Published online Dec 21, 2008. doi: 10.3748/wjg.14.7199
Revised: November 20, 2008
Accepted: November 27, 2008
Published online: December 21, 2008
AIM: To rapidly detect molecular alterations in different malignancies and investigate the possible role of Tp53, C-myc, and CCND1 genes in development of tumors in human organs and their adjacent normal tissues, as well as the possible relation between well- and poorly-differentiated tumors.
METHODS: A tissue array consisting of seven different tumors was generated. The tissue array included 120 points of esophagus, 120 points of stomach, 80 points of rectum, 60 points of thyroid gland, 100 points of mammary gland, 80 points of liver, and 80 points of colon. Expressions of Tp53, C-myc, and CCND1 were determined by RNA in situ hybridization. 3’ terminal digoxin-labeled anti-sense single stranded oligonucleotide and locked nucleic acid modifying probe were used.
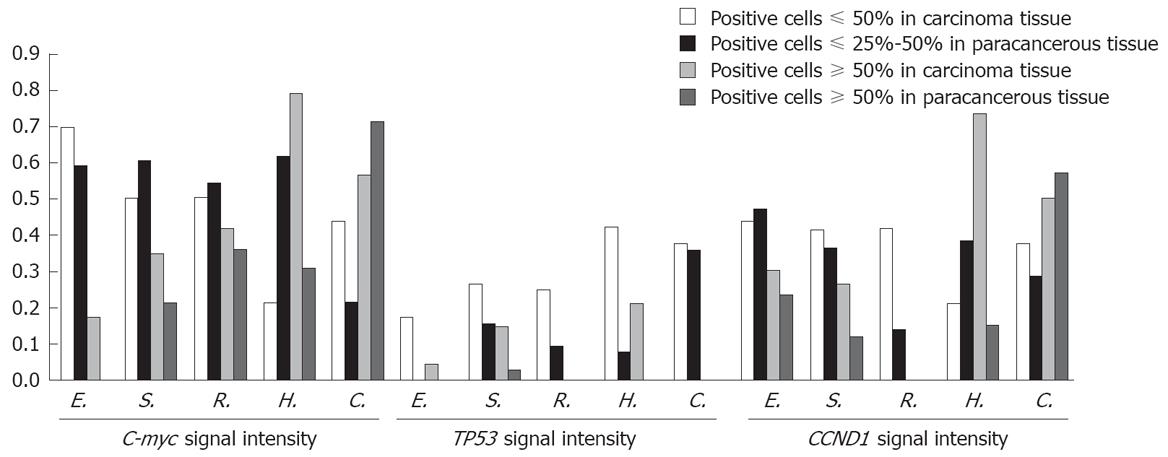
RESULTS: The expression level of Tp53 gene was higher in six different carcinoma tissue samples than in paracancerous tissue samples with the exception in colon carcinoma tissue samples (P < 0.05). The expression level of CCND1 gene was significantly different in different carcinoma tissue samples with the exception in esophagus and colon carcinoma tissue samples. The expression level of C-myc gene was different in esophagus carcinoma tissue samples (χ2 = 18.495, P = 0.000), stomach carcinoma tissue samples (χ2 = 23.750, P = 0.000), and thyroid gland tissue samples (χ2 = 10.999, P = 0.004). The intensity of signals was also different in different carcinoma tissue samples and paracancerous tissue samples.
CONCLUSION: Over-expression of the Tp53, CCND1, and C-myc genes appears to play a role in development of human cancer by regulating the expression of mRNA. Tp53, CCND1 and C-myc genes are significantly correlated with the development of different carcinomas.
-
Citation: Liu GY, Luo Q, Xiong B, Pan C, Yin P, Liao HF, Zhuang WC, Gao HZ. Tissue array for
Tp53 ,C-myc ,CCND1 gene over-expression in different tumors. World J Gastroenterol 2008; 14(47): 7199-7207 - URL: https://www.wjgnet.com/1007-9327/full/v14/i47/7199.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.7199
| Probe concentration (ng/μL) | Digest time (min) | Incubation time (h)/temperature (°C) | Chromogenic time (min) | ||
| Tissue array | Tp53 | 10 | 30 | 44/48 | 110 |
| Tissue array | C-myc | 10 | 20 | 41.5/42 | 30 |
| Tissue array | CCND1 | 10 | 20 | 36/45 | 50 |
| Histological grade | n | Tp53 positive | P value |
| Esophagus | |||
| Paracancerous tissue | 30 | 6 | P = 0.000 |
| I | 30 | 20 | |
| II | 30 | 19 | |
| III | 30 | 5 | |
| Stomach | |||
| Paracancerous tissue | 30 | 15 | P = 0.000 |
| I | 30 | 20 | |
| II | 30 | 29 | |
| III | 30 | 15 | |
| Rectum | |||
| Paracancerous tissue | 20 | 2 | P = 0.001 |
| I | 20 | 5 | |
| II | 20 | 13 | |
| III | 20 | 5 | |
| Thyroid gland | |||
| Paracancerous tissue | 20 | 2 | P = 0.023 |
| Follicular adenoma | 20 | 7 | |
| Papillary carcinoma | 20 | 10 | |
| Hepar | |||
| Paracancerous tissue | 20 | 6 | P = 0.000 |
| I | 20 | 19 | |
| II | 20 | 16 | |
| III | 20 | 15 | |
| Colon | |||
| Paracancerous tissue | 20 | 13 | P = 0.555 |
| I | 20 | 11 | |
| II | 20 | 14 | |
| III | 20 | 10 | |
| Mammary gland | |||
| Paracancerous tissue | 20 | 3 | P = 0.000 |
| Lobular hyperplasia | 20 | 4 | |
| Fibroadenoma | 20 | 4 | |
| Lobular carcinoma | 20 | 15 | |
| DCIS | 20 | 15 |
| Histological grade | n | CCND1 positive | P value |
| Esophagus | |||
| Paracancerous tissue | 30 | 19 | |
| I | 30 | 25 | P = 0.058 |
| II | 30 | 27 | |
| III | 30 | 25 | |
| Stomach | |||
| Paracancerous tissue | 30 | 15 | |
| I | 30 | 16 | P = 0.034 |
| II | 30 | 25 | |
| III | 30 | 19 | |
| Rectum | |||
| Paracancerous tissue | 20 | 1 | |
| I | 20 | 5 | P = 0.000 |
| II | 20 | 15 | |
| III | 20 | 10 | |
| Thyroid gland | |||
| Paracancerous tissue | 20 | 3 | |
| Follicular adenoma | 20 | 10 | P = 0.000 |
| Papillary carcinoma | 20 | 16 | |
| Hepar | |||
| Paracancerous tissue | 20 | 11 | |
| I | 20 | 19 | P = 0.037 |
| II | 20 | 15 | |
| III | 20 | 14 | |
| Colon | |||
| Paracancerous tissue | 20 | 13 | |
| I | 20 | 19 | P = 0.064 |
| II | 20 | 14 | |
| III | 20 | 12 | |
| Mammary gland | |||
| Paracancerous tissue | 20 | 3 | |
| Lobular hyperplasia | 20 | 4 | P = 0.001 |
| Fibroadenoma | 20 | 8 | |
| Lobular carcinoma | 20 | 10 | |
