Published online Dec 7, 2008. doi: 10.3748/wjg.14.6893
Revised: October 21, 2008
Accepted: October 28, 2008
Published online: December 7, 2008
Hereditary hemochromatosis (HH) is caused by chronic hyperabsorption of dietary iron. Progressive accumulation of excess iron within tissue parenchymal cells may lead to severe organ damage. The most prevalent type of HH is linked to mutations in the HFE gene, encoding an atypical major histocompatibility complex classImolecule. Shortly after its discovery in 1996, the hemochromatosis protein HFE was shown to physically interact with transferrin receptor 1 (TfR1) and impair the uptake of transferrin-bound iron in cells. However, these findings provided no clue why HFE mutations associate with systemic iron overload. It was later established that all forms of HH result from misregulation of hepcidin expression. This liver-derived circulating peptide hormone controls iron efflux from duodenal enterocytes and reticuloendothelial macrophages by promoting the degradation of the iron exporter ferroportin. Recent studies with animal models of HH uncover a crucial role of HFE as a hepatocyte iron sensor and upstream regulator of hepcidin. Thus, hepatocyte HFE is indispensable for signaling to hepcidin, presumably as a constituent of a larger iron-sensing complex. A working model postulates that the signaling activity of HFE is silenced when the protein is bound to TfR1. An increase in the iron saturation of plasma transferrin leads to displacement of TfR1 from HFE and assembly of the putative iron-sensing complex. In this way, iron uptake by the hepatocyte is translated into upregulation of hepcidin, reinforcing the concept that the liver is the major regulatory site for systemic iron homeostasis, and not merely an iron storage depot.
- Citation: Pantopoulos K. Function of the hemochromatosis protein HFE: Lessons from animal models. World J Gastroenterol 2008; 14(45): 6893-6901
- URL: https://www.wjgnet.com/1007-9327/full/v14/i45/6893.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.6893
Iron is essential for various physiological and metabolic pathways. However, unshielded iron is toxic, as a catalyst of free radical generation[1,2]. The adult human body contains a pool of 3-5 g of iron (about 55 mg and 44 mg per kilogram body weight in males and females, respectively), the majority of which (> 70%) is utilized by erythroid cells for heme synthesis and integration into hemoglobin[3]. A daily requirement of about 20-30 mg iron for erythropoiesis is mainly covered by recycling of the metal from senescent erythrocytes via reticuloendothelial macrophages. These cells metabolize heme and release iron into the circulation, where it is scavenged by plasma transferrin and delivered to tissues. A considerable amount of iron (about 1 g) is stored in the liver. Dietary iron absorption by duodenal enterocytes compensates for losses through bleeding or desquamation; a physiological rate of 1-2 mg/d suffices to maintain the body iron pool. This is subjected to feedback regulation and may adjust to fluctuations in iron demands.
In hereditary hemochromatosis (HH), disruption of this homeostatic loop leads to unrestricted dietary iron absorption at a rate that may reach 8-10 mg/d[4,5]. This is accompanied by a gradual increase in the saturation of transferrin with iron (from physiological 30% up to 100%), a buildup of non-transferrin-bound iron and excessive accumulation of the metal in parenchymal cells of the liver, pancreas, pituitary, heart, joints and skin. Notably, macrophages and absorptive duodenal enterocytes are spared from iron loading and exhibit increased rates of iron release. Excessive iron deposition in the liver constitutes a risk factor for fibrosis, cirrhosis and hepatocellular cancer[6-8], and may exacerbate other types of liver disease[9,10]. Iron overload may also lead to cardiomyopathy, diabetes mellitus, hypogonadism, arthritis and skin pigmentation[3]. HH is efficiently treated by phlebotomy.
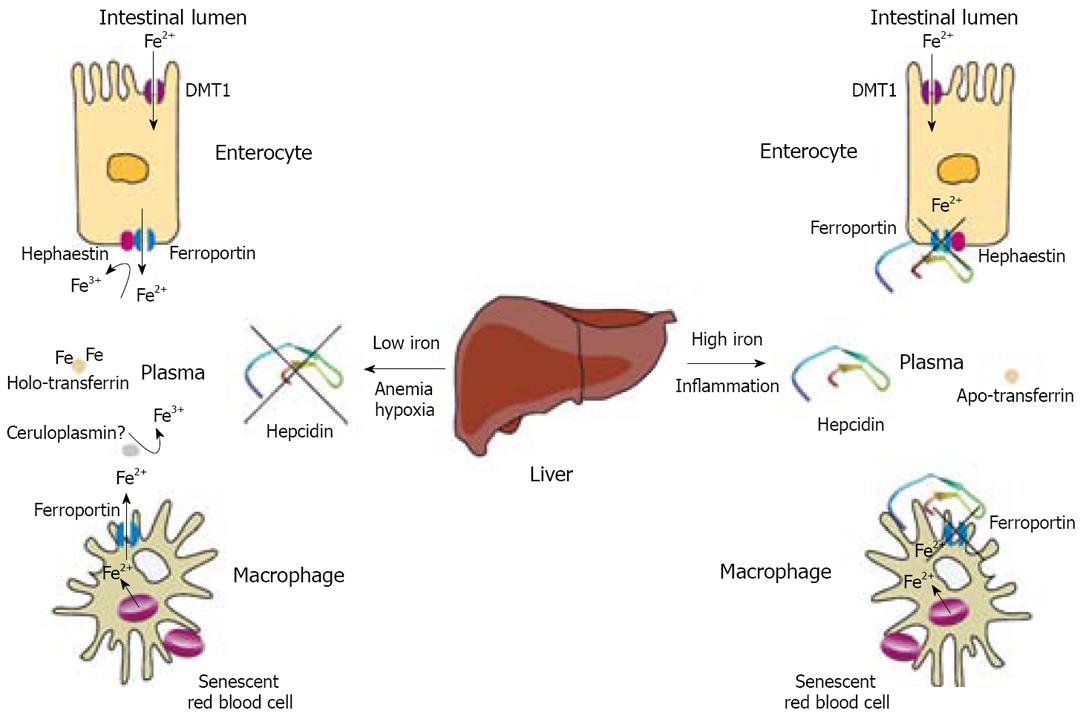
The discoveries of the divalent metal transporter (DMT1), the iron exporter ferroportin, and the iron regulatory hormone hepcidin provided a framework to understand the molecular mechanisms for systemic iron traffic and homeostasis[11,12]. DMT1 accounts for the absorption of ferrous ions across the apical membrane of duodenal enterocytes, but also for intracellular transport of transferrin-derived iron across the endosomal membrane in many cell types. Ferroportin mediates efflux of ferrous iron from enterocytes and macrophages to plasma transferrin. The transport of iron by DMT1 requires its reduction by ferric reductases (such as Dcytb or the Steap proteins), while its export by ferroportin is coupled by re-oxidation via ferroxidases (such as ceruloplasmin or hephaestin).
