, but were stained brown in the cytoplasm of only a few cells on the edge of tumors in the pSH1Si-STAT3 group (Figure 6).
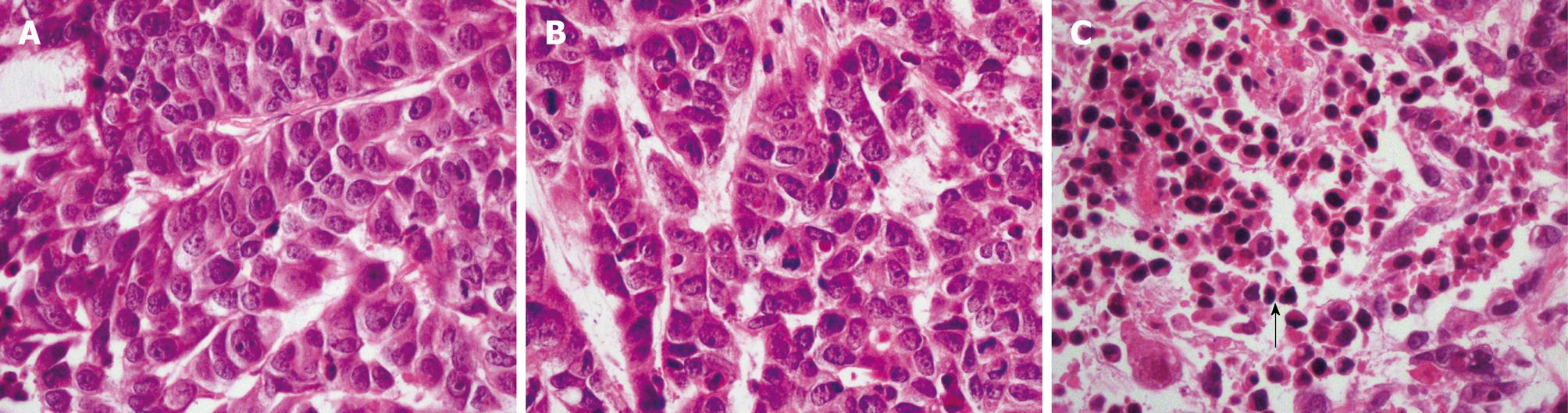
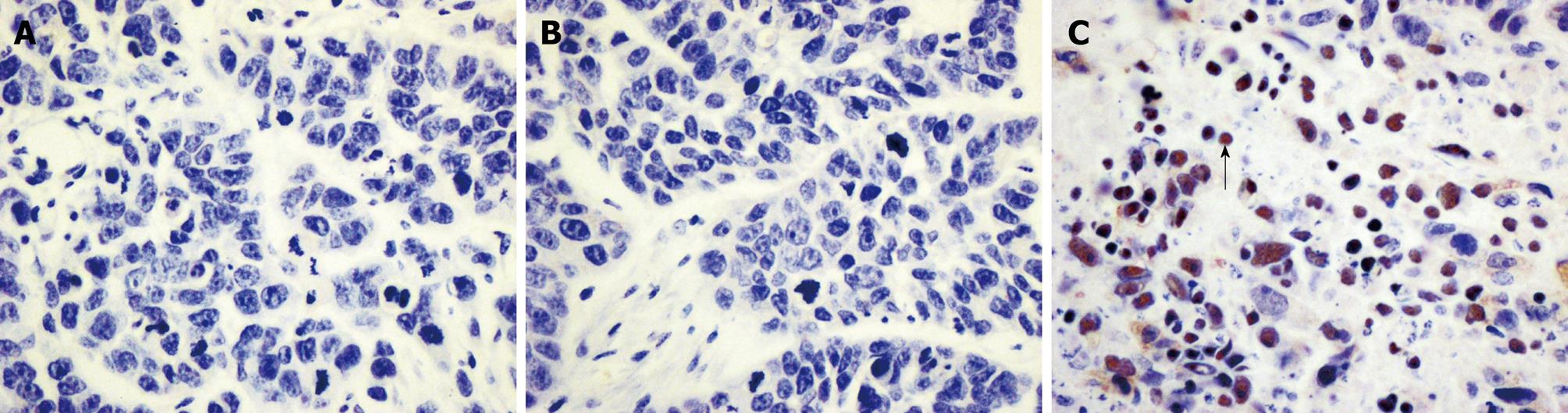
The outcome of HE staining was observed under light microscopy (Figure 7). Many apoptotic cells were observed in the pSH1Si-STAT3 group, which showed karyopyknosis and red staining of the cytoplasm. There were also apoptotic cells in the pSH1Si-STAT3 group, in which the nuclei of many cells were stained brown (Figure 8).