| DCIS | 20 | 15 |
| Histological grade | n | C-myc positive | P value |
| Esophagus | |||
| Paracancerous tissue | 30 | 17 | |
| I | 30 | 29 | P = 0.000 |
| II | 30 | 27 | |
| III | 30 | 20 | |
| Stomach | |||
| Paracancerous tissue | 30 | 28 | |
| I | 30 | 15 | P = 0.000 |
| II | 30 | 28 | |
| III | 30 | 25 | |
| Rectum | |||
| Paracancerous tissue | 20 | 16 | |
| I | 20 | 11 | P = 0.214 |
| II | 20 | 10 | |
| III | 20 | 13 | |
| Thyroid gland | |||
| Paracancerous tissue | 20 | 9 | |
| Follicular adenoma | 20 | 18 | P = 0.004 |
| Papillary carcinoma | 20 | 16 | |
| Hepar | |||
| Paracancerous tissue | 20 | 10 | |
| I | 20 | 11 | P = 0.813 |
| II | 20 | 10 | |
| III | 20 | 8 | |
| Colon | |||
| Paracancerous tissue | 20 | 10 | |
| I | 20 | 15 | P = 0.305 |
| II | 20 | 14 | |
| III | 20 | 11 | |
| Mammary gland | |||
| Paracancerous tissue | 20 | 7 | |
| Lobular hyperplasia | 20 | 12 | P = 0.133 |
| Fibroadenoma | 20 | 12 | |
| Lobular carcinoma | 20 | 10 | |
| DCIS | 20 | 15 |
Over-expression or amplification of a particular oncogene was first described in a tumor. Subsequently, other tumors were evaluated, mostly in the order of their perceived importance with rare tumors neglected. Therefore, it may take several years from the discovery of a potentially important molecular alteration to the definition of different primary tumors where this specific alteration may play a role in the development of such tumors. Tissue microarray has the potential to greatly facilitate analysis of alterations in different tumors. Tp53 is a specific protein produced by the most commonly mutated gene in human cancer that suppresses the growth of tumors[1]. Like other tumor-suppressor genes, Tp53 normally controls cell growth. If Tp53 is physically lost or not in effect (because of its inactivation), it may permit cells to divide without restraint[2]. The level of Tp53 has a prognostic (predictive) value for tumors. For example, breast cancer patients with a high level of Tp53 after mastectomy are at a higher risk for cancer recurrence than those with a low level of Tp53[3]. The buildup of Tp53 within cancer cells is a sign that Tp53 is not working properly to suppress the growth of tumors[4]. CCND1 forms a holoenzyme with a cyclin-dependent kinase (CDK), either CDK4 or CDK6 that phosphorylates the retinoblastoma gene product of pRb. Since the phosphorylation of pRb results in the release of E2F transcription factors, freeing them to stimulate transcription of growth-promoting target genes, over-expression of CCND1 promotes tumor progression through the G1 phase of cell cycle in cells grown on a substratum[5,6]. Over-expression of CCND1 has been reported in a variety of human tumors including cancers of the lung, head, neck, and bladder[7]. It was reported the over-expression rate of CCND1 is 30%-73% in patients with breast carcinoma[8,9]. Whether the expression of CCND1 can serve as a prognostic indicator of tumors has also been investigated, but the conclusions are contradictory in breast carcinoma[10]. C-myc gene is an important member of the myc gene family, can translocate and regulate a variety of substances, enable an unlimited cell proliferation, immortalize cell life, and is involved in tumor development[11,12].
At present, studies about Tp53, CCND1 and C-myc are mainly focused on Caucasian (white race) patients but not on Asians. Since the carcinogenesis of some organ carcinomas might show discrepancies among different races (Caucasian and Asian), detailed information on over-expression and amplification of the three genes in Chinese patients with carcinoma and its correlation with pathological parameters is needed. To more clearly address the importance of over-expression and expression of Tp53, CCND1 and C-myc genes in human cancer, we used in situ hybridation technique, which can clearly distinguish stromal from carcinoma components, and decrease the loss of such components in RNA extraction procedure. This approach to the specific location of genes on chromosomes is a technique for the hybridization of DNA and RNA “in situ”. This procedure can isolate or synthesize “in vitro” specific radioactive RNA or DNA (known as probes), and then anneal them to chromosomes treated in such a manner that their basic double stranded DNA has been “melted” or dissociated. The relation between Tp53 and CCND1, C-myc mRNA expressions was also discussed in this study.