The ferroportin-mediated efflux of iron from enterocytes and macrophages defines a key regulatory checkpoint for iron homeostasis. This process is negatively controlled by hepcidin, a cysteine-rich peptide hormone that binds to ferroportin and promotes its internalization and lysosomal degradation[13]. Hepcidin is synthesized in hepatocytes as a pro-peptide, which undergoes proteolytic processing to form a bioactive molecule of 25 amino acids[14]. The mature peptide is secreted into plasma and orchestrates homeostatic responses to iron, erythropoiesis, hypoxia and inflammation. An increase in hepcidin levels, commonly encountered following dietary iron intake or in inflammation[15,16], impairs iron absorption by duodenal enterocytes and promotes retention of the metal within macrophages (Figure 1), limiting its availability for erythropoiesis. Excessive hepcidin expression, in response to prolonged inflammation, contributes to the anemia of chronic disease[17]. On the other hand, low hepcidin levels triggered by iron deficiency, hypoxia or phlebotomy[18] facilitate duodenal iron absorption and iron release from macrophages (Figure 1). Importantly, HH patients fail to mount an appropriate upregulation of hepcidin expression, despite high transferrin saturation and elevated body iron stores[19,20]. Thus, HH is largely based on the loss of feedback control in dietary iron absorption due to defects in the hepcidin pathway.
Juvenile hemochromatosis, a rare but severe form of hereditary iron overload results from genetic inactivation of the hepcidin gene[21] or mutations in hemojuvelin (HJV) associated with profound hepcidin deficiency[22]. The most prevalent form of HH is linked to mutations in HFE[23], while another less common but phenotypically indistinguishable HH subtype is caused by mutations in transferrin receptor 2 (TfR2)[24]. Iron overload patients with either HFE or TfR2 mutations exhibit inappropriately decreased hepcidin levels or blunted hepcidin responses[19,20,25,26]. Similar results were obtained with mouse models of iron overload, bearing targeted disruptions of the HFE[27-30], HJV[31,32] or TfR2[33] genes. These findings suggest that HFE, HJV and TfR2 are upstream regulators of hepcidin expression.
Hepcidin is transcriptionally activated by distinct iron-and cytokine-dependent pathways. The latter is mediated by IL-6 (and IL-1) via STAT3[34-36]. The iron-dependent pathway is less well characterized and involves proximal and distal promoter elements[37,38]. The lack of hepcidin expression, accompanied by iron overload, in mice carrying a hepatocyte-specific disruption of SMAD4[39] has linked iron-sensing with bone morphogenetic protein (BMP) signaling. In fact, BMP-2, -4 and -9 are potent inducers of hepcidin transcription, while hemojuvelin stimulates this pathway as a BMP co-receptor[40-42]. The CCAAT/enhancer-binding protein α (C/EBPα) appears necessary for basal hepcidin transcription[43].
Hepcidin expression is suppressed in anemia by a mechanism that requires erythropoietic activity[44,45]. At least in thalassemia patients, the silencing of hepcidin is mediated by overexpression of growth differentiation factor 15 (GDF15), a member of the transforming growth factor β (TGFβ) superfamily[46]. Erythropoietin (EPO) directly reduces the binding of C/EBPα to the hepcidin promoter via EPO receptor signaling[47]. Hepcidin is also negatively regulated by hypoxia[18]. Experiments in mice with hepatic disruption of HIF-1α provided evidence for the involvement of this transcription factor in the underlying pathway[48]. However, other reports suggested that the hypoxic downregulation of hepcidin is HIF-independent[49,50] and involves oxidative stress-mediated repression of C/EBPα and STAT3[49], or inhibition of 2-oxoglutarate dependent oxygenases[50]. Recent work revealed that the transmembrane serine protease TMPRSS6 negatively regulates signaling to hepcidin[51-55], by a yet unknown mechanism.
What is the role of HFE in hepcidin regulation?
The HFE gene was elucidated by linkage disequilibrium and haplotype analysis from a large group of HH patients[23], culminating lengthy efforts to map the hemochromatosis locus. It encodes an atypical major histocompatibility complex (MHC) classIprotein, which is processed via the Golgi network to the cell surface, following interaction with β2-microglobulin. Structural analysis revealed that in contrast to typical MHC classIhomologues, HFE formed a smaller groove between the α1 and α2 subunits, which was predicted to preclude peptide antigen presentation[56]. The majority of HH patients carry an HFE C282Y substitution. This abrogates a disulphide bridge and prevents the association of HFE with β2-microglobulin, a necessary step for its processing and transport to the plasma membrane[57,58]. Unprocessed HFE C282Y undergoes proteasomal degradation following retention in the endoplasmic reticulum (ER), which promotes ER stress[59]. An HFE H63D mutation may also lead to HH, especially in the compound heterozygous state with C282Y. Homozygosity for the HFE C282Y genotype is highly prevalent (1:200) in populations of Northern European ancestry; however, the clinical penetrance is lower and remains a matter of debate[4-6,60,61]. It appears that HH is a multifactorial disease and the development of iron overload in individuals bearing disease-associated HFE mutations requires the contribution of additional, yet incompletely understood environmental, genetic and/or epigenetic factors[62]. Nevertheless, mice with either targeted disruption of the HFE[63,64] or β2-microglobulin[65,66] genes, or expressing orthologues of the HFE C282Y[67] or H63D[68] mutants, develop progressive iron overload, the degree of which depends on the genetic background of the animals[69-71]. Collectively, these findings underlie the significance of HFE in the control of body iron homeostasis.
