Published online Feb 28, 2008. doi: 10.3748/wjg.14.1175
Revised: December 5, 2007
Published online: February 28, 2008
AIM: To elucidate the localization of RhoA in gastric SGC-7901 cancer cells and its translocation by lysophosphatidic acid (LPA) and/or 8-chlorophenylthio-cAMP (CPT-cAMP).
METHODS: Immunofluorescence microscopy was used to determine the localization of RhoA. Western blotting was used to detect both endogenous and exogenous RhoA in different cellular compartments (membrane, cytosol, nucleus) and the translocation of RhoA following treatment with LPA, CPT-cAMP, or CPT-cAMP + LPA.
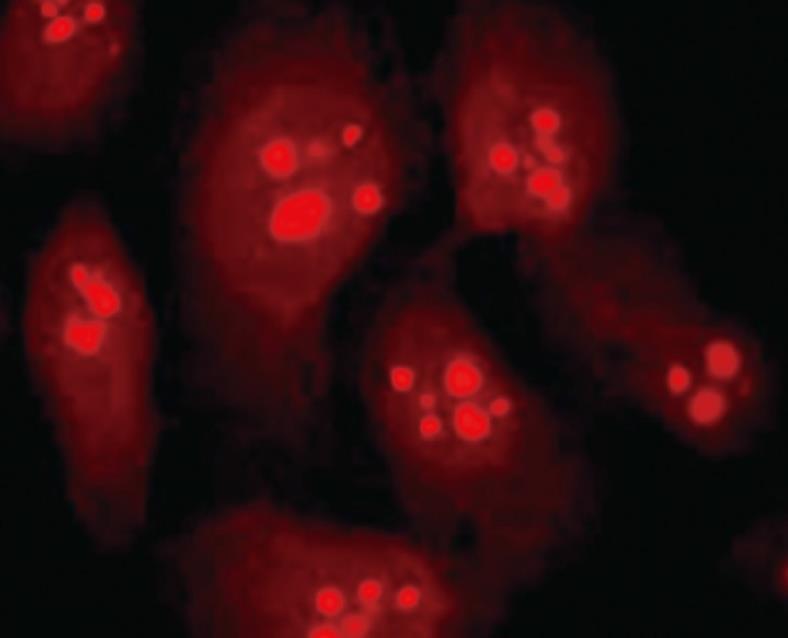
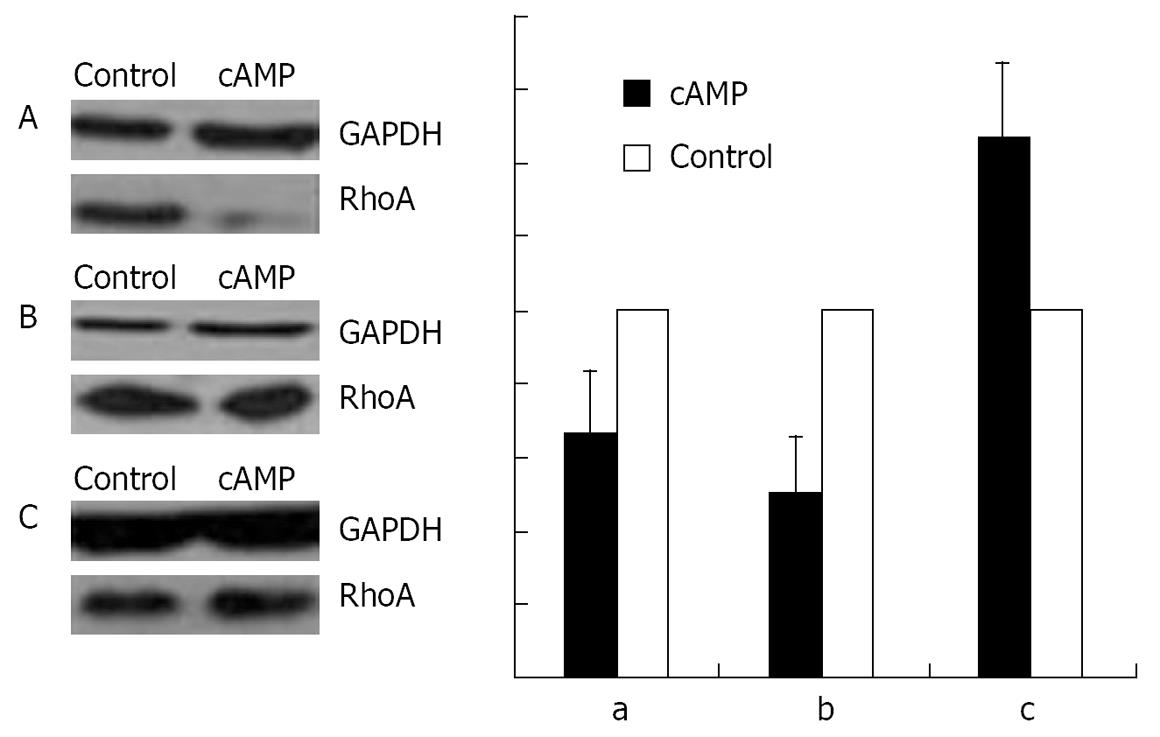
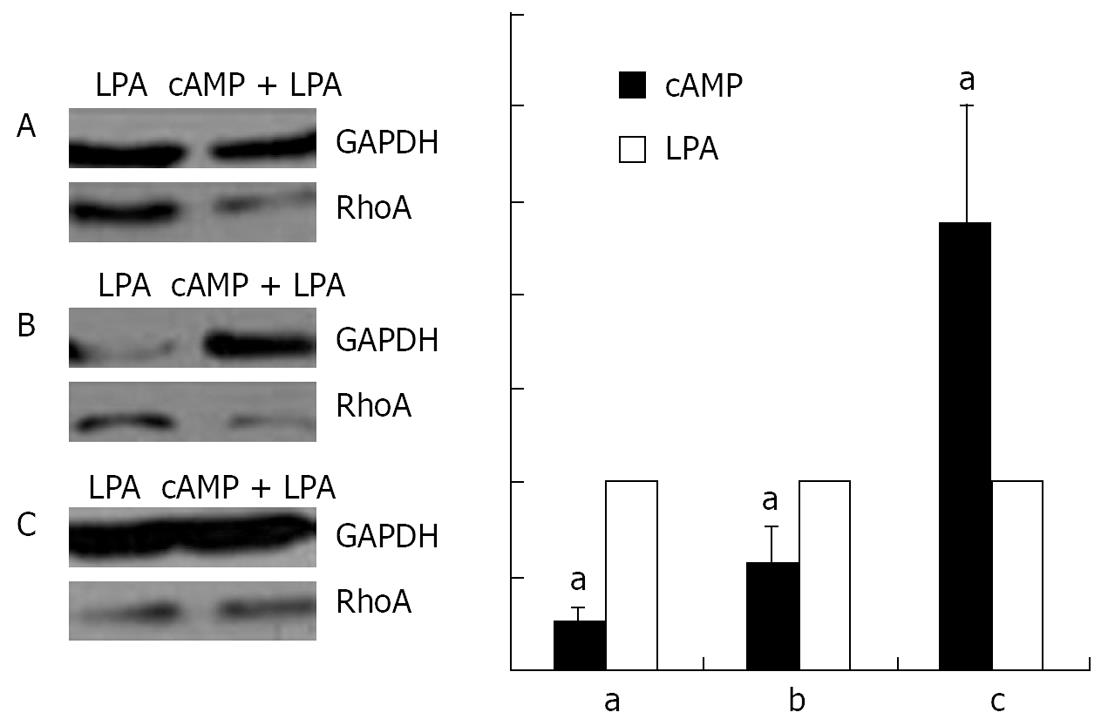
RESULTS: Immunofluorescence staining revealed endogenous RhoA to be localized in the membrane, the cytosol, and the nucleus, and its precise localization within the nucleus to be the nucleolus. Western blotting identified both endogenous and exogenous RhoA within different cellular compartments (membrane, cytosol, nucleus, nucleolus). After stimulation with LPA, the amount of RhoA within membrane and nuclear extracts increased, while it decreased in the cytosol fractions. After treatment with CPT-cAMP the amount of RhoA within the membrane and the nuclear extracts decreased, while it increased within the cytosol fraction. Treatment with a combination of both substances led to a decrease in RhoA in the membrane and the nucleus but to an increase in the cytosol.