A total of 620 primary tumor tissue samples from 7 different tumors and 20 normal tissue samples were snap-frozen and stored at -70°C. All patients were Chinese and underwent operation at Xiamen University Hospital during 2000-2006. Tissue blocks measuring approximately 1.5 cm × 1.5 cm × 0.3 cm of grossly apparent carcinoma and non-pathologic organs were fixed in phosphate-buffered saline (PBS) containing 4% paraformaldehyde (1‰ DEPC, pH 7.4) for 24 h, dehydrated through gradient ethanol, and embedded in paraffin. A hematoxylin and eosin (HE)-stained section was made from each block to define the representative tumor region. Representative areas in different lesions were carefully selected on HE-stained sections and marked on individual paraffin blocks. Tissue cylinders with a diameter of 1-mm were then punched from tumor areas in each “donor” tissue block and put into a recipient paraffin block using a custom-made precision instrument. Five-mm sections of the resulting multiple tumor tissue microarray blocks were transferred to glass slides using the paraffin sectioning aid system [adhesive-coated slides (PSA-CS4x), adhesive tape, and UV lamp; Instru-medics, Inc., Hackensack, NJ], supporting the cohesion of 0.6-mm array elements. The final TMA consisted of 640 1-mm diameter TMA cores each spaced at 0.8 mm between core centers. A section stained with HE was reviewed to confirm the presence of morphologically representative areas in the original lesions.
Anti-sense probes matched the corresponding sequence. Locked nucleic acid (LNA) was modified to increase the stability and sensitivity of probes. The sequences of probes are 5'-CAGGACAGGCACAAACACGCACCT*CAAAGCTGTTCCGTCCAGTAGATTAC-3Dig (Tp53), 5'-CCTCCTCGCACTTCTGTTCCTCGCAGACCT*CCAGCATCCAGGT GGCGACGATCTTCCG-3Dig (CCND1), 5'- CTTCCTCATCTTCTTGTTCCTCCTCAGAG T*CGCTGCTG GTGGTGGGCGGTGTC-3Dig (C-myc). The positive probe was 30T. Asterisk indicates that the LNA modifying site and 3' terminal were labeled with digoxigenin. All probes were synthesized by Sangon (Shanghai).
Hybridization procedures were performed in this study based on the instructions of RISH kits (Cybrdi USA) with some modifications. The glassware was washed, rinsed in distilled deionized water, and autoclaved before use. Gloves were worn when the glassware and slides were handled to prevent RNase contamination on the tissue. Because of the differences in tissues and probes, we performed different pilot-experiments to achieve the best results (Table 1). Deparaffinized sections were mounted on Denhardt-coated glass slides and treated with pepsin (0.25 mg/mL in DEPC H2O-HCl) for 25-30 min in a 37°C water bath. The treated sections were then processed for in situ hybridization at 42°C-45°C for 36-48 h. The hybridization mixture contained the labeled oligonucleotide probe, 50% formamide, 10 mmol/L Tris-HCl, 1 mmol/L vanadyl-ribonucleoside complex (Sigma 94740), 1 mmol/L CTAB (Sigma 855820, pH 7.0), 0.15 mol/L Nacl, 1 mmol/L EDTA (pH 7.0), 1 × Denhardt’s mixture and 10% dextran sulfate. After hybridization, the slides were washed three times, 30 min each time, in 0.1 mol/L TBS at room temperature, then treated with TBS (100 mmol/L Tris, pH 7.5, 150 mmol/L NaCl) containing a 1% blocking reagent (Roche) and 0.03% Triton X-100 for 30 min at room temperature and incubated for 30 min with antidioxigenin alkaline phosphataseconjugated antibodies (Roche) diluted at 1:500 in TBS containing 0.03% Triton X-100 and a 1% blocking reagent. After washed three times, 15 min each time, in TBS and 0.05% Tween, the slides were rinsed in a DAP-buffer (100 mmol/L Tris, pH 9.5, 100 mmol/L NaCl, 50 mmol/L MgCl2) and subsequently hybridization signals were visualized using nitroblue tetrazolium and 5-brom-4-chlor-3-indolyl phosphate as substrates [DAP-buffer in 10% PVA(Sigma 341584)].
All cases were first grouped to calculate the percentages of positive and negative cases. χ2 contingency test was used to evaluate the differences among groups. Analyses were performed using the statistical package SPSS 10.0 (SPSS, Chicago, IL). P < 0.05 was considered statistically significant.
The tissue micro-array technology is substantially different from the traditional multi-tissue blocks, which are often used in pathology laboratories for antibody testing. The most important advantages of tissue micro-array technology include increased capacity, negligible damage to the original tissue blocks, precise positioning of tissue specimens and possibility of automatic construction and analysis of arrays. In this study, we chose 4% paraformaldehyde in phosphate-buffered saline (1‰ DEPC PBS) as a fixation agent, which can decrease degradation of RNA and result in a good morphology. RISH analysis showed that 80%-95% of tumor samples were interpretable. RISH-related weak hybridization, background, and tissue damage were responsible for about one-sixth of the non-informative cases.