Biochemical[72,73] and crystallographic[74] studies revealed that HFE interacts with TfR1 (Kd about 60 nmol/L) and competes for the binding of transferrin to its receptor, which has a Kd of about 1 nmol/L[75]. However, considering that the physiological concentration of plasma diferric holotransferrin is about 5 μmol/L[76], HFE is unlikely to affect the rate of TfR1 endocytosis in vivo. In transfected cell lines, overexpressed HFE reduced the efficiency of the transferrin cycle[77] and promoted an iron-deficient phenotype[78-81], without or with co-expression of β2-microglobulin[82]. Notably, a similar phenotype was observed with an HFE W81A mutant that is unable to bind to TfR1, suggesting that the HFE-mediated decrease of intracellular iron levels is independent of the HFE/TfR1 interaction[83].
The above data did not shed much light on how HFE controls systemic iron homeostasis and rather created some confusion. The immunohistochemical detection of HFE in precursor enterocytes of the intestinal crypts[84] and its association with TfR1 in these cells[85] laid the foundation for the “crypt-programming model”[86]. This postulates that iron absorption is regulated by signals that are sensed by precursor enterocytes, which undergo maturation and migrate along the crypt-villus axis. An iron deficient status in the crypt cells would program mature enterocytes to absorb more dietary iron from the lumen. According to the crypt-programming model, HFE would serve to promote iron retention within crypt cells, possibly by increasing the uptake of plasma transferrin[87] and/or inhibiting iron efflux[88]. The model is supported by the iron deficient status manifested in duodenal biopsies from HH patients[89,90]. Experimental evidence has been provided that HFE may also facilitate iron accumulation[91] or retention[92] within macrophages, which are likewise iron-deficient in HH patients[93]. In the pre-hepcidin era, these findings have highlighted the enterocytes and macrophages as possible primary sites of the HFE regulatory function. Nonetheless, HFE is expressed in multiple cell types, including hepatocytes[94], the major producers of hepcidin.
Definite clues as to the site of HFE regulatory function in the context of systemic iron homeostasis were recently provided by experiments with genetically engineered mice, bearing a targeted, tissue-specific disruption of the HFE gene. The technology is based on the generation of animals carrying a “floxed”HFE allele, surrounded by loxP sites, which are specific targets of the Cre recombinase. Crossing of “floxed”HFE mice with a transgenic line expressing the Cre recombinase under the control of the villin promoter resulted in intestinal-specific disruption of HFE in the progeny[95]. Importantly, mice lacking HFE expression in the intestine did not show any signs of abnormal iron metabolism, at least with regard to liver iron content, serum iron parameters and serum ferritin levels. Moreover, they exhibited physiological expression of the mRNAs encoding liver hepcidin and the intestinal iron transporters DMT1 and ferroportin[95]. By showing that intestinal HFE expression is dispensable for the regulation of body iron homeostasis, these data challenge the validity of the “crypt-programming model” and raise the possibility for a critical role of HFE in the liver.
In a follow-up study, “floxed”HFE mice were crossed with transgenic animals expressing the Cre recombinase under the control of either the hepatocyte-specific albumin promoter, or the macrophage-specific lysozyme M promoter[96]. While HFE ablation in macrophages did not affect body iron metabolism, the disruption of HFE in hepatocytes recapitulated the hemochromatosis phenotype of null HFE-/-mice. Thus, mice lacking HFE expression in hepatocytes exhibited hyperabsorption of dietary iron, increased serum iron, transferrin saturation and iron deposition in the liver[96]. Taken together, the tissue-specific knock-out experiments demonstrate that hepatocyte HFE is necessary to promote appropriate hepcidin responses and thereby prevent iron overload.
These data also corroborate clinical findings, showing that the iron status of recipients of a liver transplant was largely dependent on the HFE genotype of the donors[97,98]. Nevertheless, a contribution of macrophage HFE to hepcidin regulation cannot be completely ruled out. While macrophages are dispensable for hepcidin expression in response to iron or inflammatory signals[99,100], bone marrow transplantation from wild type mice into irradiated HFE-/-counterparts corrected iron parameters and significantly increased hepcidin levels in the recipients[101]. Conceivably, this could be the result of intercellular communication and signaling to hepatocytes and/or HFE-mediated autocrine production of hepcidin in macrophages[102].
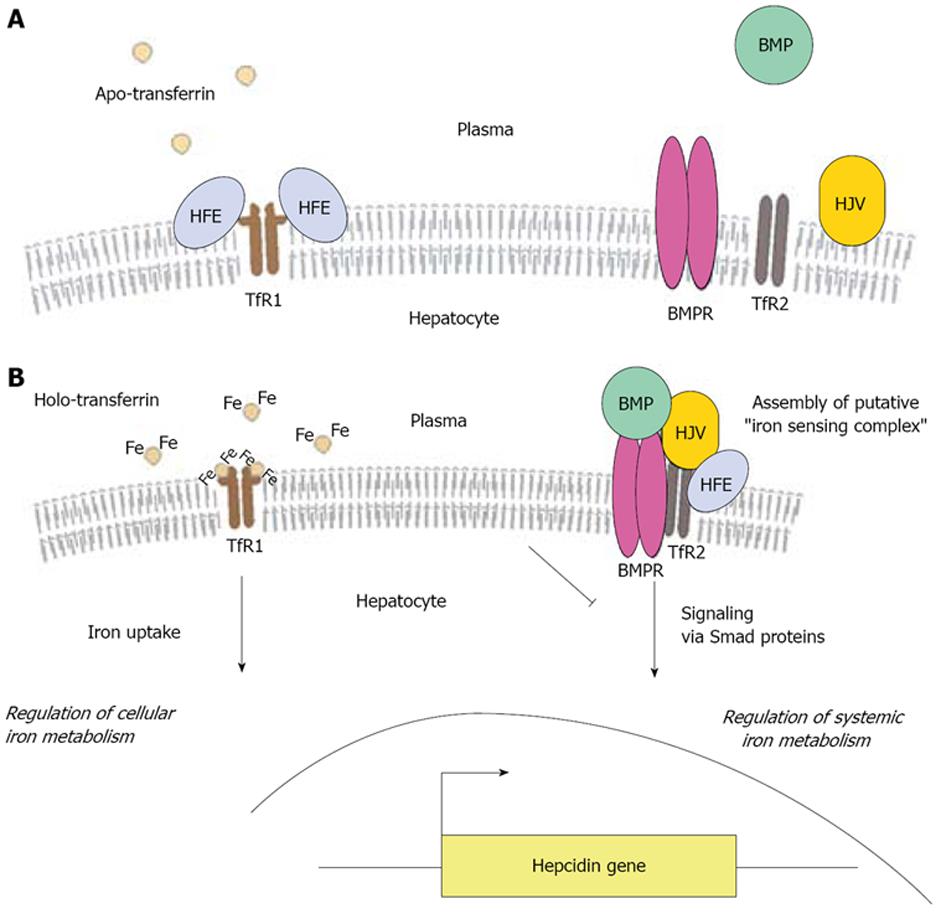
How does HFE modulate signaling to hepcidin? Biochemical work showed that HFE not only interacts with TfR1, but also with TfR2[103]. Moreover, the HFE/TfR2 interaction leads to an increase in TfR2 levels[104]. TfR2 is primarily expressed in hepatocytes[105] and stabilized by diferric holo-transferrin[106,107]. While TfR1 mediates cellular iron uptake from circulating transferrin, TfR2 is thought to function as an upstream regulator of hepcidin, and possibly an iron sensor[14]. A testable prediction arising from the capacity of HFE to interact with both TfR1 and TfR2 would be that the choice of its binding partner is regulated by transferrin and, furthermore, this event is crucial for signaling to hepcidin.