CONCLUSION: In SGC-7901 cells RhoA was found to be localized within the membrane, the cytosol, and the nucleus. Within the nucleus its precise localization could be demonstrated to be the nucleolus. Stimulation with LPA caused a translocation of RhoA from the cytosol towards the membrane and the nucleus; treatment with CPT-cAMP caused the opposite effect. Furthermore, pre-treatment with CPT-cAMP was found to block the effect of LPA.
- Citation: Tao Y, Chen YC, Li YY, Yang SQ, Xu WR. Localization and translocation of RhoA protein in the human gastric cancer cell line SGC-7901. World J Gastroenterol 2008; 14(8): 1175-1181
- URL: https://www.wjgnet.com/1007-9327/full/v14/i8/1175.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.1175
RhoA, with molecular mass of 21 kDa, is the most extensively studied member of the Rho GTPase family belonging to the Ras superfamily of small G proteins. The Rho GTPase family consists of at least 11 members sharing more than 50% identity, the best known of them being Cdc42, Rac1, and RhoA. RhoA has been reported to regulate many biological activities including the formation of stress fibers and focal adhesions[1].
Like other small G proteins, RhoA is inactive in its GDP-bound and active in its GTP-bound form, these two forms can be converted by GDP/GTP exchange or GTPase reactions[2–5]. The former and latter reactions are regulated by GDP/GTP exchange protein and GTPase activating protein, respectively. Two types of GDP/GTP exchange proteins are known: one is a stimulatory type named GDS, and the other is an inhibitory type named GDI. GDS stimulates the dissociation of GDP from and the subsequent binding of GTP to each substrate small G protein, whereas GDI inhibits both reactions[6–15]. In addition to the action of GDS and GDI regulating the GDP/GTP exchange reaction, both GDP/GTP exchange proteins regulate the translocation of each substrate small G protein between the cytosol and membranes.
Lysophosphatidic acid (LPA), a commonly used RhoA activator, is a serum-born phospholipid with hormone and growth factor-like properties, which is formed by and released from activated platelets and which can be generated by the action of secretory phospholipase A2[1617]. Previous studies showed that it is able to induce the translocation of RhoA from the cytosol to the membrane accompanied with the activation of RhoA[18], probably via conversion of the GDP-bound into the GTP-bound form.
cAMP is a well-known classical secondary messenger, and a cAMP-dependent protein kinase A (PKA) phosphorylates RhoA in its C-terminal region on serine residue 188[19]. The cAMP signal mediates down-regulation of RhoA activity[20]. It was reported that cAMP could induce the translocation of RhoA from the membrane to the cytosol accompanied with the inactivation of RhoA[19]. Moreover, translocation of RhoA was shown to be caused by stabilizing the RhoA/Rho GDIs complex by phosphorylating GTP-bound RhoA[21]. Phosphorylation enhances translocation of RhoA to the cytosol through increased interaction of GTP-bound RhoA with Rho-GDI, possibly terminating its activity.
We here describe RhoA to be localized in the nucleus of SGC-7901 cells and a cross talk between LPA/RhoA-mediated and cAMP/PKA mediated signal transduction pathways[22]. Whether there is interaction between LPA induced and cAMP induced RhoA distributions in different cellular compartments deserves further investigation. Our experiments were designed to explore the localization of RhoA and to investigate the effects of a treatment with LPA, cAMP, and cAMP + LPA.
The human gastric cancer cell line SGC-7901 and human cervix cancer cell line HeLa were provided by the Institute of Cell Biology (Shanghai, China).
Dulbecco's Modified Eagle Media (DMEM) was purchased from GIBCO (Grand Island, NY), new-born calf serum (NBCS) from Minhai Bio-engineering C (Lanzhou, China), the antibody against RhoA from Santa Cruz Biotechnology (Santa Cruz, CA), the antibody against GAPDH (glyceraldehyde phosphate dehydrogenase) from Kangcheng (China), the horseradish peroxidase (HRP)-conjugated secondary antibody from Jackson ImmunoResearch Laboratories (West Grove, PA), the cell transfection reagent Lipofectamin 2000, TRIzol reagent, and reverse transcription kit from Invitrogen (Carlsbad, CA), the cellular permeable cAMP analog 8-chlorophenylthio-cAMP (CPT-cAMP) from Calbiochem, LPA and nuclear fluorochrome Hoechst 33342 from Sigma (St. Louis, Missouri), electrochemiluminescence (ECL) reagents from Amersham Biosciences (Buckinghamshire, England), and pGEmol/L vector from Promega (USA). Plasmid DNA constructs of RhoA pcDNAEE-wt (wild type of RhoA), pcDNAEE-63L (a constitutive active form of RhoA), and pcDNAEE-N19 (an inactive mutant of RhoA) were kind gifts from Dr. Renate Pilz, University of California, San Diego.