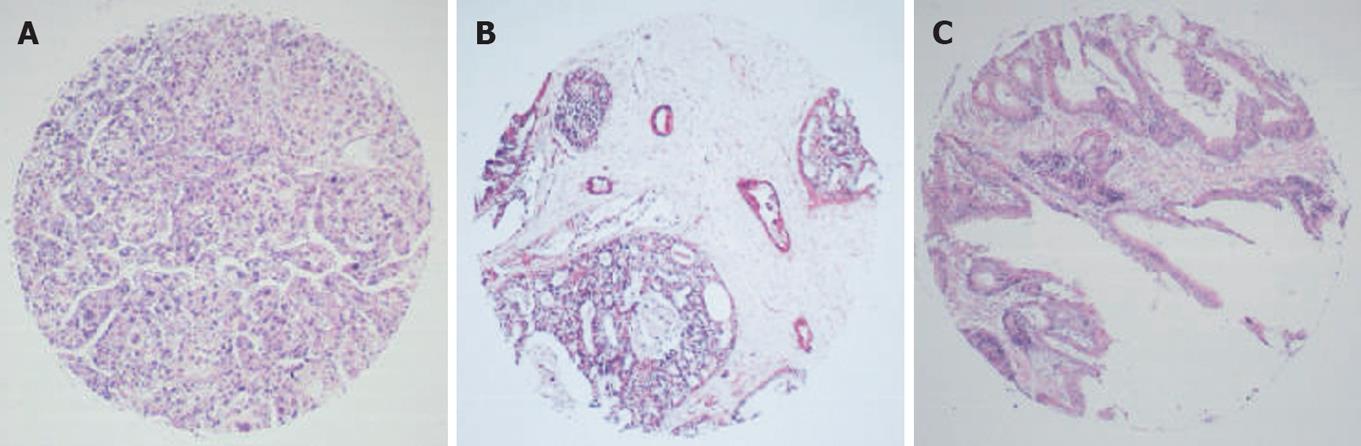
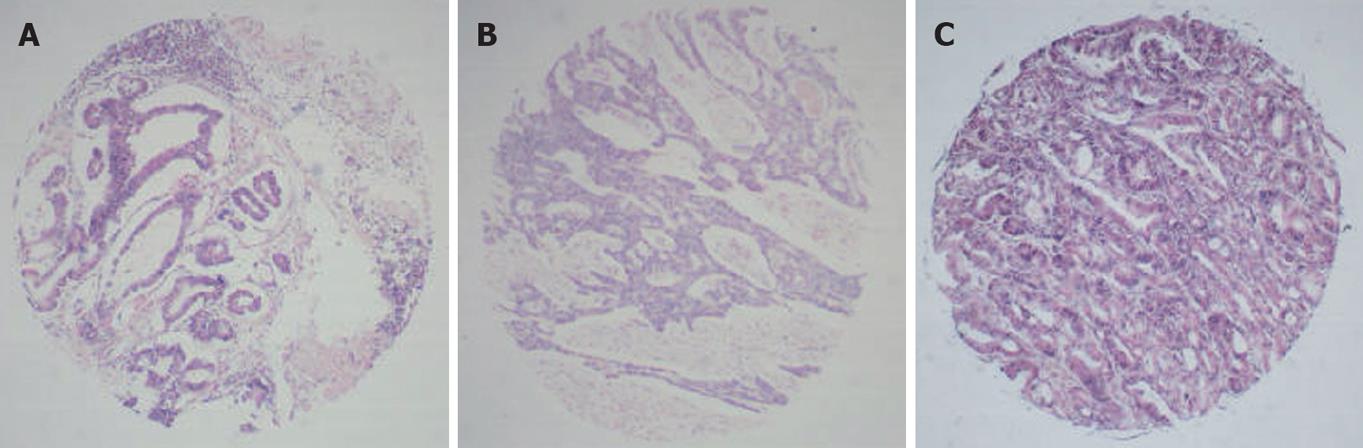
RNA in situ hybridization (RNA-ISH) was used to detect specific RNAs in situ. Most protocols using 4% paraformaldehyde as a fixation agent increased the probe permeability, and hybridization was performed in a buffer containing 50% formamide. The typical results of ISH were observed as amethyst dots on arrays, locating in cytoplasm or cytoblasts (Figures 1, 2 and 3). RNA analysis and quantification required completely intact, non-degraded RNA samples to produce optimal results. Vanadium oxide ions and formation of complex nucleoside could protect RNA degradation from RNase. Cetyltrimethyl ammonium bromide (CTAB) could stabilize the Oligo probe and target sequence formation of double-stranded structures, thus improving the re-annealing speed.
Two major factors, probe accessibility and affinity to the targeted RNA molecules, were found to affect the hybridization efficiency. Poor probe hybridization efficiency was found to be one of the major drawbacks of RNA-targeted in situ hybridization. The monomer containing LNA greatly improved the stability and sensitivity of RNA-targeted in situ hybridization. The six array elements resulting in 640 points are shown in Figures 1 and 2.
A total of 640 samples were studied for Tp53, CCND1 and C-myc mRNA expression, with non-radioactive in situ hybridization. ISH results were expressed as intensity and percentage based upon the signal intensity of positive staining and the number of stained cells within the sample, respectively. Tumor was graded according to the World Health Organization System. Normal tissues were also obtained from patients and a tumor free area in the same specimen served as a control. The presence of occasional tumor cells without detectable over-expression might be attributed to the truncated cells that had lost their genetic material during sectioning or tissue pretreatment before hybridization.