This hypothesis was explored in a recent study, based on the idea to induce or abolish HFE/TfR1 interactions in vivo[108]. To this end, transgenic mice were engineered for expression of TfR1 mutants that prevent the binding of either transferrin (R654A) or HFE (L622A). In light of the early embryonic lethality of TfR1-/-mice[109], indicating an utmost importance for the interaction of TfR1 with transferrin, a TfR1 R654A cDNA was integrated by homologous recombination into the heterologous ROSA26 locus, maintaining endogenous wild type TfR1 expression (thus, the transgenic product did not disrupt the transferrin cycle, excluding abnormalities of erythropoiesis). In contrast, the L622A mutation was introduced by homologous recombination within the TfR1 locus (“knock-in”).
TfR1 R654A, that is unable to bind to transferrin, would be expected to constitutively associate with HFE. Transgenic mice expressing TfR1 R654A developed iron overload, associated with decreased hepcidin mRNA levels, closely resembling the HFE-/-phenotype. On the other hand, TfR1 L622A, that is unable to bind to HFE, would be expected to be highly efficient in the uptake of transferrin-bound iron. Interestingly, transgenic mice expressing TfR1 L622A developed a mild hypochromic microcytic anemia, and exhibited decreased serum iron and elevated hepcidin mRNA levels. These results suggest that HFE stimulates hepcidin expression when it is free of TfR1. In support of this notion, the hepatocyte-specific transgenic overexpression of an HFE cDNA in HFE-/-mice substantially induced hepcidin mRNA expression to the extent that it not only corrected hepatic iron overload, but also promoted hypochromic microcytic anemia.
A model accommodating the above findings postulates that under low serum iron conditions, hepatocyte HFE is predominantly bound to TfR1 (Figure 2A). An increase in transferrin saturation triggers the release of HFE from TfR1 and concomitantly stabilizes TfR2[106,107]. In that way, TfR1 becomes accessible for the binding and endocytosis of holo-transferrin, resulting in cellular iron uptake. At the same time, HFE associates with stabilized TfR2 and possibly other proteins, such as hemojuvelin and BMPs and their receptor (BMPR), to form a putative iron signaling complex that induces hepcidin transcription via Smad proteins (Figure 2B). Thus, an increase in the iron content of the hepatocyte is indirectly translated into a systemic regulatory response via hepcidin. Iron-dependent degradation of TfR1 mRNA by iron regulatory proteins[110] would terminate this process in a feedback loop. According to this model, HFE serves to sense alterations in transferrin saturation.
Considering that a number of HH patients with HFE C282Y mutations[25] and some HFE-/-mice[29] express normal (or close to normal) basal hepcidin mRNA levels but exhibit blunted hepcidin responses to dietary iron, it is conceivable that the role of HFE is somehow restricted to the fine-tuning of iron-dependent signaling to hepcidin. Along these lines, BMP-2, -4 and -9 can induce hepcidin mRNA transcription in HFE-/-and TfR2-/-hepatocytes[41]. Several reports have also shown that HFE is dispensable for signaling to hepcidin via the inflammatory pathway[29,41,111,112], even though opposing views exist[113].
Recent animal studies[95,96,108] have not entirely solved the mystery of HFE function, but have significantly advanced our understanding on how this protein regulates systemic iron homeostasis. First, they uncovered HFE as a hepatocyte iron sensor, necessary to prevent iron overload and sufficient to control hepcidin expression (at least at the mRNA level). And second, they demonstrated that HFE-dependent signaling to hepcidin is regulated by the interaction of HFE with TfR1.
Several outstanding issues remain to be addressed. For example, the proposed function of HFE as a sensor of transferrin saturation requires experimental validation. The functional significance of the interaction between HFE and TfR2, as well as the role and composition of the putative iron-sensing complex await further investigation. It will be interesting to explore a potential functional redundancy between HFE and classical MHC classImolecules with regard to iron regulation, considering that mice lacking such molecules develop iron overload[114]. Conversely, the proposed capacity of HFE to engage into immune responses, following recognition by cytotoxic T lymphocytes[115], deserves additional attention, especially in light of immunological abnormalities of HH patients[116]. A possible connection between the unfolded protein response, caused by defective processing of HFE C282Y, and the hepcidin pathway would not be totally unexpected[117]. Finally, it will be important to examine whether HFE may also affect the maturation of hepcidin; this would necessitate analytical methods for direct measurement of the peptide in plasma[118,119] and in cell culture.
