Published online Oct 21, 2008. doi: 10.3748/wjg.14.6044
Revised: August 11, 2008
Accepted: August 18, 2008
Published online: October 21, 2008
AIM: To investigate the prevalence and genotype distribution of Torque teno virus (TTV) in patients with different liver diseases and chronic renal failure treated at a referral hospital in North India.
METHODS: Whereas prevalence of TTV was based on amplification of conserved region of ORF2 of TTV genome, the genotyping of TTV was carried out using restriction fragment length polymorphism (RFLP) procedure on the N22 region of ORF1.
RESULTS: TTV-DNA was detected in 137 of 513 (26.7%) patients with liver diseases and 38 of 65 (58.5%) patients with chronic renal failure. TTV was also detected in 27% of healthy controls. The sequence analysis of the PCR product from 10 randomly selected cases failed to show a significant sequence divergence when compared with that of the TRM1 isolate of TTV genotype 1. The results of genotyping in 55 randomly selected patients showed the presence of genotype 1 (G1) in 53 (96.4%) and genotype 2 (G2) in 2 cases (3.6%), respectively. Other genotypes were not identified in this patient subgroup, suggesting that G1 is predominant in this area. The results of genotyping by RFLP were also supported by phylogenetic tree analysis, where G1 was found to be the major genotype.
CONCLUSION: These results indicate that TTV is moderately present in Indian patients, with G1 to be the major genotype in North India. The pathogenicity and etiological role of TTV in different diseases is still a question mark and warrant further studies.
- Citation: Irshad M, Singh S, Irshad K, Agarwal SK, Joshi YK. Torque teno virus: Its prevalence and isotypes in North India. World J Gastroenterol 2008; 14(39): 6044-6051
- URL: https://www.wjgnet.com/1007-9327/full/v14/i39/6044.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.6044
| Disease group | Number of samples | TTV-DNA positivity n (%) |
| Acute viral hepatitis (AVH) | 126 | 29 (23.0) |
| Chronic viral hepatitis (CVH) | 111 | 26 (23.4) |
| Cirrhosis (CIR) | 132 | 46 (34.8) |
| Fulminant hepatic failure (FHF) | 51 | 15 (29.4) |
| Hepatocellular carcinoma (HCC) | 93 | 21 (22.6) |
| Chronic renal failure (CRF) | 65 | 38 (58.5) |
| Healthy controls | 100 | 27 (27.0) |
| Diseases group | TTV-DNA+ samples | ALT level (IU/L) | |||
| 50-200 | 201-400 | 401-600 | > 600 | ||
| AVH | 29 | 17 (59) | 8 (28) | 0 | 4 (14) |
| CVH | 26 | 18 (69) | 6 (23) | 0 | 2 (8) |
| CIR | 46 | 35 (76) | 19 (41) | 0 | 2 (4) |
| FHF | 15 | 2 (13) | 6 (40) | 1 (7) | 6 (40) |
| HCC | 21 | 8 (38) | 5 (24) | 3 (14) | 5 (24) |
| Restrictionenzymes used | Number of cases treated | Digestion noticed | Possible TTV genotypes | Genotypes prevalence |
| Nde I | 55 | 53 | G1 | 53 (96.4) |
| Pst I | 55 | 2 | G2 | 2 (3.6) |
| Mse I | 55 | 0 | - | 0 |
| Nla III | 55 | 0 | - | 0 |
| Each sample was treated with four restriction enzymes (Nde I, Pst I, Mse I, Nla III) to determine genotyping of TTV following criteria mentioned in material and methods | ||||
| The DNA remaining undigested with REs indicated absence of corresponding genotypes | ||||
| There was no effect of Mse I and Nla III on TTV-DNA on digestion with these enzymes | ||||
Torque teno virus (TTV), formerly known as transfusion-transmitted virus, is a small, non-enveloped, icosahedral, single-stranded, circular-DNA virus[1] approximately 30 nm in diameter. TTV contains a 3.6-3.9 kb DNA genome of minus polarity[2-4] and belongs to a novel virus family, Circoviridae[3]. At least 40 TTV genotypes from 5 major phylogenetic groups have been identified[5]. They display over 30% nucleotide diversity[6]. TTVs are ubiquitous in nature and have been demonstrated in high proportion of serum samples from healthy individuals where they persist overtime[3,7-9]. A tissue culture system that supports efficient replication of TTV is not available[10], and this has delayed the study of both TTV genome replication and TTV gene expression. Antibodies reacting with two proteins encoded by the 2.8-kb mRNA have been detected in TTV-infected individuals[11,12]. The first is the large protein encoded by TTV ORF1 (the ORF1 protein), which is predicted to be 736 amino acids in length and initiates from a methionine at nucleotide (nt) 581 (O1AUG). The second protein is encoded by the 2.8-kb mRNA in ORF2, initiating at a methionine in the position of nt 354 (O2AUG) and extending for 117 amino acids[13].