SGC-7901 cells were cultured in DMEM supplied with 10% NBCS. The medium was changed every two days and the cells were subcultured at confluence. For transfection, the cells were subcultured the day before the process. The seeding amount of cells was adjusted to attain a density of 80%-90% confluence on the day of transfection. Transfection was performed according to the instruction of the manufacturer.
The cells grown on cover slips were incubated to reveal nuclei with 0.2 &mgr;mol/L Hoechst 33 342 for 10 min before fixed with freshly prepared 40 g/L paraformaldehyde in PBS at 4°C overnight (o/n). After being penetrated with 30 mL/L Triton X-100 and blocked with 30 g/L BSA, the cells were incubated with primary antibodies at 4°C o/n, and then with FITC- or TRITC-conjugated second antibodies for 1 h at room temperature (RT), with three washes after each incubation. The morphologic changes of the cells were analyzed by fluorescence microscopy.
According to the method from Jeffrey C. Chen et al[23], four 100 mm plates of cells were prepared, one of them considered as control, and the others treated with LPA (1 &mgr;mol/L, 15 min), CPT-cAMP (250 &mgr;mol/L, 30 min), or CPT-cAMP (250 &mgr;mol/L, 30 min) followed by an incubation with LPA (1 &mgr;mol/L, 15 min). Cells were extracted by Dounce homogenization in 10 mmol/L Hepes pH 7.5, 2 mmol/L EDTA, 1 mmol/L MgCl2 (HEM buffer). The homogenate was centrifuged at 500 g at 4°C for 5 min to obtain nuclear proteins, and the supernatant was centrifuged at 37 000 g at 4°C for 30 min. The supernatant and pellet from the second centrifugation are referred as cytosol and membranes, respectively, with the membrane preparation washed twice in HEM buffer to remove contaminating cytosol. Protein concentrations were determined and equal amounts of protein from each preparation (30 &mgr;g) were subjected to SDS-PAGE/Western blotting using mouse monoclonal anti-RhoA (1:200) and GAPDH (1:5000) antibodies.
Nucleoli were purified using a procedure adapted from previously published protocols[24]. For a typical experiment, 1 × 108 SGC-7901 cells were plated onto 245 mm Petri dishes in DMEM containing 10% NBCS. Cells were incubated in a humidified atmosphere containing 5% CO2 at 37°C. At 80% confluence, cells were washed with cold PBS, and scraped off on ice. Cells were collected by centrifugation at 500 g at 4°C for 5 min. Cells were resuspended in 5 mL hypotonic buffer (10 mmol/L Tris-HCl pH 7.4, 10 mmol/L NaCl, and 1 mmol/L MgCl2) and incubated on ice for 30 min. Cell lysis was performed by addition of a final concentration of 0.3% Nonidet P-40 (Roche Applied Science, Mannheim, Germany) and homogenization was performed using a 0.4 mm clearance Dounce homogenizer (Kimblr/Kontes, Owens, IL) for 15 times on ice. Nuclei were collected by centrifugation at 228 g at 4°C for 5 min and resuspended in 3 mL 0.25 mol/L sucrose-containing 10 mmol/L MgCl2. Nuclei were centrifuged at 1430 g at 4°C for 5 min through a 0.35 mol/L sucrose cushion containing 0.5 mmol/L MgCl2 and resuspended in 3 mL 0.35 mol/L sucrose-containing 0.5 mmol/L MgCl2 and then sonicated on ice for ten bursts of 10 s with 1 min intervals between them. Nuclei were then purified by centrifugation at 2800 g at 4°C for 10 min through a 0.88 mol/L sucrose cushion containing 0.5 mmol/L MgCl2. Nuclei were washed with 500 &mgr;L 0.35 mol/L sucrose containing 0.05 mmol/L MgCl2 twice, and centrifuged at 2800 g at 4°C for 10 min. Purified nucleoli were resuspended in 50 &mgr;L 0.35 mol/L sucrose containing 0.05 mmol/L MgCl2 for further analyses.
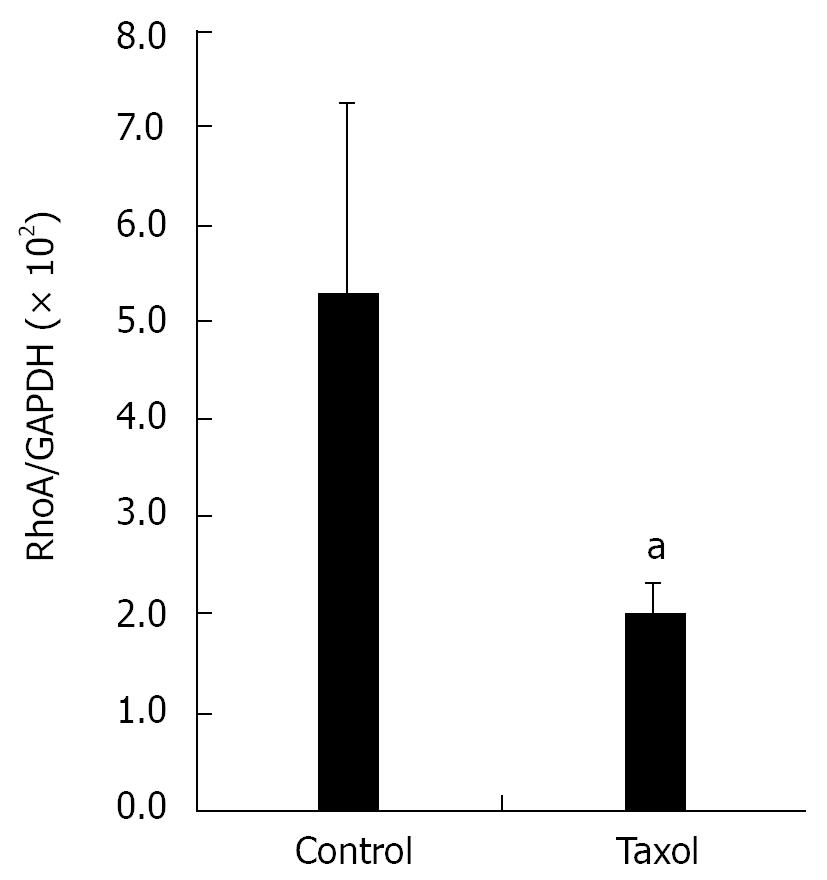
Hela cells were seeded on four 100 mm plates and treated with 5 &mgr;g/mL Taxol for 12 h. Cells in mitosis phase were collected by “shake off”, and the cells left on plates considered in interphase were scraped. Cells were extracted with 500 &mgr;L RIPA (50 mmol/L Tris-HCl pH 7.4, 150 mmol/L NaCl, 1 mmol/L PMSF, 1 mmol/L EDTA, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS), incubated on ice for 30 min, and then centrifuged at 14 000 g at 4°C for 15 min to obtain total protein extracts. Protein concentrations were determined and equal amounts of protein from each preparation (30 &mgr;g) were subjected to SDS-PAGE/Western blotting using mouse monoclonal anti-RhoA (1:200) and GAPDH (1:5000) antibodies.