Peer reviewers: Debbie Trinder, Professor, School of Medicine and Pharmacology, University of Western Australia, Fremantle Hospital, PO 480, Fremantle 6950, Australia; Alberto Piperno, Professor, Department of Clinical Medicine and Prevention, University of Milano-Bicocca, Via Pergolesi 33, Monza 20052, Italy
S- Editor Zhong XY L- Editor Logan S E- Editor Ma WH
| 1. | Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285-297. |
| 2. | Papanikolaou G, Pantopoulos K. Iron metabolism and toxicity. Toxicol Appl Pharmacol. 2005;202:199-211. |
| 3. | Andrews NC. Disorders of iron metabolism. N Engl J Med. 1999;341:1986-1995. |
| 4. | Pietrangelo A. Hereditary hemochromatosis--a new look at an old disease. N Engl J Med. 2004;350:2383-2397. |
| 5. | Beutler E. Hemochromatosis: genetics and pathophysiology. Annu Rev Med. 2006;57:331-347. |
| 7. | Ramm GA, Ruddell RG. Hepatotoxicity of iron overload: mechanisms of iron-induced hepatic fibrogenesis. Semin Liver Dis. 2005;25:433-449. |
| 8. | Kowdley KV. Iron, hemochromatosis, and hepatocellular carcinoma. Gastroenterology. 2004;127:S79-S86. |
| 9. | Pietrangelo A. Hemochromatosis gene modifies course of hepatitis C viral infection. Gastroenterology. 2003;124:1509-1523. |
| 10. | Sebastiani G, Walker AP. HFE gene in primary and secondary hepatic iron overload. World J Gastroenterol. 2007;13:4673-4689. |
| 11. | Andrews NC, Schmidt PJ. Iron homeostasis. Annu Rev Physiol. 2007;69:69-85. |
| 12. | De Domenico I, McVey Ward D, Kaplan J. Regulation of iron acquisition and storage: consequences for iron-linked disorders. Nat Rev Mol Cell Biol. 2008;9:72-81. |
| 13. | Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090-2093. |
| 14. | Nemeth E, Ganz T. Regulation of iron metabolism by hepcidin. Annu Rev Nutr. 2006;26:323-342. |
| 15. | Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811-7819. |
| 16. | Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, Ganz T. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271-1276. |
| 17. | Weiss G, Goodnough LT. Anemia of chronic disease. N Engl J Med. 2005;352:1011-1023. |
| 18. | Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, Devaux I, Beaumont C, Kahn A, Vaulont S. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest. 2002;110:1037-1044. |
| 19. | Gehrke SG, Kulaksiz H, Herrmann T, Riedel HD, Bents K, Veltkamp C, Stremmel W. Expression of hepcidin in hereditary hemochromatosis: evidence for a regulation in response to the serum transferrin saturation and to non-transferrin-bound iron. Blood. 2003;102:371-376. |
| 20. | Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, Subramaniam VN, Powell LW, Anderson GJ, Ramm GA. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet. 2003;361:669-673. |
| 21. | Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, Loukopoulos D, Camaschella C. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21-22. |
| 22. | Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77-82. |
| 23. | Feder JN, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy DA, Basava A, Dormishian F, Domingo R Jr, Ellis MC, Fullan A. A novel MHC class I-like gene is mutated in patients with hereditary haemochromatosis. Nat Genet. 1996;13:399-408. |
| 24. | Camaschella C, Roetto A, Cali A, De Gobbi M, Garozzo G, Carella M, Majorano N, Totaro A, Gasparini P. The gene TFR2 is mutated in a new type of haemochromatosis mapping to 7q22. Nat Genet. 2000;25:14-15. |
| 25. | Piperno A, Girelli D, Nemeth E, Trombini P, Bozzini C, Poggiali E, Phung Y, Ganz T, Camaschella C. Blunted hepcidin response to oral iron challenge in HFE-related hemochromatosis. Blood. 2007;110:4096-4100. |
| 26. | Nemeth E, Roetto A, Garozzo G, Ganz T, Camaschella C. Hepcidin is decreased in TFR2 hemochromatosis. Blood. 2005;105:1803-1806. |
| 27. | Ahmad KA, Ahmann JR, Migas MC, Waheed A, Britton RS, Bacon BR, Sly WS, Fleming RE. Decreased liver hepcidin expression in the Hfe knockout mouse. Blood Cells Mol Dis. 2002;29:361-366. |
| 28. | Muckenthaler M, Roy CN, Custodio AO, Minana B, deGraaf J, Montross LK, Andrews NC, Hentze MW. Regulatory defects in liver and intestine implicate abnormal hepcidin and Cybrd1 expression in mouse hemochromatosis. Nat Genet. 2003;34:102-107. |
| 29. | Constante M, Jiang W, Wang D, Raymond VA, Bilodeau M, Santos MM. Distinct requirements for Hfe in basal and induced hepcidin levels in iron overload and inflammation. Am J Physiol Gastrointest Liver Physiol. 2006;291:G229-G237. |
| 30. | Ludwiczek S, Theurl I, Bahram S, Schumann K, Weiss G. Regulatory networks for the control of body iron homeostasis and their dysregulation in HFE mediated hemochromatosis. J Cell Physiol. 2005;204:489-499. |
| 31. | Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115:2187-2191. |
| 32. | Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115:2180-2186. |
| 33. | Kawabata H, Fleming RE, Gui D, Moon SY, Saitoh T, O’Kelly J, Umehara Y, Wano Y, Said JW, Koeffler HP. Expression of hepcidin is down-regulated in TfR2 mutant mice manifesting a phenotype of hereditary hemochromatosis. Blood. 2005;105:376-381. |
| 34. | Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108:3204-3209. |
| 35. | Pietrangelo A, Dierssen U, Valli L, Garuti C, Rump A, Corradini E, Ernst M, Klein C, Trautwein C. STAT3 is required for IL-6-gp130-dependent activation of hepcidin in vivo. Gastroenterology. 2007;132:294-300. |