There are many different genes and various signal transduction pathways involved in initiation and development of human HCC. The STAT3 and JAK/STAT3 signal transduction pathways are of considerable interest in tumors[78], and many studies[910] have detected the activation of STAT3 in many hepatocellular cell lines and HCC, and found that abnormal activation of STAT3 is related to the initiation and development of human liver cancer. It may be an effective strategy for HCC therapy to inhibit the abnormal expression of STAT3. At present, there are many methods[11–13] of targeting the STAT3 gene, which include inhibiting indirectly activation of STAT3 by receptor antagonist, neutralizing antibody, protein tyrosine kinase inhibitor and other agents, or blocking directly the functions of STAT3 by antisense oligonucleotides, decoy oligonucleotides, and RNAi. RNAi is a potent tool in tumor gene therapy, because of its high performance, specificity, convenient manipulation and low toxicity.
Our previous studies have demonstrated that the recombination plasmid pSH1Si-STAT3, which targets the STAT3 gene, dramatically inhibits the expression of STAT3 mRNA and protein in the HCC cell line SMMC7721 in vitro and proliferation of HCC cells, and induces apoptosis of HCC cells[6]. Therefore, we designed this experiment in order to verify the therapeutic effect of recombination plasmid pSH1-siRNA-STAT3 in HCC in vivo. The present study indicated that body weight of mice was increased, and tumor volume was decreased. Tumor weight also declined in the pSH1Si-STAT3 group, and there were marked differences compared with that in the mock-treated and pSH1Si-Scramble groups. RT-PCR, Western blotting and immunohistochemistry showed that expression of mRNA and protein was markedly down-regulated, and there were many apoptotic cells, as shown by HE staining and TUNEL screening. These results verified that the recombination plasmid pSH1Si-STAT3 has a significant inhibitory effect on tumors in HCC-bearing nude mice.
At the same time, we detected the mRNA and protein levels of STAT3-related genes, and found that the expression levels of VEGF, survivin and c-myc were down-regulated clearly, but expression of caspase3 and p53 in pSH1Si-STAT3 was up-regulated. Compared with the mock-treated and pSH1Si-Scramble groups, the differences in expression were significant. Hence, we can infer that inhibiting the expression of STAT3 by siRNA in HCC may inhibit the activation of its target genes, recover the activity of some important anti-oncogenes, restrict the proliferation of tumor cells, and increase apoptosis. Therefore, it plays an important antineoplastic role.
The fate of c-myc attracted our attention. c-myc was not down-regulated by inhibition of the STAT3 gene in vitro[6], but it was down-regulated in vivo. This may be related to the changed regulation mechanism of HCC cells after they have been removed from the body. It may also indicate that c-myc is not regulated directly by STAT3, but it is regulated by other genes that are targeted by STAT3. Hence, this mechanism of regulation requires further research.