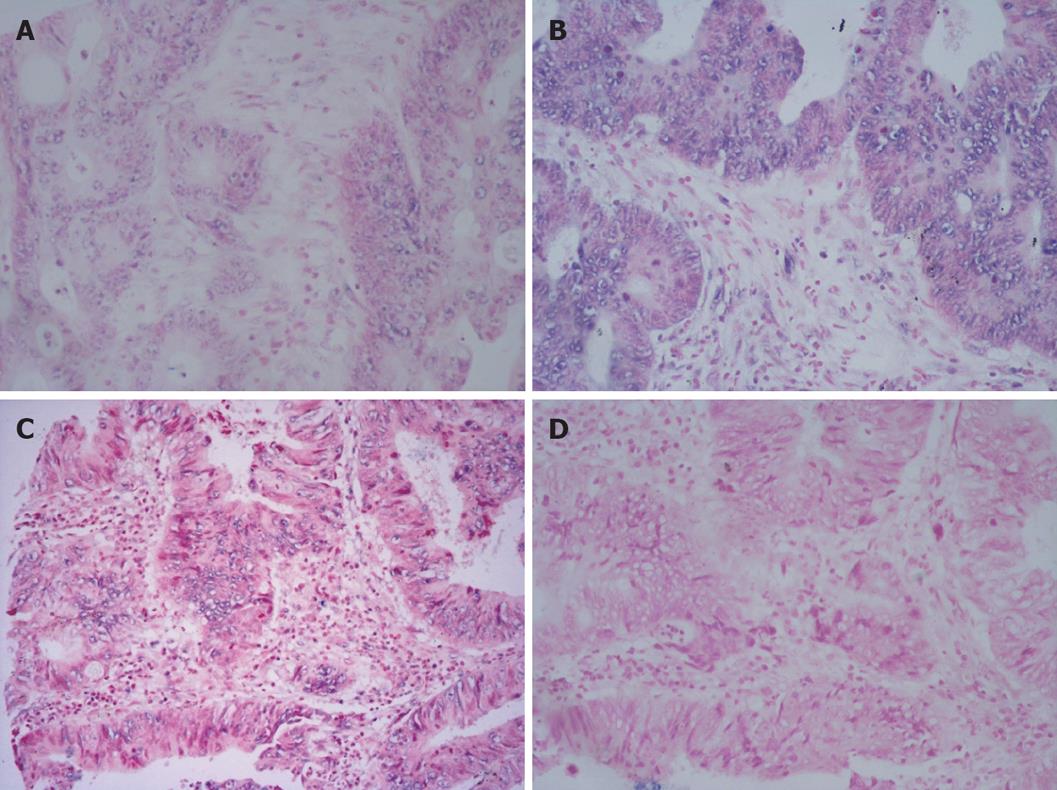
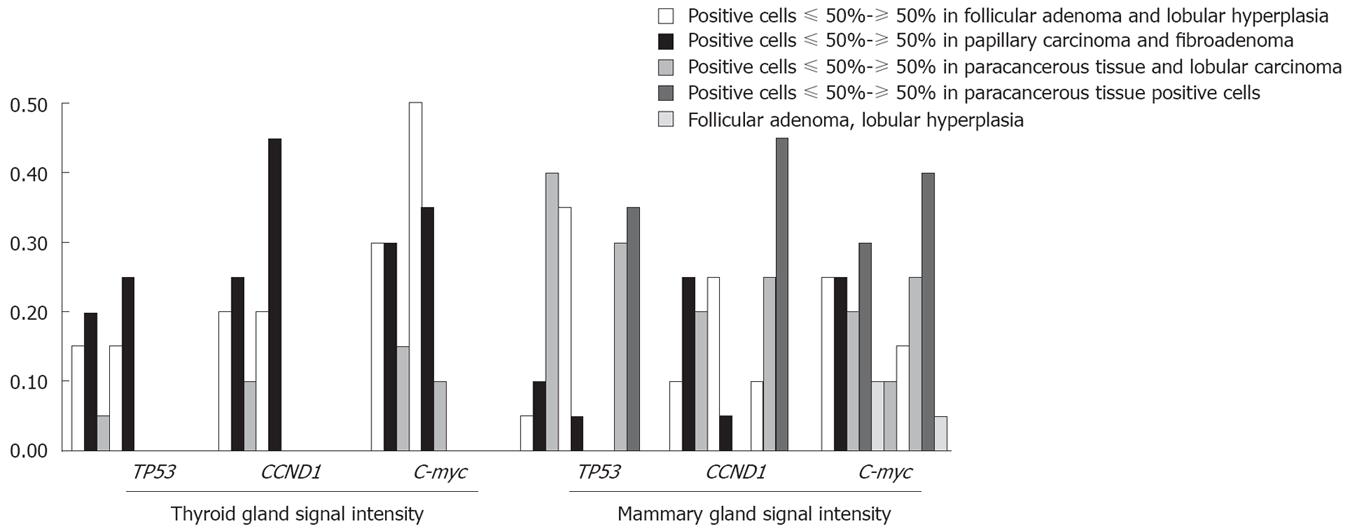
Tp53 was over-expressed in different tumors. The over-expression frequencies of Tp53 in these tumors are shown in Table 2. A significant difference was observed in carcinomatous and paracancerous tissue samples, including those of esophagus, stomach, rectum, thyroid gland, liver, mammary gland. Tp53 was over-expressed in almost all tumor cells within an array element. Our data indicate that expression of Tp53 in tumor tissues may play a role in cell carcinomatous change. Intracellular levels of Tp53 were elevated due to the increased stability and higher steady state of the protein, which may permit cells to divide without restraint. The positive expression rate of Tp53 was 48.9% (44/90) in carcinoma tissue samples and 20% (6/30) in normal adjacent tissue samples. In most cases, carcinomatous tissue samples had stronger signals than paracancerous tissue samples (Figures 4 and 5). Stronger positive dots (positive cells > 50%) were observed in carcinomatous tissue samples.