SDS-PAGE gels of different concentrations were cast according to the molecular size of target proteins. Sample proteins were run on 10% SDS-polyacrylamide gels, and were blotted onto polyvinyl difluoride (PVDF) membrane. The PVDF membrane was blocked with 3% (w/v) bovine serum albumin (BSA) in TBS-T for 1 h at room temperature (RT). Membranes were incubated with the primary antibody at 4°C o/n, and with the secondary antibody for 1 h at RT, with three washes after each incubation. Electrochemiluminescence reagents were used to show the positive bands on the membrane. Briefly, same volumes of solution A and solution B were mixed and added to the protein side of the membrane. The incubation was 5 min at RT. The exposure time of the first film was 15 s. The exposure time of the second film was adjusted according to the intensity of the signal on the first film. The bands on film were analyzed with GeneSnap/Gene Tool software from Syngene (Cambridge, UK).
To detect the difference in expression of RhoA gene between interphase and mitosis phase in HeLa cells, total RNA of interphase and mitosis cells was isolated using TRIzol reagent. cDNA was synthesized in a 20 &mgr;L reaction containing 4 &mgr;g of total RNA and SuperScript™II RT according to the manufacturer's instructions. Primers designed for amplification of the human RhoA gene and GAPDH gene were forward 5’-GGCTGCCATCCGGAA GAAA-3’ and reverse 5’-CACAAGACAAGGCACCCAGA-3’ and forward 5’-GGATTTGGTCGTATTGGG-3’, and reverse 5’-GGAAGATGGTGATGGGAT T-3’, respectively. The 25 &mgr;L reaction contained 0.5 &mgr;L cDNA, 10 pmol/L of each primer, 2 mmol/L MgCl2, 2.5 &mgr;L of 200 nmol/L dNTP, and 1 unit of Taq DNA polymerase. The thermal cycle profile for PCR was 94°C for 5 min followed by 35 cycles of 30 s at 94°C, 30 s at annealing temperature (63.8°C forRhoA and 55°C for GAPDH), and 30 s at 72°C with a final 10 min incubation at 72°C. The amplified fragments of the target genes were cloned into pGEmol/L vector and DNA sequencing was performed by Gene Co. Ltd. (Shanghai, China).
For real-time quantitative PCR, standard curves were prepared by serial dilution of pGEM-RhoA and pGEM-GAPDH vector. Reactions of 25 &mgr;L included 1 &mgr;L of 1:1500 SYBR Green I, 0.5 &mgr;L cDNA, 10 pmol of each primer, 2 mmol/L MgCl2, 2.5 &mgr;L 200 nmol/L dNTP, and 1 unit of Taq DNA polymerase. To compensate for variations of input RNA amounts and efficiency of reverse transcription, an endogenous 'housekeeping' gene (GAPDH) was also quantified and used to normalize the results. The thermal cycle profile for PCR was 94°C for 2 min followed by 35 cycles of 30 s at 94°C, 30 s at annealing temperature (63.8°C for RhoA and 55°C for GAPDH) and 30 s at 72°C. After the cycling of PCR assay was completed, the samples were subjected to a temperature ramp (from 45°C-95°C at 0.5°C/2 s) with continuous fluorescence monitoring for melting curve analysis. Analyses were done in triplicates and all reactions were repeated six times independently to ensure the reproducibility of the results. Data were analyzed by Rotor-Gene real-time analysis software (Rotor-Gene 2000, CR, Australia). For each sample the amplification plot and the corresponding dissociation curves were examined. To obtain standardized quantitative results, external controls consisting of cDNA plasmid standards were constructed.
Data were expressed as mean ± SE. Statistic significance was examined with Wilcoxon signed-rank test and Student’s t test, respectively. Probability below 0.05 (P < 0.05) was considered as significant.
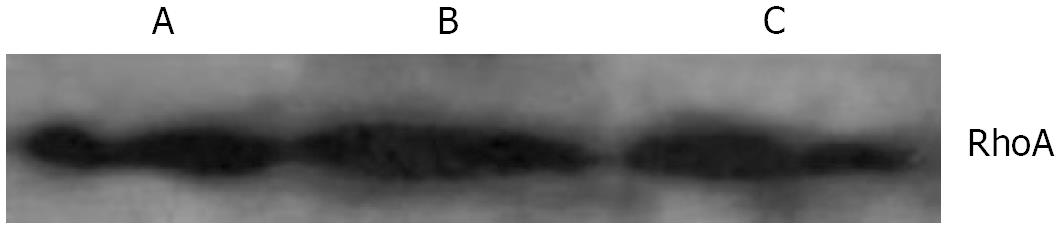
With immunofluorescence staining (Figure 1) and Western blotting (Figure 2) endogenous RhoA was detected in the membrane, the cytosol, and the nucleus of SGC-7901 cells. As shown by Western blotting (Figure 3) exogenous RhoA was also localized in the membrane, the cytosol, and the nucleus of SGC-7901 cells.