Compared with Lipofectamine 2000 that we have used previously in vitro[6], which is characterized by high transfection efficiency and low toxicity, but high cost, the electrotransfection process used here in vivo, has the advantage of convenient operation, low cost, high efficiency and no toxicity. The results of the experiments demonstrated that the two methods of transfection obtained fine potency. Our study indicated that recombination plasmid pSH1-siRNA-STAT3 had significant anti-neoplastic activity in vitro and in vivo. A recombination plasmid that carries the same sequence of siRNA-STAT3 has a therapeutic effect on prostate tumor and melanoma[1415], which leads us to conclude that one siRNA oligonucleotide drug may treat a variety of tumors. At the same time, one tumor can also be treated by several siRNA oligonucleotide drugs, and an optimal therapeutic effect may be obtained if multiple oncogenes are inhibited.
In our study, there were some changes after transfection in the pSH1Si-Scramble group, for example, changes in cell morphology and modification of gene expression. Although there was no significant difference compared with the mock-treated group, it suggested that the expression vectors that carried the scrambled sequence had some unknown effects on the cells, the mechanism of which needs further research.
We used intratumoral injection together with electrotransfection in our in vivo experiment, and obtained favorable results. Compared with intra-abdominal and intravenous injection, intratumoral injection is targeted and localized, allows direct injection of drugs into the feeding artery of the tumor, reduces the drug dose, and lengthens the dosage interval. However, this is an invasive procedure that requires skillful operators and special equipment, therefore, its application is limited. There are also potential dangers in the electrotransfection method, such as tissue injury or fibrosis where the electrode makes contact[16]. As a result, the key issues in the clinical application of RNAi are to develop vectors of high transfection efficiency and low toxicity, and to establish effective routes of administration.
In conclusion, the recombination plasmid pSH1-siRNA-STAT3 inhibited the growth of HCC cells in vivo, induced apoptosis, and had significant antineoplastic efficacy. RNAi that targets STAT3 has a clear therapeutic effect in HCC. Therefore, STAT3 may become an important target of biological therapy in HCC, which brings hope of clinical therapy using RNAi oligonucleotide drugs.
Hepatocarcinogenesis involves the mutation of multiple genes and alteration of the relevant signaling pathways. In recent years, signal transducer and activator of transcription 3 (STAT3) and related signaling pathways in liver carcinogenesis have attracted increasing attention. It has been shown that constitutively activated STAT3 is detected in many hepatocellular carcinoma (HCC) cell lines and tissues. This suggests that STAT3 is a promising molecular target for HCC gene therapy. Inhibition of abnormal expression of STAT3 may be an effective strategy for biological therapy of HCC.
Some antineoplastic drugs, such as STAT3 inhibitors, tyrosine kinase inhibitor AG-490, micromolecular peptide and antisense oligodeoxynucleotides, have been used to target STAT3 in vitro and in vivo for HCC, however their efficacy is not satisfactory because of their poor specificity, rapid degradation, unsuitable molecular size, immunogenicity, poor ability to cross cell membranes, and need of special drug delivery mechanisms. RNA interference (RNAi) is a regulatory mechanism in most eukaryotic cells, which uses small double-stranded RNA (dsRNA) molecules to direct homology-dependent control of gene activity. As a result of the potency and specificity of RNAi, it has become an important tool to control gene expression through the introduction of siRNA.
Studies of targeting in vitro and in vivo over-expressed genes in HCC by RNAi, including c-myc, survivin, CCR1 and SMYD3, have been reported. However, there has been still no report about targeting STAT3 by RNAi in HCC. In the present study, the authors used a DNA-vector-based RNAi approach to block STAT3 expression in HCC cells and tumor-bearing nude mice, to determine the role of constitutively activated STAT3 during HCC pathogenesis, and to explore the role and molecular mechanism of targeting STAT3 in HCC therapy.
By investigating the effect of silencing STAT3 expression by RNAi on the growth suppression and apoptosis of HCC cells, and the growth inhibition of HCC in tumor-bearing nude mice, this study may provides a new strategy for biological therapy of HCC by targeting STAT3. It may also offer a theoretical and experimental foundation for the clinical application of synthetic dsRNA-based RNAi.