| 36. | Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353-358. |
| 37. | Truksa J, Peng H, Lee P, Beutler E. Different regulatory elements are required for response of hepcidin to interleukin-6 and bone morphogenetic proteins 4 and 9. Br J Haematol. 2007;139:138-147. |
| 38. | Truksa J, Lee P, Peng H, Flanagan J, Beutler E. The distal location of the iron responsive region of the hepcidin promoter. Blood. 2007;110:3436-3437. |
| 39. | Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399-409. |
| 40. | Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531-539. |
| 41. | Truksa J, Peng H, Lee P, Beutler E. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL-6. Proc Natl Acad Sci USA. 2006;103:10289-10293. |
| 42. | Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117:1933-1939. |
| 43. | Courselaud B, Pigeon C, Inoue Y, Inoue J, Gonzalez FJ, Leroyer P, Gilot D, Boudjema K, Guguen-Guillouzo C, Brissot P. C/EBPalpha regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Cross-talk between C/EBP pathway and iron metabolism. J Biol Chem. 2002;277:41163-41170. |
| 44. | Pak M, Lopez MA, Gabayan V, Ganz T, Rivera S. Suppression of hepcidin during anemia requires erythropoietic activity. Blood. 2006;108:3730-3735. |
| 45. | Vokurka M, Krijt J, Sulc K, Necas E. Hepcidin mRNA levels in mouse liver respond to inhibition of erythropoiesis. Physiol Res. 2006;55:667-674. |
| 46. | Tanno T, Bhanu NV, Oneal PA, Goh SH, Staker P, Lee YT, Moroney JW, Reed CH, Luban NL, Wang RH. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat Med. 2007;13:1096-1101. |
| 47. | Pinto JP, Ribeiro S, Pontes H, Thowfeequ S, Tosh D, Carvalho F, Porto G. Erythropoietin mediates hepcidin expression in hepatocytes through EPOR signaling and regulation of C/EBPalpha. Blood. 2008;111:5727-5733. |
| 48. | Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, Nizet V, Johnson RS. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs). J Clin Invest. 2007;117:1926-1932. |
| 49. | Choi SO, Cho YS, Kim HL, Park JW. ROS mediate the hypoxic repression of the hepcidin gene by inhibiting C/EBPalpha and STAT-3. Biochem Biophys Res Commun. 2007;356:312-317. |
| 50. | Braliou GG, Verga Falzacappa MV, Chachami G, Casanovas G, Muckenthaler MU, Simos G. 2-Oxoglutarate-dependent oxygenases control hepcidin gene expression. J Hepatol. 2008;48:801-810. |
| 51. | Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EM. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320:1088-1092. |
| 52. | Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM, Strouse JJ, Markianos K. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA). Nat Genet. 2008;40:569-571. |
| 53. | Folgueras AR, de Lara FM, Pendas AM, Garabaya C, Rodriguez F, Astudillo A, Bernal T, Cabanillas R, Lopez-Otin C, Velasco G. Membrane-bound serine protease matriptase-2 (Tmprss6) is an essential regulator of iron homeostasis. Blood. 2008;112:2539-2545. |
| 54. | Guillem F, Lawson S, Kannengiesser C, Westerman M, Beaumont C, Grandchamp B. Two nonsense mutations in the TMPRSS6 gene in a patient with microcytic anemia and iron deficiency. Blood. 2008;112:2089-2091. |
| 55. | Melis MA, Cau M, Congiu R, Sole G, Barella S, Cao A, Westerman M, Cazzola M, Galanello R. A mutation in the TMPRSS6 gene, encoding a transmembrane serine protease that suppresses hepcidin production, in familial iron deficiency anemia refractory to oral iron. Haematologica. 2008;93:1473-1479. |
| 56. | Lebron JA, Bennett MJ, Vaughn DE, Chirino AJ, Snow PM, Mintier GA, Feder JN, Bjorkman PJ. Crystal structure of the hemochromatosis protein HFE and characterization of its interaction with transferrin receptor. Cell. 1998;93:111-123. |
| 57. | Feder JN, Tsuchihashi Z, Irrinki A, Lee VK, Mapa FA, Morikang E, Prass CE, Starnes SM, Wolff RK, Parkkila S. The hemochromatosis founder mutation in HLA-H disrupts beta2-microglobulin interaction and cell surface expression. J Biol Chem. 1997;272:14025-14028. |
| 58. | Waheed A, Parkkila S, Zhou XY, Tomatsu S, Tsuchihashi Z, Feder JN, Schatzman RC, Britton RS, Bacon BR, Sly WS. Hereditary hemochromatosis: effects of C282Y and H63D mutations on association with beta2-microglobulin, intracellular processing, and cell surface expression of the HFE protein in COS-7 cells. Proc Natl Acad Sci USA. 1997;94:12384-12389. |
| 59. | de Almeida SF, Fleming JV, Azevedo JE, Carmo-Fonseca M, de Sousa M. Stimulation of an unfolded protein response impairs MHC class I expression. J Immunol. 2007;178:3612-3619. |
| 60. | Allen KJ, Gurrin LC, Constantine CC, Osborne NJ, Delatycki MB, Nicoll AJ, McLaren CE, Bahlo M, Nisselle AE, Vulpe CD. Iron-overload-related disease in HFE hereditary hemochromatosis. N Engl J Med. 2008;358:221-230. |
| 61. | Waalen J, Beutler E. Iron-overload-related disease in HFE hereditary hemochromatosis. N Engl J Med. 2008;358:2293-2294; author reply 2294-2295. |
| 62. | Beutler E. Iron storage disease: facts, fiction and progress. Blood Cells Mol Dis. 2007;39:140-147. |
| 63. | Zhou XY, Tomatsu S, Fleming RE, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt EM, Ruddy DA, Prass CE. HFE gene knockout produces mouse model of hereditary hemochromatosis. Proc Natl Acad Sci USA. 1998;95:2492-2497. |
| 64. | Bahram S, Gilfillan S, Kuhn LC, Moret R, Schulze JB, Lebeau A, Schumann K. Experimental hemochromatosis due to MHC class I HFE deficiency: immune status and iron metabolism. Proc Natl Acad Sci USA. 1999;96:13312-13317. |