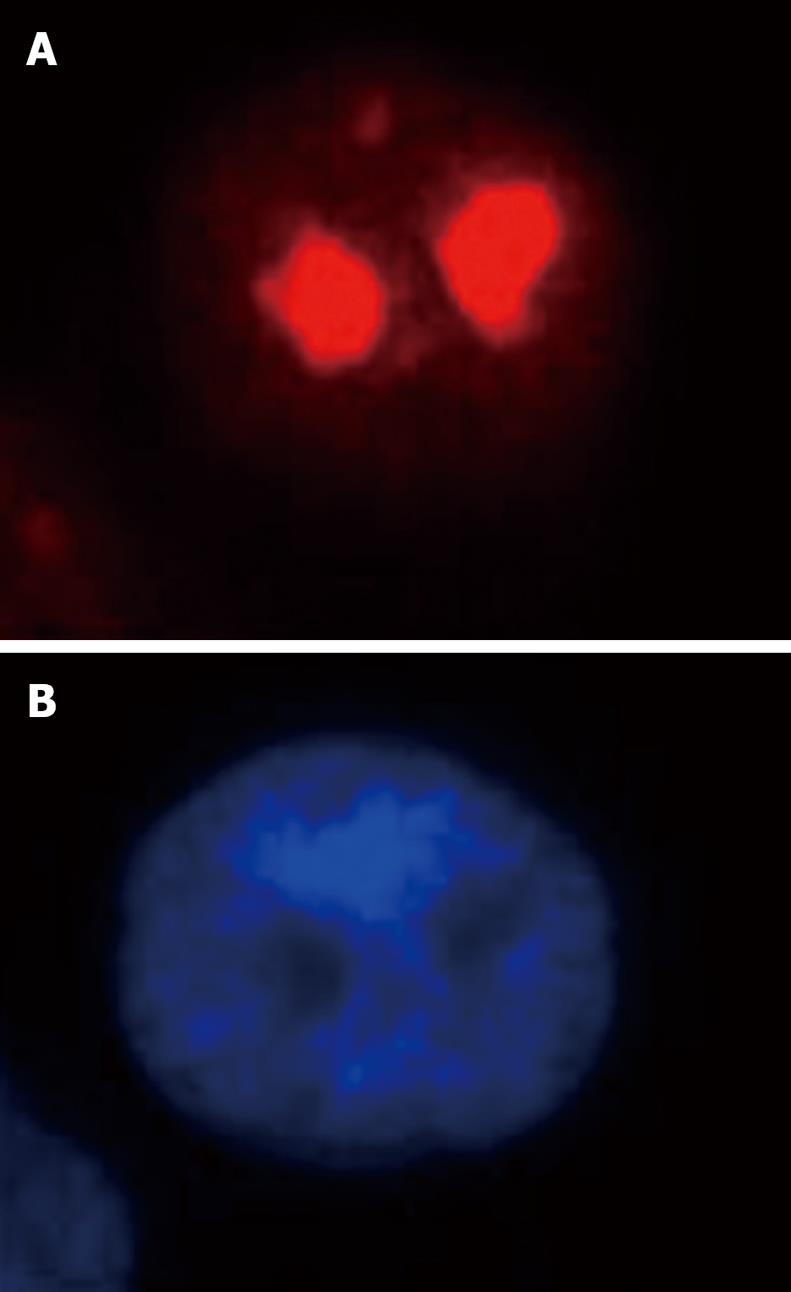
Both immunofluorescence staining (Figure 4) and Western blotting (Figure 5) indicated the nucleolus as the precise localization of RhoA within SGC-7901 interphase nucleus.
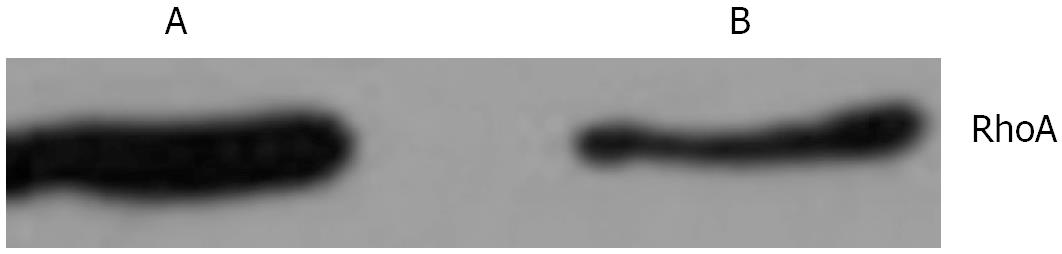
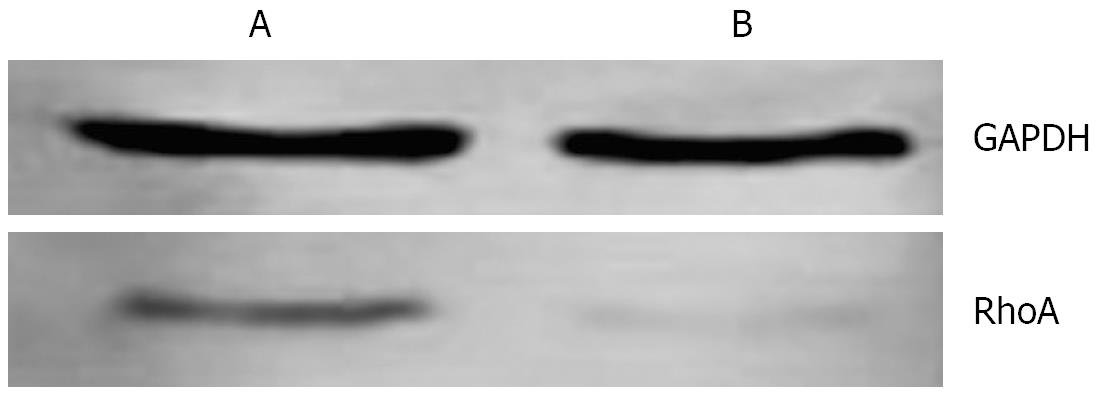
In mitotic active Hela cells, Western blotting indicated that the expression of RhoA protein in interphase (Figure 6A) was higher than that in mitosis phase (Figure 6B). Consistent with this finding, the amounts of RhoA transcripts were also found to be higher in interphase (Figure 7A) than in mitosis phase (Figure 7B).