CCND1 ISH signals, located exclusively in nuclei, were variable in terms of staining intensity and proportion of positive nuclei among the cells in individual cases. The over-expression frequencies of all tumors are shown in Table 3. In this study, a significant difference was found in carcinomatous and paracancerous tissue samples, including those of stomach, rectum, thyroid gland, liver, and mammary gland. Our data indicate that CCND1 expression was significantly associated with carcinomatous change. CCND1 was expressed only in one paracancerous tissue sample of rectum, but in 4 carcinomatous tissue samples. In most cases, carcinomatous tissue samples had stronger signals than paracancerous tissue samples (Figures 4 and 5).
The over-expression frequencies of C-myc gene in all tumors are shown in Table 4. In this study, a significant difference was found in carcinomatous and paracancerous tissue samples, including those of esophagus, stomach, and thyroid gland. The expression of C-myc gene in carcinomatous and paracancerous tissues samples was not significantly associated with the histological grade of tumors. The expression of C-myc mRNA was heterogeneous in breast tumor tissue samples, with no predominant morphologic subtype in the high or low categories(χ2 = 7.062, P = 0.133). However, breast carcinoma tissue samples had stronger signals (positive cells > 75 %) than normal adjacent tissue samples. The positive expression rate of C-myc mRNA was 62.5% (25/40) in carcinomatous tissue samples, 60% (12/20) in lobular hyperplasia samples, and 60% (12/20) in fibroadenoma tissue samples.
In our study, the positive expression rate of the three probes was not significantly different in colon carcinoma and paracancerous tissue samples, which was 58.3%, 75.0%, 66.7% and 65.0%, 65.0%, 50.0%, respectively (χ2 =2.083, P = 0.555; χ2 = 7.273, P = 0.064; χ2 = 3.627, P = 0.305), suggesting that tumor grade is not related with the gene expression level. The signal intensity was different in colon. Hybridization signals (positive cells > 75%) were always observed both in CCND1 and in C-myc. Carcinoma tissue was associated with weaker signals (positive cells < 50%). However, an opposite tendency was found in thyroid and mammary glands.
The correlation coefficients of Tp53-CCND1, CCND1-C-myc and Tp53-C-myc were 0.653 (t = 3.76, P = 0.001), 0.737 (t = 4.753, P = 0.000) and 0.459 (t = 2.253, P = 0.036), respectively. If this finding was validated by an additional analysis in a larger population, these gene ratios could be used as prognostic markers in diagnostic biopsies.
In this study, we used the TMA technology because it allows analysis of a large number of samples and markers. A major concern for the TMA technique is the extent to which tumor heterogeneity may affect the validity of results. This issue has been addressed in TMA studies, which demonstrated that all previous findings from large sections could be fully reproduced[13,14]. The data on Tp53, CCND1, and C-myc, RNA in situ hybridization most commonly studied in associated tumors, are consistent with the reported findings[13,14]. In this study, the positive expression rates of Tp53, CCND1, and C-myc RNA were higher than those in previous reports[13,14], confirming the usefulness of the TMA approach. In our study, all tissue samples were fixed in phosphate-buffered saline (1‰ DEPC PBS) containing 4% paraformaldehyde that can prevent mRNA degradation from RNase and result in good morphology, indicating that tissue micro-array may be powerful in identification of different types of tumor with a particular molecular alteration.
Northern blot, dot blot or PCR-based approach has been used in detecting the expression of Tp53, CCND1 and C-myc mRNA in different tumors, but just a few reports are available on in situ hybridization. Some normal tissuses are dominated by adipose cells, differing greatly from tumor tissue in its epithelial cellularity. Normal and tumor tissues cannot be rigorously compared using techniques involving RNA extraction from total tissue. Therefore, conclusions such as ‘increased expression’ may be more difficult to make from studies with Northern blot, dot blot and PCR-based techniques requiring RNA extraction from tissues not fastidiously microdissected for selection of tumor cells. In this study, a more sensitive hybridization mixture decreased RNA degradation, thus accelerating oligonucleotide probe-RNA annealing. The signal intensity can be increased and low abundance RNA can be detected using a locked nucleic acid modifier to increase its stability and sensitivity[15-17]. The non-specificity signal can be decreased and the specificity can be increased using a different temperature and probe concentration. A strong hybridization signal appears in the transmitting tissue of pistil a few hours after 10% PVA (MW, 70-100 kDa) is used[18-20].
Our results reveal that the expression of Tp53 was higher in six different carcinoma samples than in their adjacent normal adjacent tissue samples with the exception in colon tissue samples. In this study, the expression of Tp53 was observed in 4 cases of lobular hyperplasia and in 20 cases of fibroadenoma. However, the positive expression rate of Tp53 was 75% (30/40) in breast carcinoma samples and 75% (30/40) in lobular carcinoma tissue samples, suggesting that determination of Tp53 gene by RISH contributes to the diagnosis of carcinoma and distinguishes DCIS from atypical hyperplasia. Moreover, carcinoma tissue often has a stronger signal than paracancerous tissue. In the present study, the signal (positive cells > 50%) was stronger in liver and thyroid gland carcinoma tissue samples than in follicular adenoma tissue samples. Originally, TP53 was thought to be an oncogene because over 50% of cancer cells tested showed a high level of Tp53 protein. However, all of them are mutated forms of Tp53. It was reported that the Tp53 gene acquires frequent mutations during the development of human malignancies including cancer of colon, breast, and lungs[21]. As described earlier, intracellular regulation of Tp53 expression can occur at the level of mRNA or Tp53 protein. A recent study indicated that even a brief reactivation of endogenous Tp53 in Tp53-deficient tumors can lead to a complete tumor regression[22].