| 65. | de Sousa M, Reimao R, Lacerda R, Hugo P, Kaufmann SH, Porto G. Iron overload in beta 2-microglobulin-deficient mice. Immunol Lett. 1994;39:105-111. |
| 66. | Rothenberg BE, Voland JR. beta2 knockout mice develop parenchymal iron overload: A putative role for class I genes of the major histocompatibility complex in iron metabolism. Proc Natl Acad Sci USA. 1996;93:1529-1534. |
| 67. | Levy JE, Montross LK, Cohen DE, Fleming MD, Andrews NC. The C282Y mutation causing hereditary hemochro-matosis does not produce a null allele. Blood. 1999;94:9-11. |
| 68. | Tomatsu S, Orii KO, Fleming RE, Holden CC, Waheed A, Britton RS, Gutierrez MA, Velez-Castrillon S, Bacon BR, Sly WS. Contribution of the H63D mutation in HFE to murine hereditary hemochromatosis. Proc Natl Acad Sci USA. 2003;100:15788-15793. |
| 69. | Fleming RE, Holden CC, Tomatsu S, Waheed A, Brunt EM, Britton RS, Bacon BR, Roopenian DC, Sly WS. Mouse strain differences determine severity of iron accumulation in Hfe knockout model of hereditary hemochromatosis. Proc Natl Acad Sci USA. 2001;98:2707-2711. |
| 70. | Levy JE, Montross LK, Andrews NC. Genes that modify the hemochromatosis phenotype in mice. J Clin Invest. 2000;105:1209-1216. |
| 71. | Sproule TJ, Jazwinska EC, Britton RS, Bacon BR, Fleming RE, Sly WS, Roopenian DC. Naturally variant autosomal and sex-linked loci determine the severity of iron overload in beta 2-microglobulin-deficient mice. Proc Natl Acad Sci USA. 2001;98:5170-5174. |
| 72. | Feder JN, Penny DM, Irrinki A, Lee VK, Lebron JA, Watson N, Tsuchihashi Z, Sigal E, Bjorkman PJ, Schatzman RC. The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci USA. 1998;95:1472-1477. |
| 73. | Lebron JA, West AP Jr, Bjorkman PJ. The hemochromatosis protein HFE competes with transferrin for binding to the transferrin receptor. J Mol Biol. 1999;294:239-245. |
| 74. | Bennett MJ, Lebron JA, Bjorkman PJ. Crystal structure of the hereditary haemochromatosis protein HFE complexed with transferrin receptor. Nature. 2000;403:46-53. |
| 75. | West AP Jr, Bennett MJ, Sellers VM, Andrews NC, Enns CA, Bjorkman PJ. Comparison of the interactions of transferrin receptor and transferrin receptor 2 with transferrin and the hereditary hemochromatosis protein HFE. J Biol Chem. 2000;275:38135-38138. |
| 76. | Ponka P, Beaumont C, Richardson DR. Function and regulation of transferrin and ferritin. Semin Hematol. 1998;35:35-54. |
| 77. | Roy CN, Penny DM, Feder JN, Enns CA. The hereditary hemochromatosis protein, HFE, specifically regulates transferrin-mediated iron uptake in HeLa cells. J Biol Chem. 1999;274:9022-9028. |
| 78. | Riedel HD, Muckenthaler MU, Gehrke SG, Mohr I, Brennan K, Herrmann T, Fitscher BA, Hentze MW, Stremmel W. HFE downregulates iron uptake from transferrin and induces iron-regulatory protein activity in stably transfected cells. Blood. 1999;94:3915-3921. |
| 79. | Muckenthaler M, Richter A, Gunkel N, Riedel D, Polycarpou-Schwarz M, Hentze S, Falkenhahn M, Stremmel W, Ansorge W, Hentze MW. Relationships and distinctions in iron-regulatory networks responding to interrelated signals. Blood. 2003;101:3690-3698. |
| 80. | Corsi B, Levi S, Cozzi A, Corti A, Altimare D, Albertini A, Arosio P. Overexpression of the hereditary hemochromatosis protein, HFE, in HeLa cells induces and iron-deficient phenotype. FEBS Lett. 1999;460:149-152. |
| 81. | Gross CN, Irrinki A, Feder JN, Enns CA. Co-trafficking of HFE, a nonclassical major histocompatibility complex class I protein, with the transferrin receptor implies a role in intracellular iron regulation. J Biol Chem. 1998;273:22068-22074. |
| 82. | Wang J, Chen G, Pantopoulos K. The haemochromatosis protein HFE induces an apparent iron-deficient phenotype in H1299 cells that is not corrected by co-expression of beta 2-microglobulin. Biochem J. 2003;370:891-899. |
| 83. | Zhang AS, Davies PS, Carlson HL, Enns CA. Mechanisms of HFE-induced regulation of iron homeostasis: Insights from the W81A HFE mutation. Proc Natl Acad Sci USA. 2003;100:9500-9505. |
| 84. | Parkkila S, Waheed A, Britton RS, Feder JN, Tsuchihashi Z, Schatzman RC, Bacon BR, Sly WS. Immunohistochemistry of HLA-H, the protein defective in patients with hereditary hemochromatosis, reveals unique pattern of expression in gastrointestinal tract. Proc Natl Acad Sci USA. 1997;94:2534-2539. |
| 85. | Waheed A, Parkkila S, Saarnio J, Fleming RE, Zhou XY, Tomatsu S, Britton RS, Bacon BR, Sly WS. Association of HFE protein with transferrin receptor in crypt enterocytes of human duodenum. Proc Natl Acad Sci USA. 1999;96:1579-1584. |
| 87. | Trinder D, Olynyk JK, Sly WS, Morgan EH. Iron uptake from plasma transferrin by the duodenum is impaired in the Hfe knockout mouse. Proc Natl Acad Sci USA. 2002;99:5622-5626. |
| 88. | Davies PS, Enns CA. Expression of the hereditary hemochromatosis protein HFE increases ferritin levels by inhibiting iron export in HT29 cells. J Biol Chem. 2004;279:25085-25092. |
| 89. | Pietrangelo A, Rocchi E, Casalgrandi G, Rigo G, Ferrari A, Perini M, Ventura E, Cairo G. Regulation of transferrin, transferrin receptor, and ferritin genes in human duodenum. Gastroenterology. 1992;102:802-809. |
| 90. | Pietrangelo A, Casalgrandi G, Quaglino D, Gualdi R, Conte D, Milani S, Montosi G, Cesarini L, Ventura E, Cairo G. Duodenal ferritin synthesis in genetic hemochromatosis. Gastroenterology. 1995;108:208-217. |