TTV belongs to the group of Anelloviruses that are widely diverse. Indeed, based on their heterogeneity, Anelloviruses are currently classified into two species, each subdivided into numerous genotypes. Thus, TTV, the first anellovirus species identified[1], is currently sub-divided into approximately 40 genotypes, which cluster in five clearly distinct phylogenetic groups designated from 1 to 5[1,6,10,14]. TTV is transmitted parenterally through transfusion with blood or blood products, but the natural route of its transmission is still unknown[15,16].
TTV is found in the plasma of > 80% of the human population worldwide. Co-infection of single individuals with multiple TTV isolates is frequent[17]. The epidemiology and pathogenic potential of TTV is poorly understood. In several studies, however, the viral genome has been detected at comparable prevalence rates in the blood of healthy persons and patients and this led to the hypothesis that TTV might be essentially non-pathogenic in nature[18].
TTV can be transmitted by parenteral route, although its role in causing post-transfusion hepatitis has not been established. The majority of individuals who become TTV-DNA-positive after blood transfusion usually have normal ALT and do not develop chronic hepatitis, although TTV viremia frequently persists for several years. Patients who develop chronic hepatitis are invariably coinfected with HBV or HCV and chronic hepatitis is closely correlated with HBV or HCV infection. This raises the possibility that TTV is merely an innocent bystander rather than a primary hepatitis virus[19].
Although TTV appears to be widespread in the general population of several geographical regions, its prevalence in many areas is still unknown. The reports on status of TTV available from India are very preliminary and therefore there is a need of extensive studies to understand the endemicity, epidemiology and etiological potential of TTV infection in various diseases. Also, very little is known about the genotyping of TTV strains circulating in this country. Thus, the present study was undertaken to elucidate the prevalence and detect genotypes distribution of TTV in patients with liver and renal diseases in North India.
Five hundred and seventy eight adult patients of both sexes were included. There were 126 patients with acute viral hepatitis (AVH, age range: 21-48 years), 111 patients with chronic viral hepatitis (CVH, age range: 19-48 years), 132 patients with liver cirrhosis (CIR, age range: 34-57 years), 51 patients with fulminant hepatic failure (FHF, age range: 28-46 years), 93 patients with hepatocellular carcinoma (HCC, age range: 24-71 years) and 65 patients with chronic renal failure (CRF, age range: 20-74 years)[19]. All these patients attended either the Outpatient Department or were admitted to the Liver and Renal Units of All India Institute of Medical Sciences, New Delhi, from June 2001 to March 2008. They were evaluated clinically and biochemically and their sera were tested for various markers and parameters. The diagnosis of different types of diseases was based on accepted clinical, biochemical and histological criteria as outlined elsewhere[20].
AVH was diagnosed when patients exhibited overt jaundice and/or increased alanine aminotransferase levels (at least 3 times above the normal value) documented at least twice at a 1-wk interval without any history of pre-existing liver disease. None of the patients had a past history of alcohol intake or were using any drug or had clinical or serological evidence of autoimmune diseases or biliary infection. The patients with CVH and liver cirrhosis were diagnosed based on histopathological criteria established by the International Study Group on Chronic Hepatitis[21]: All of them had persistent elevation of transaminases (at least twice the upper limit of the normal range) for more than six months and histologic evidence of chronic hepatitis on liver biopsy at the beginning of follow-up. FHF was diagnosed if the patients developed hepatic encephalopathy within 4 wk from the onset of acute hepatitis, as outlined elsewhere[20]. The diagnosis of HCC was based on histological criteria. CRF was diagnosed using criteria as detailed elsewhere[22]. One hundred age- and sex-matched healthy subjects were used as controls.