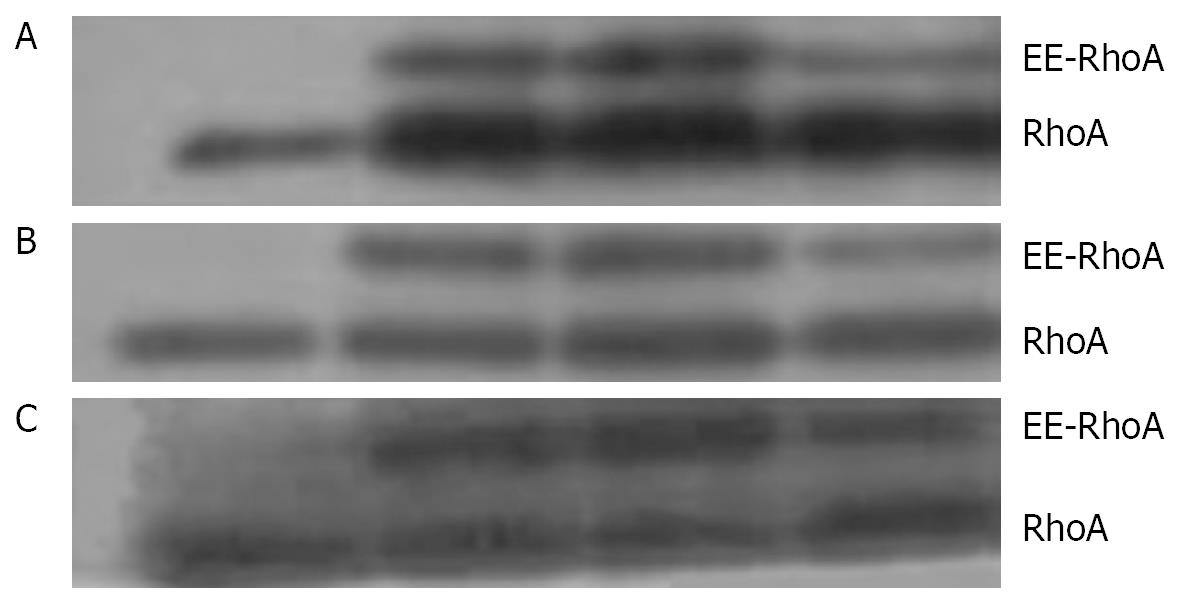
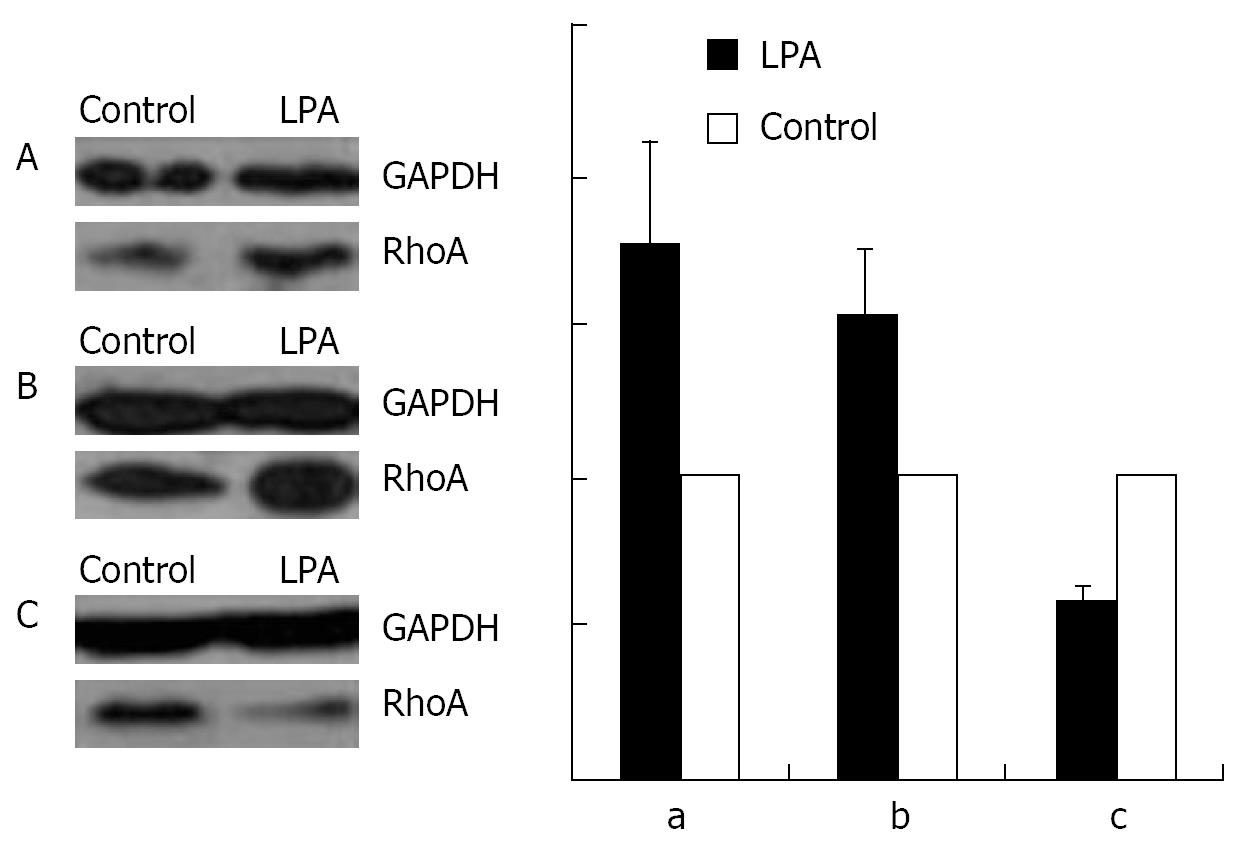
As shown by Western blotting, a stimulation with 1 &mgr;mol/L LPA at 37°C for 15 min causes an increase in the amount of RhoA in the membrane (Figure 8A) and the nuclear (Figure 8B), but a decrease in the cytosol fraction (Figure 8C).
An incubation with 100 &mgr;mol/L cAMP at 37°C for 30 min led to a decrease in the amounts of RhoA protein in membrane (Figure 9A) and nuclear (Figure 9B) fractions, but to an increase in cytosol preparations (Figure 9C).
Stimulation with 100 &mgr;mol/L cAMP at 37°C for 30 min followed by an incubation with 1 &mgr;mol/L LPA at 37°C for 15 min was found to cause a decrease in the amounts of RhoA protein in the membrane (Figure 10A) and the nuclear (Figure 10B), but an increase in the cytosol (Figure 10C) preparations.
RhoA acts as a molecular switch in the cells, regulates signal transduction from cell surface receptors to intracellular target molecules, and is involved in a variety of biological processes, including cell morphology[25], motility[26], cytokinesis[2728], smooth muscle contraction[2930], and tumor progression[3132]. It was widely accepted that the subcellular localization of RhoA was in the membrane and cytosol, and mainly in the latter one[3334]. Recently, our laboratory reported the existence of RhoA in the nuclei of SGC-7901 cells[22]. In this study, we not only confirm the nuclear localization of RhoA; we were able to localize RhoA proteins more precisely, namely within the nucleolus.
Protein import into the cell nucleus is a typical transport process between the cytosol and the nucleus. It occurs through nuclear pore complexes (NPCs)[35–39]. These elaborate proteinaceous structures act not only as molecular sieves, allowing free diffusion of ions and small molecules, but also mediate the active transport of proteins and ribonucleoprotein particles. In order to enter the nucleus, proteins larger than about 60 kDa generally require a specific nuclear localization signal (NLS). Protein molecules can bind with transporting protein such as importin via NLS and get into the nucleus through ATP dependent process. However, the molecular mass of RhoA is 21 kDa and there is no NLS with its protein structure. Our primary experiment did not detect the combination of RhoA with nuclear transporting proteins such as importin α and importin β (data not shown). So, it was reasonable for us to presume that the molecular mechanism of RhoA protein transportation to the nucleus was not through the classic transportation pathway, but diffusion.