| 91. | Montosi G, Paglia P, Garuti C, Guzman CA, Bastin JM, Colombo MP, Pietrangelo A. Wild-type HFE protein normalizes transferrin iron accumulation in macrophages from subjects with hereditary hemochromatosis. Blood. 2000;96:1125-1129. |
| 92. | Drakesmith H, Sweetland E, Schimanski L, Edwards J, Cowley D, Ashraf M, Bastin J, Townsend AR. The hemochromatosis protein HFE inhibits iron export from macrophages. Proc Natl Acad Sci USA. 2002;99:15602-15607. |
| 93. | Moura E, Noordermeer MA, Verhoeven N, Verheul AF, Marx JJ. Iron release from human monocytes after erythrophagocytosis in vitro: an investigation in normal subjects and hereditary hemochromatosis patients. Blood. 1998;92:2511-2519. |
| 94. | Zhang AS, Xiong S, Tsukamoto H, Enns CA. Localization of iron metabolism-related mRNAs in rat liver indicate that HFE is expressed predominantly in hepatocytes. Blood. 2004;103:1509-1514. |
| 95. | Vujic Spasic M, Kiss J, Herrmann T, Kessler R, Stolte J, Galy B, Rathkolb B, Wolf E, Stremmel W, Hentze MW. Physiologic systemic iron metabolism in mice deficient for duodenal Hfe. Blood. 2007;109:4511-4517. |
| 96. | Vujic Spasic M, Kiss J, Herrmann T, Galy B, Martinache S, Stolte J, Grone HJ, Stremmel W, Hentze MW, Muckenthaler MU. Hfe acts in hepatocytes to prevent hemochromatosis. Cell Metab. 2008;7:173-178. |
| 97. | Wigg AJ, Harley H, Casey G. Heterozygous recipient and donor HFE mutations associated with a hereditary haemochromatosis phenotype after liver transplantation. Gut. 2003;52:433-435. |
| 98. | Crawford DH, Fletcher LM, Hubscher SG, Stuart KA, Gane E, Angus PW, Jeffrey GP, McCaughan GW, Kerlin P, Powell LW. Patient and graft survival after liver transplantation for hereditary hemochromatosis: Implications for pathogenesis. Hepatology. 2004;39:1655-1662. |
| 99. | Montosi G, Corradini E, Garuti C, Barelli S, Recalcati S, Cairo G, Valli L, Pignatti E, Vecchi C, Ferrara F. Kupffer cells and macrophages are not required for hepatic hepcidin activation during iron overload. Hepatology. 2005;41:545-552. |
| 100. | Lou DQ, Lesbordes JC, Nicolas G, Viatte L, Bennoun M, Van Rooijen N, Kahn A, Renia L, Vaulont S. Iron- and inflammation-induced hepcidin gene expression in mice is not mediated by Kupffer cells in vivo. Hepatology. 2005;41:1056-1064. |
| 101. | Makui H, Soares RJ, Jiang W, Constante M, Santos MM. Contribution of Hfe expression in macrophages to the regulation of hepatic hepcidin levels and iron loading. Blood. 2005;106:2189-2195. |
| 102. | Theurl I, Theurl M, Seifert M, Mair S, Nairz M, Rumpold H, Zoller H, Bellmann-Weiler R, Niederegger H, Talasz H. Autocrine formation of hepcidin induces iron retention in human monocytes. Blood. 2008;111:2392-2399. |
| 103. | Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281:28494-28498. |
| 104. | Chen J, Chloupkova M, Gao J, Chapman-Arvedson TL, Enns CA. HFE modulates transferrin receptor 2 levels in hepatoma cells via interactions that differ from transferrin receptor 1-HFE interactions. J Biol Chem. 2007;282:36862-36870. |
| 105. | Kawabata H, Yang R, Hirama T, Vuong PT, Kawano S, Gombart AF, Koeffler HP. Molecular cloning of transferrin receptor 2. A new member of the transferrin receptor-like family. J Biol Chem. 1999;274:20826-20832. |
| 106. | Johnson MB, Enns CA. Diferric transferrin regulates transferrin receptor 2 protein stability. Blood. 2004;104:4287-4293. |
| 107. | Robb A, Wessling-Resnick M. Regulation of transferrin receptor 2 protein levels by transferrin. Blood. 2004;104:4294-4299. |
| 108. | Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7:205-214. |
| 109. | Levy JE, Jin O, Fujiwara Y, Kuo F, Andrews NC. Transferrin receptor is necessary for development of erythrocytes and the nervous system. Nat Genet. 1999;21:396-399. |
| 110. | Pantopoulos K. Iron metabolism and the IRE/IRP regulatory system: an update. Ann N Y Acad Sci. 2004;1012:1-13. |
| 111. | Lee P, Peng H, Gelbart T, Beutler E. The IL-6- and lipopolysaccharide-induced transcription of hepcidin in HFE-, transferrin receptor 2-, and beta 2-microglobulin-deficient hepatocytes. Proc Natl Acad Sci USA. 2004;101:9263-9265. |
| 112. | Frazer DM, Wilkins SJ, Millard KN, McKie AT, Vulpe CD, Anderson GJ. Increased hepcidin expression and hypoferraemia associated with an acute phase response are not affected by inactivation of HFE. Br J Haematol. 2004;126:434-436. |
| 113. | Roy CN, Custodio AO, de Graaf J, Schneider S, Akpan I, Montross LK, Sanchez M, Gaudino A, Hentze MW, Andrews NC. An Hfe-dependent pathway mediates hyposideremia in response to lipopolysaccharide-induced inflammation in mice. Nat Genet. 2004;36:481-485. |
| 114. | Cardoso EM, Macedo MG, Rohrlich P, Ribeiro E, Silva MT, Lemonnier FA, de Sousa M. Increased hepatic iron in mice lacking classical MHC class I molecules. Blood. 2002;100:4239-4241. |
| 115. | Rohrlich PS, Fazilleau N, Ginhoux F, Firat H, Michel F, Cochet M, Laham N, Roth MP, Pascolo S, Nato F. Direct recognition by alphabeta cytolytic T cells of Hfe, a MHC class Ib molecule without antigen-presenting function. Proc Natl Acad Sci USA. 2005;102:12855-12860. |
| 116. | Porto G, De Sousa M. Iron overload and immunity. World J Gastroenterol. 2007;13:4707-4715. |
| 117. | de Almeida SF, de Sousa M. The unfolded protein response in hereditary haemochromatosis. J Cell Mol Med. 2008;12:421-434. |