From each of the above patients, 6-10 mL of venous blood was drawn and aliquoted in plain tubes without anticoagulant. Serum was separated after centrifugation and then stored at -70°C until further analysis. Repeated freezing and thawing of serum was avoided as far as possible. These serum samples were used to analyze liver function tests and routine hematogram.
Viral DNA was extracted from 200 μL of sera stored at -20°C using QiAmp Mini Elute viral spin kit (Qiagen, Germany) and following the manufacturer’s instructions. The DNA was eluted in 50 μL of elution buffer supplied with the kit. TTV-DNA (conserved region of ORF2) was detected by nested PCR using primers NS1 (sense) 5'-GGGTGCCGAAGGTGAGTTTAC-3' (175-195), NS2 (anti-sense) 5'-GCGGGGCACGAA-GCACAGAAG-3' (474-494), NS3 (sense) 5'-AGTTTA-CACACCGAAGTCAAG-3' (189-209) and NS4 (anti-sense) 5'-AGCACAGAAGCAAGATGATTA-3' (463-483) as described by Biagini et al[23], 1999 (Accession No. AB008394). Briefly, 10 μL of DNA was used for the amplification in a 50 μL reaction mixture containing 10 × PCR buffer, 25 pmol/μL of each primer, 10 mmol/L of each dNTPs, and 1.5 U Taq polymerase (Qiagen). Each of the 35 cycles of the 1st round and 25 cycles for the 2nd round of amplification consisted in an initial denaturation step at 95°C for 5 min and a cycling denaturation at 94°C for 30 s, annealing at 60°C for 45 s, extension at 74°C for 45 s, with a final extension for 3 min at 74°C.
Genotyping of TTV was done by amplifying the N22 region using specific primers as described by Okamoto et al[4], 1999. Briefly, a first round of amplification was performed with sense primer NG059 5'-ACAGAC-AGAGGAGAAGGCAACATG-3' and anti-sense primer NG063 5'-CTGGCATTTTACCATTTCCAAAGTT-3' for 10 min at 95°C (initial denaturation) followed by 35 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 1 min, and extension at 74°C for 1 min with a final extension at 74°C for 5 min. The second round of PCR was performed using sense primer NG061 5'-GGCAACATGYTRTGGATAGACTGG-3' and anti-sense primer NG063 5'-CTGGCATTTT-ACCATTTCCAAAGTT-3' following same conditions as used for first round amplification. Three microliters of the PCR product were electrophoresed on 2% agarose gel and stained by ethidium bromide to observe as 295 bp product of ORF2 region and 271 bp product of N22 region.
PCR products of ORF2 were recovered from 2% agarose gels after staining with ethidium bromide and visualized under UV and were purified with QIAquick Gel Extraction Kit, Qiagen, Germany. They were sequenced in both directions using an automated DNA sequencer at M/s Lab India. The same set of primers was used for both sequencing and amplification by PCR.