The nucleolus of eukaryotic cells was first described in the early 19th century and was discovered in the 1960s to be the site of ribosome synthesis. It owned a variety of roles of cell activity. rRNA transcription, rRNA processing, and ribosome assembly have been clearly established as major functions of the nucleolus. As research progresses, many other functions of the nucleolus have been recognized, including gene expression, processing of nuclear export of certain mRNAs, processing of U6 RNA, one of the spliceosomal small nuclear RNAs, and processing of tRNA precursors. Recently, the plurifunctional nucleolus concept has been widely accepted.
According to the concept of plurifunctional nucleolus, it is reasonable to speculate that RhoA will share, at least, part of the function of the nucleolus. A previous study suggested a role for major nucleolar proteins in the nucleocytoplasmic transport of ribosomal components, and transient exposure of shuttling proteins to the cytoplasm may provide a mechanism for cytoplasmic regulation of nuclear activities[40]. The expression of some nucleolar proteins, such as B23, changes during cell cycle. In this experiment, the results showed that the amount RhoA protein also changed during cell cycle, exhibiting high amount in S phase, and the change of RhoA mRNA expression was the same as that of the protein. This suggested that RhoA probably played a role in regulation of cell mitogenesis. In this way, our results obviously provide a new insight into investigating some unknown functions of RhoA according to its new distribution.
In our previous study[22], with immunofluorescent microscopy, we did not detect significant translocation of RhoA in SGC-790 treated with RhoA activator LPA or RhoA activation inhibiting reagent cAMP. Since some other studies reported RhoA translocation according to its activity with Western blot assay, in this experiment, we applied different methods to separate cellular components and western blotting to investigate RhoA translocation, especially in nucleus. With these more concise and sensitive methods, we showed that after the treatments with LPA, cAMP, and cAMP + LPA, nucleus translocation of RhoA was the same as its membrane translocation. Furthermore, since it was reported that RhoA in membrane represented its active form[41] and our results showed that RhoA moved into the nucleus and onto the membrane during the stimulation of LPA, we supposed that RhoA in nucleus is also in the active status.
In conclusion, our results indicated that the nucleolus provided a new compartment for RhoA in SGC-7901 cells and that the activation of RhoA had an obvious effect on its translocation, stimulating the distribution of this protein not only in membrane but also in nucleus.
Until now, more than 100 small G proteins have been found. Among them, only Ran/TC4, Rap1, and H-Ras were found to be located within the nucleus. However, the nuclear localization of these small G proteins was closely related to the proliferation and the malignant change of the cells.
Both Rap1 and H-Ras are members of Ras family. Active Rap1A and Rap1B were located within the nucleus of epithelium-originated cancer cells, and the distribution was related to the malignancy of the cells. Active H-Ras was found to be located within the nucleus of both transformed cells and NDEA-induced hepatic tumor cells of mice.
Through immunofluorescence microscopy and Western blotting, RhoA has been found to be located in the nucleus of the gastric cancer cell line SGC-7901. The phenomenon was confirmed by confocal microscope observation of vital cells transfected with plasmid encoding GFP-tagged RhoA. However, the concise localization of the protein within the nucleus was not clear. Furthermore, whether the activation of RhoA is related to its nuclear localization or not was not elucidated.
Since RhoA was found to be located on the nucleolus of the cells and some nucleolar proteins played important role in controlling cell mitosis, it deserves to study the role of nucleolar RhoA in controlling cell mitosis.
Small G proteins are GTP/GDP binding proteins with molecular size around 20 kD. Each small G protein has two forms: GTP-binding active form and GDP-binding inactive form. When it changes from GDP-binding form to GTP-binding form, it is activated. Through this change cycle, small G protein performs as molecular switch. RhoA is an important member of Rho small G protein family. It has been found that RhoA played an important role in regulating diverse biological activities of cells.
The conclusion of this study was as follows: The localization of RhoA in the SGC-7901 cells was in membrane, cytosol, and nucleus. The novel finding of this study is showing that the precise localization of RhoA protein was in the nucleolus.
| 1. | Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389-399. |
| 2. | McCormick F. ras GTPase activating protein: signal transmitter and signal terminator. Cell. 1989;56:5-8. |
| 3. | Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: a conserved switch for diverse cell functions. Nature. 1990;348:125-132. |
| 5. | Takai Y, Kaibuchi K, Kikuchi A, Kawata M. Small GTP-binding proteins. Int Rev Cytol. 1992;133:187-230. |
| 6. | Sasaki T, Kikuchi A, Araki S, Hata Y, Isomura M, Kuroda S, Takai Y. Purification and characterization from bovine brain cytosol of a protein that inhibits the dissociation of GDP from and the subsequent binding of GTP to smg p25A, a ras p21-like GTP-binding protein. J Biol Chem. 1990;265:2333-2337. |
| 7. | Matsui Y, Kikuchi A, Araki S, Hata Y, Kondo J, Teranishi Y, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for smg p25A, a ras p21-like GTP-binding protein. Mol Cell Biol. 1990;10:4116-4122. |
| 8. | Ueda T, Kikuchi A, Ohga N, Yamamoto J, Takai Y. Purification and characterization from bovine brain cytosol of a novel regulatory protein inhibiting the dissociation of GDP from and the subsequent binding of GTP to rhoB p20, a ras p21-like GTP-binding protein. J Biol Chem. 1990;265:9373-9380. |