The manuscript by Li et al investigates the effects of silencing STAT3 on the growth of HCC in vivo. The authors present interesting and convincing data that silencing of STAT3 suppresses tumor growth in vivo. This is a good paper with very interesting data.
| 1. | Gartel AL, Kandel ES. RNA interference in cancer. Biomol Eng. 2006;23:17-34. |
| 2. | Wu X, Fan J, Wang X, Zhou J, Qiu S, Yu Y, Liu Y, Tang Z. Downregulation of CCR1 inhibits human hepatocellular carcinoma cell invasion. Biochem Biophys Res Commun. 2007;355:866-871. |
| 3. | Salvi A, Arici B, Portolani N, Giulini SM, De Petro G, Barlati S. In vitro c-met inhibition by antisense RNA and plasmid-based RNAi down-modulates migration and invasion of hepatocellular carcinoma cells. Int J Oncol. 2007;31:451-460. |
| 4. | Chen LB, Xu JY, Yang Z, Wang GB. Silencing SMYD3 in hepatoma demethylates RIZI promoter induces apoptosis and inhibits cell proliferation and migration. World J Gastroenterol. 2007;13:5718-5724. |
| 5. | Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE Jr. Stat3 as an oncogene. Cell. 1999;98:295-303. |
| 6. | Li J, Piao YF, Jiang Z, Ding BJ. Silencing of STAT3 expression by siRNA suppresses the growth of human hepatocellular carcinoma cells and regulates genes related to growth control. Shijie Huaren Xiaohua Zazhi. 2007;15:2101-2107. |
| 7. | Kijima T, Niwa H, Steinman RA, Drenning SD, Gooding WE, Wentzel AL, Xi S, Grandis JR. STAT3 activation abrogates growth factor dependence and contributes to head and neck squamous cell carcinoma tumor growth in vivo. Cell Growth Differ. 2002;13:355-362. |
| 8. | Isomoto H, Kobayashi S, Werneburg NW, Bronk SF, Guicciardi ME, Frank DA, Gores GJ. Interleukin 6 upregulates myeloid cell leukemia-1 expression through a STAT3 pathway in cholangiocarcinoma cells. Hepatology. 2005;42:1329-1338. |
| 9. | Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48-54. |
| 10. | Fattovich G, Stroffolini T, Zagni I, Donato F. Hepatocellular carcinoma in cirrhosis: incidence and risk factors. Gastroenterology. 2004;127:S35-S50. |
| 11. | Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63:1270-1279. |
| 12. | Jing N, Li Y, Xiong W, Sha W, Jing L, Tweardy DJ. G-quartet oligonucleotides: a new class of signal transducer and activator of transcription 3 inhibitors that suppresses growth of prostate and breast tumors through induction of apoptosis. Cancer Res. 2004;64:6603-6609. |
| 13. | Turkson J, Kim JS, Zhang S, Yuan J, Huang M, Glenn M, Haura E, Sebti S, Hamilton AD, Jove R. Novel peptidomimetic inhibitors of signal transducer and activator of transcription 3 dimerization and biological activity. Mol Cancer Ther. 2004;3:261-269. |
| 14. | Gao L, Zhang L, Hu J, Li F, Shao Y, Zhao D, Kalvakolanu DV, Kopecko DJ, Zhao X, Xu DQ. Down-regulation of signal transducer and activator of transcription 3 expression using vector-based small interfering RNAs suppresses growth of human prostate tumor in vivo. Clin Cancer Res. 2005;11:6333-6341. |
| 15. | Zhang L, Gao L, Zhao L, Guo B, Ji K, Tian Y, Wang J, Yu H, Hu J, Kalvakolanu DV. Intratumoral delivery and suppression of prostate tumor growth by attenuated Salmonella enterica serovar typhimurium carrying plasmid-based small interfering RNAs. Cancer Res. 2007;67:5859-5864. |