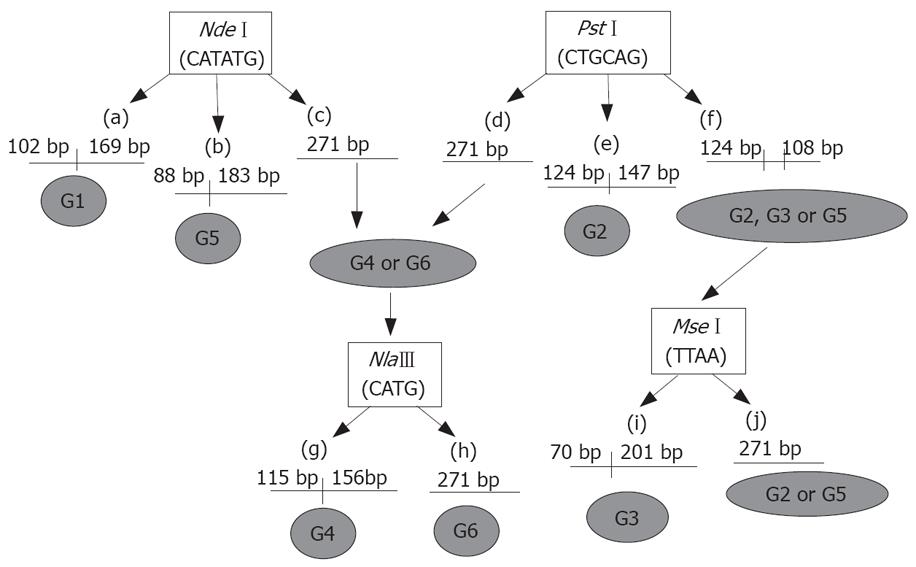
Restriction digestion was carried out overnight using 10 μL of the second round PCR product of the N22 region and 10 × enzyme buffer according to the manufacturer’s instructions. Reactions were carried out with 20 U each of NdeI, PstI (New England Biolabs, MA, USA), NlaIII (MBI Fermantas, Canada) at 37°C. Similarly, 20 U of MseI (MBI Fermantas, Canada) were used for digestion at 65°C. The digested PCR products were electrophoresed on 2% agarose gel and stained with ethidium bromide. The RFLP pattern was then evaluated under UV light[24]. The procedure using restriction digestion enzymes for genotyping by RFLP is shown schematically in Figure 1. Briefly, restriction digestion pattern with NdeI producing two fragments of 102 bp and 169 bp shows the presence of genotype 1 (G1). Production of fragments of 88 bp and 183 bp with NdeI shows the presence of genotype 5 (G5). Similarly, a digestion pattern with PstI producing two fragments of 124 bp and 147 bp size shows the presence of genotype 2 (G2). Restriction digestion with NlaIII produces two fragments of 115 bp and 156 bp if genotype 4 (G4) is present. Digestion pattern of MseI, producing two fragments of 70 bp and 201 bp length, shows the presence of genotype 3 (G3). Genotype 6 (G6) does not contain site of any of the above restriction enzymes.
DNA sequences derived from TTV ORF2 positive samples were compared to an online database for the best possible match using the BLAST (Basic Local Alignment Search Tool) program of National Center for Biotechnology Information (http://www.ncbi.nlm.nic.gov) and CLUSTALX program[25]. Phylogenetic tree was constructed using the neighbor-joining method in CLUSTALX program and PHYLIP version 3.5[26]. The data set was bootstrap re-sampled 1000 times to ascertain support for major branches of the tree.
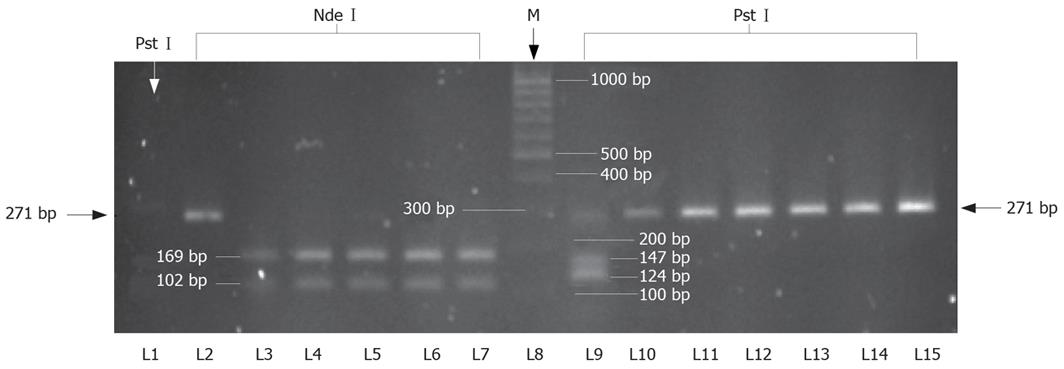
Presence of TTV in sera samples from healthy persons and patients with different liver and renal diseases was detected using PCR amplifying the conserved region of ORF2 (107 to 715 nt) (Accession No. AB008394)[23]. TTV-DNA was detected in 175 of 578 patients, giving an overall prevalence of 30.3% (Table 1). Similarly, it was detected in 27 of 100 healthy persons (prevalence of 27%). Typical TTV amplicons of 295 bp are shown in Figure 2. The break-up of TTV prevalence in different disease groups shown its presence in 29 of the 126 (23.0%) patients with AVH, 26 of 111 (23.4%) with CVH, 46 of 132 (34.8%) with CIR, 15 of 51 (29.4%) with FHF, 21 of 93 (22.6%) with HCC and 38 of 65 (58.5%) with CRF.