| 9. | Fukumoto Y, Kaibuchi K, Hori Y, Fujioka H, Araki S, Ueda T, Kikuchi A, Takai Y. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene. 1990;5:1321-1328. |
| 10. | Hiraoka K, Kaibuchi K, Ando S, Musha T, Takaishi K, Mizuno T, Asada M, Menard L, Tomhave E, Didsbury J. Both stimulatory and inhibitory GDP/GTP exchange proteins, smg GDS and rho GDI, are active on multiple small GTP-binding proteins. Biochem Biophys Res Commun. 1992;182:921-930. |
| 11. | Rubin EJ, Gill DM, Boquet P, Popoff MR. Functional modification of a 21-kilodalton G protein when ADP-ribosylated by exoenzyme C3 of Clostridium botulinum. Mol Cell Biol. 1988;8:418-426. |
| 12. | Chardin P, Boquet P, Madaule P, Popoff MR, Rubin EJ, Gill DM. The mammalian G protein rhoC is ADP-ribosylated by Clostridium botulinum exoenzyme C3 and affects actin microfilaments in Vero cells. EMBO J. 1989;8:1087-1092. |
| 13. | Yamamoto T, Kaibuchi K, Mizuno T, Hiroyoshi M, Shirataki H, Takai Y. Purification and characterization from bovine brain cytosol of proteins that regulate the GDP/GTP exchange reaction of smg p21s, ras p21-like GTP-binding proteins. J Biol Chem. 1990;265:16626-16634. |
| 14. | Kaibuchi K, Mizuno T, Fujioka H, Yamamoto T, Kishi K, Fukumoto Y, Hori Y, Takai Y. Molecular cloning of the cDNA for stimulatory GDP/GTP exchange protein for smg p21s (ras p21-like small GTP-binding proteins) and characterization of stimulatory GDP/GTP exchange protein. Mol Cell Biol. 1991;11:2873-2880. |
| 15. | Mizuno T, Kaibuchi K, Yamamoto T, Kawamura M, Sakoda T, Fujioka H, Matsuura Y, Takai Y. A stimulatory GDP/GTP exchange protein for smg p21 is active on the post-translationally processed form of c-Ki-ras p21 and rhoA p21. Proc Natl Acad Sci USA. 1991;88:6442-6446. |
| 16. | Xie Y, Gibbs TC, Meier KE. Lysophosphatidic acid as an autocrine and paracrine mediator. Biochim Biophys Acta. 2002;1582:270-281. |
| 17. | Moolenaar WH. Lysophosphatidic acid signalling. Curr Opin Cell Biol. 1995;7:203-210. |
| 18. | Kranenburg O, Poland M, Gebbink M, Oomen L, Moolenaar WH. Dissociation of LPA-induced cytoskeletal contraction from stress fiber formation by differential localization of RhoA. J Cell Sci. 1997;110:2417-2427. |
| 19. | Lang P, Gesbert F, Delespine-Carmagnat M, Stancou R, Pouchelet M, Bertoglio J. Protein kinase A phosphorylation of RhoA mediates the morphological and functional effects of cyclic AMP in cytotoxic lymphocytes. EMBO J. 1996;15:510-519. |
| 20. | Dong JM, Leung T, Manser E, Lim L. cAMP-induced morphological changes are counteracted by the activated RhoA small GTPase and the Rho kinase ROKalpha. J Biol Chem. 1998;273:22554-22562. |
| 21. | Laudanna C, Campbell JJ, Butcher EC. Elevation of intracellular cAMP inhibits RhoA activation and integrin-dependent leukocyte adhesion induced by chemoattractants. J Biol Chem. 1997;272:24141-24144. |
| 22. | Chen Y, Wang Y, Yu H, Wang F, Xu W. The cross talk between protein kinase A- and RhoA-mediated signaling in cancer cells. Exp Biol Med (Maywood). 2005;230:731-741. |
| 23. | Chen JC, Zhuang S, Nguyen TH, Boss GR, Pilz RB. Oncogenic Ras leads to Rho activation by activating the mitogen-activated protein kinase pathway and decreasing Rho-GTPase-activating protein activity. J Biol Chem. 2003;278:2807-2818. |
| 24. | Scherl A, Coute Y, Deon C, Calle A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ. Functional proteomic analysis of human nucleolus. Mol Biol Cell. 2002;13:4100-4109. |
| 25. | Paterson HF, Self AJ, Garrett MD, Just I, Aktories K, Hall A. Microinjection of recombinant p21rho induces rapid changes in cell morphology. J Cell Biol. 1990;111:1001-1007. |
| 26. | Takaishi K, Kikuchi A, Kuroda S, Kotani K, Sasaki T, Takai Y. Involvement of rho p21 and its inhibitory GDP/GTP exchange protein (rho GDI) in cell motility. Mol Cell Biol. 1993;13:72-79. |
| 27. | Kishi K, Sasaki T, Kuroda S, Itoh T, Takai Y. Regulation of cytoplasmic division of Xenopus embryo by rho p21 and its inhibitory GDP/GTP exchange protein (rho GDI). J Cell Biol. 1993;120:1187-1195. |
| 28. | Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M. CYK-4: A Rho family gtpase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol. 2000;149:1391-1404. |
| 29. | Hirata K, Kikuchi A, Sasaki T, Kuroda S, Kaibuchi K, Matsuura Y, Seki H, Saida K, Takai Y. Involvement of rho p21 in the GTP-enhanced calcium ion sensitivity of smooth muscle contraction. J Biol Chem. 1992;267:8719-8722. |
| 30. | Gong MC, Iizuka K, Nixon G, Browne JP, Hall A, Eccleston JF, Sugai M, Kobayashi S, Somlyo AV, Somlyo AP. Role of guanine nucleotide-binding proteins--ras-family or trimeric proteins or both--in Ca2+ sensitization of smooth muscle. Proc Natl Acad Sci USA. 1996;93:1340-1345. |
| 31. | Perona R, Esteve P, Jimenez B, Ballestero RP, Ramon y Cajal S, Lacal JC. Tumorigenic activity of rho genes from Aplysia californica. Oncogene. 1993;8:1285-1292. |
| 32. | Prendergast GC, Khosravi-Far R, Solski PA, Kurzawa H, Lebowitz PF, Der CJ. Critical role of Rho in cell transformation by oncogenic Ras. Oncogene. 1995;10:2289-2296. |
| 34. | Narumiya S. The small GTPase Rho: cellular functions and signal transduction. J Biochem. 1996;120:215-228. |
| 35. | Feldherr CM, Kallenbach E, Schultz N. Movement of a karyophilic protein through the nuclear pores of oocytes. J Cell Biol. 1984;99:2216-2222. |
| 36. | Jarnik M, Aebi U. Toward a more complete 3-D structure of the nuclear pore complex. J Struct Biol. 1991;107:291-308. |
| 38. | Forbes DJ. Structure and function of the nuclear pore complex. Annu Rev Cell Biol. 1992;8:495-527. |
| 40. | Borer RA, Lehner CF, Eppenberger HM, Nigg EA. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56:379-390. |
| 41. | Kranenburg O, Poland M, Gebbink M, Oomen L, Moolenaar WH. Dissociation of LPA-induced cytoskeletal contraction from stress fiber formation by differential localization of RhoA. J Cell Sci. 1997;110:2417-2427. |