At present, the expression of CCND1 has been investigated in several differences tumors, showing that patients with positive expression of CCND1 usually have a poor prognosis compared to those with negative expression of CCND1 in lung cancer, head and neck squamous cell carcinoma, and bladder cancer[23]. Although positive expression of CCND1 can serve as a poor prognostic factor or is associated with a worse prognosis, the expression of CCND1 is not correlated with the prognosis of cancer patients[24]. Furthermore, intensive investigations have been done on the correlation between CCND1 expression and patient survival in Caucasian females with breast carcinoma, but systematic investigations on alteration of CCND1 in Asian females are rare[23,24]. Whether CCND1 expression, clinicopathological parameters, survival rate and other prognostic markers are associated with cell cycle is not clear. In our study, a significant difference was found in positive expression rate of CCND1 between different tumors except for tumors of esophagus (χ2 = 7.500, P = 0.058) and colon (χ2 = 7.273, P = 0.064). These results agree with the conclusions of other studies[23,24].
The expression of CCND1 plays an important role in the early staging of carcinogenesis in Caucasian females with breast carcinoma. In the present study, 4 patients had lobular hyperplasia, 25 had breast carcinoma, suggesting that expression of CCND1 also plays an important role in Chinese patients with breast carcinoma. CCND1 was over-expressed in gastric adenocarcinoma tissue samples.
The C-myc oncogene is amplified or over-expressed in different human cancers. Experiments in vivo have also causally linked aberrant expression of this gene to the development and progression of cancer in different body sites[25]. However, several critical issues regarding the significance of C-myc in human cancer still remain obscure. C-myc is essential for tumor development, since it regulates factors necessary for the growth of tumors lending a new potential target to anti-angiogenic cancer therapies. Our study showed that the expression of C-myc was significantly different in carcinoma and its adjacent normal tissue samples. In our study, 18 patients had follicular adenoma and 16 had papillary carcinoma. The signal intensity of C-myc was also similar in follicular adenoma and papillary carcinoma patients with no strong signal occurred in paracancerous tissue samples, indicating that determination of C-myc gene by RISH can contribute to the diagnosis of carcinoma and distinguish carcinoma from follicular adenoma.
C-myc gene over-expression is associated with a poor prognosis of breast cancer patients[26]. The abnormal expression of C-myc mRNA and over-expression of C-myc in tissue array are listed in Table 4. The prognostic value for the over-expression of C-myc mRNA or protein is inconsistent and conflicting[26]. In our study, c-myc gene expression had no significant difference in breast lobular hyperplasia, fibroadenoma, lobular and ductal carcinoma tissue samples and their adjacent normal tissue samples (χ2 = 7.062, P = 0.133), and C-myc mRNA was highly expressed in 50% (75%) of high grade breast carcinoma tissue samples.
Our data show that the three genes were not differently expressed in colon carcinoma tissue samples, which is not consistent with the data reported by Deming[27] using the relatively insensitive Southern blot technique, suggesting that the three genes are not important factors for colon carcinogenesis and not significantly correlated with DCC deleted in colon cancer (DCC) and mutated in colon cancer (MCC) genes.
In normal cells, Tp53 gene is activated due to DNA damage and increases the transcription of p21 inhibiting the activity of cyclin/CDK complex and preventing cells from entering S phase. Mutation of Tp53 gene reflected by positive expression of mutated type Tp53 protein is one of the main causes for malignant transformation. In this study, CCND1 was correlated with Tp53 expression. We believe that although Tp53 could induce the expression of p21 and inhibit the function of CCND1/CDK complex in normal cells, abnormal expression of CCND1 does not necessarily respond to the dysfunction of wild-type Tp53 protein or positive expression of mutated type p53 in carcinoma cells. A pathway independent of Tp53 mutation may exist in carcinoma, which could also result in CCND1 over-expression. Over-expression of CCND1 may play an important role in the carcinogenesis of tumors without cooperation of Tp53 mutation[28].
In conclusion, it is necessary to analyze the lower grade tumors and premalignant lesions with the same tools to determine whether the expressions of Tp53, CCND1 and C-myc are different, and to compare the development and progression of tumors. Only a subtle deregulation of the expression of C-myc is sufficient to allow genomic instability. Whether gene expression precedes or follows its over-expression in the course of cancer needs to be further studied.
The Tp53, CCND1 and C-myc oncogenes are amplified or over-expressed in many types of human cancer. Experiments in vivo have also causally linked aberrant expression of these genes to the development and progression of cancer in different body sites. However, several critical issues regarding the significance of Tp53, CCND1 and C-myc in human cancer still remain obscure. The frequency of amplification, over-expression of mRNA and protein in breast cancer is about 50%-100%. Whether the expression of these genes is altered at the cytogenetic level in different types of human carcinoma remains unclear.
The development and progression of cancer in different body sites are very intricate and involve the expression of multiple cytokines and oncogenes such as Tp53, CCND1 and C-myc.
Few studies are available on the correlation of Tp53, CCND1, C-myc with seven kinds of tissue. To study the expression of oncogene during the development of cancer, we constructed tissue microarrays consisting of samples from 7 types of tumor. Using scrupulous R & ISH and LNA modifying probe, we achieved high expressions of these genes in different tumor tissues. C-myc mRNA was highly expressed in 50% of high grade breast carcinomas, which was much higher than the reported data (22%) by real-time RT-PCR.
The relationship between oncogene and tumor clarifies the mechanism of tumor and provides an important molecular basis for closer observation of the nature of tumor, which can be widely applied in the diagnosis, treatment, and prognosis of cancer.
The authors studied the expression of Tp53, C-myc, and CCND1 in different tumors using tissue microarrays. The study was well designed. The findings are reliable and can be widely applied in the diagnosis, treatment, and prognosis of cancer.
Peer reviewer: Xin-Yuan Guan, Professor, Department of Clinical Oncology, The University of Hong Kong, Room L10-56, 10/F, Laboratory Block, 21 Sassoon Road, Pokfulam, Hong Kong, China
S- Editor Tian L L- Editor Wang XL E- Editor Lin YP
| 1. | Rogel A, Popliker M, Webb CG, Oren M. p53 cellular tumor antigen: analysis of mRNA levels in normal adult tissues, embryos, and tumors. Mol Cell Biol. 1985;5:2851-2855. |
| 2. | Mercer WE, Avignolo C, Baserga R. Role of the p53 protein in cell proliferation as studied by microinjection of monoclonal antibodies. Mol Cell Biol. 1984;4:276-281. |
| 3. | Davidoff AM, Humphrey PA, Iglehart JD, Marks JR. Genetic basis for p53 overexpression in human breast cancer. Proc Natl Acad Sci USA. 1991;88:5006-5010. |
| 4. | Sasano H, Goukon Y, Nishihira T, Nagura H. In situ hybridization and immunohistochemistry of p53 tumor suppressor gene in human esophageal carcinoma. Am J Pathol. 1992;141:545-550. |
| 5. | Ligueros M, Jeoung D, Tang B, Hochhauser D, Reidenberg MM, Sonenberg M. Gossypol inhibition of mitosis, cyclin D1 and Rb protein in human mammary cancer cells and cyclin-D1 transfected human fibrosarcoma cells. Br J Cancer. 1997;76:21-28. |
| 6. | Zwijsen RM, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJ. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405-415. |
| 7. | Michalides RJ, van Veelen NM, Kristel PM, Hart AA, Loftus BM, Hilgers FJ, Balm AJ. Overexpression of cyclin D1 indicates a poor prognosis in squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 1997;123:497-502. |
| 8. | Zhang SY, Caamano J, Cooper F, Guo X, Klein-Szanto AJ. Immunohistochemistry of cyclin D1 in human breast cancer. Am J Clin Pathol. 1994;102:695-698. |
| 10. | Weinstein IB. Relevance of cyclin D1 and other molecular markers to cancer chemoprevention. J Cell Biochem Suppl. 1996;25:23-28. |
| 11. | Augenlicht LH, Wadler S, Corner G, Richards C, Ryan L, Multani AS, Pathak S, Benson A, Haller D, Heerdt BG. Low-level c-myc amplification in human colonic carcinoma cell lines and tumors: a frequent, p53-independent mutation associated with improved outcome in a randomized multi-institutional trial. Cancer Res. 1997;57:1769-1775. |
| 12. | Aulmann S, Bentz M, Sinn HP. C-myc oncogene amplification in ductal carcinoma in situ of the breast. Breast Cancer Res Treat. 2002;74:25-31. |
| 13. | Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med. 1998;4:844-847. |
| 14. | Battifora H. The multitumor (sausage) tissue block: novel method for immunohistochemical antibody testing. Lab Invest. 1986;55:244-248. |
| 15. | Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA). RNA. 2006;12:913-920. |
| 16. | Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27-29. |
| 17. | Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834-838. |
| 18. | Pontius BW, Berg P. Rapid renaturation of complementary DNA strands mediated by cationic detergents: a role for high-probability binding domains in enhancing the kinetics of molecular assembly processes. Proc Natl Acad Sci USA. 1991;88:8237-8241. |
| 19. | Pontius BW, Berg P. Renaturation of complementary DNA strands mediated by purified mammalian heterogeneous nuclear ribonucleoprotein A1 protein: implications for a mechanism for rapid molecular assembly. Proc Natl Acad Sci USA. 1990;87:8403-8407. |
| 20. | De Block M, Debrouwer D. RNA-RNA in situ hybridization using digoxigenin-labeled probes: the use of high-molecular-weight polyvinyl alcohol in the alkaline phosphatase indoxyl-nitroblue tetrazolium reaction. Anal Biochem. 1993;215:86-89. |
| 21. | Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661-665. |
| 22. | Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656-660. |
| 23. | Lam WW, Fielding R, Ho EY. Predicting psychological morbidity in Chinese women after surgery for breast carcinoma. Cancer. 2005;103:637-646. |
| 24. | Luini A, Gatti G, Ballardini B, Zurrida S, Galimberti V, Veronesi P, Vento AR, Monti S, Viale G, Paganelli G. Development of axillary surgery in breast cancer. Ann Oncol. 2005;16:259-262. |
| 25. | Chana JS, Grover R, Wilson GD, Hudson DA, Forders M, Sanders R, Grobbelaar AO. The clinical significance of c-myc oncogene expression in melanomas of the scalp. Br J Plast Surg. 1998;51:191-194. |
| 26. | Balch CM, Soong SJ, Gershenwald JE, Thompson JF, Reintgen DS, Cascinelli N, Urist M, McMasters KM, Ross MI, Kirkwood JM. Prognostic factors analysis of 17,600 melanoma patients: validation of the American Joint Committee on Cancer melanoma staging system. J Clin Oncol. 2001;19:3622-3634. |
| 27. | Deming SL, Nass SJ, Dickson RB, Trock BJ. C-myc amplification in breast cancer: a meta-analysis of its occurrence and prognostic relevance. Br J Cancer. 2000;83:1688-1695. |
| 28. | Jin M, Inoue S, Umemura T, Moriya J, Arakawa M, Nagashima K, Kato H. Cyclin D1, p16 and retinoblastoma gene product expression as a predictor for prognosis in non-small cell lung cancer at stages I and II. Lung Cancer. 2001;34:207-218. |