The relation of TTV-DNA with ALT level in sera in these disease groups is shown in Table 2. The majority of cases with liver diseases had low levels of ALT, suggesting a very benign role of TTV in causing liver cell necrosis. The raised level of ALT up to the level of more than 600 IU was detected only in FHF cases, where TTV infection may not be the major cause.
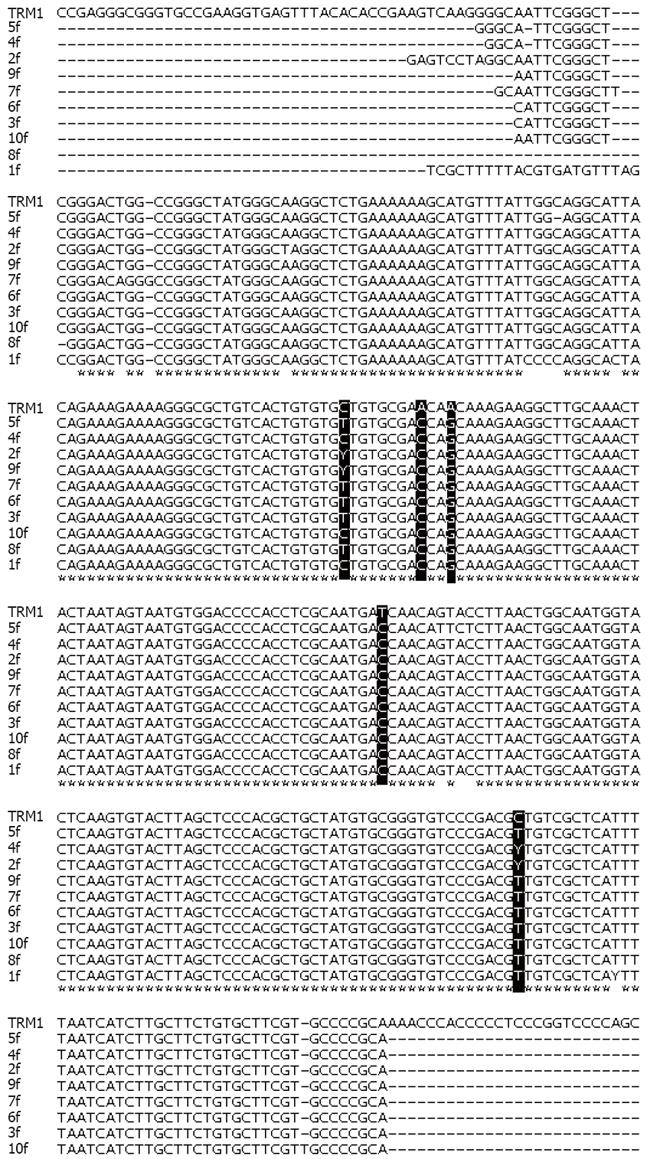
The PCR products from 10 sera were processed for DNA sequencing. The sequence of TTV-DNA from each case was compared to the corresponding region of ORF2 from the TRM1 isolate of genotype 1 (Accession No. AB026345), gathered from the published literature. The full length sequence of amplicons compared to the published sequence of this same region is shown in Figure 3. The results of sequencing indicate the substitution of C by T or Y at nt position 632, A by C at 640 position, substitution of A by G at position 643, T by C at position 696 and at 770 C is being substituted by T or Y. Significant sequence divergence involving more than 2 bp was not observed. Sequencing of additional cases is underway.
Characterization of the TTV genotypes prevalent in the current study population was conducted in 55 cases by RFLP analysis of N22 nucleotide sequence belonging to ORF1. Digestion of the N22 amplicon with NdeI produced fragments of 169 and 102 bp, corresponding to genotype 1, in 53 of 55 cases. Digestion with PstI produced fragments of 124 and 147 bp, corresponding to genotype 2, in 2 of 55 cases. The digestion with other restriction enzymes, NlaIII and MseI, could not produce any fragments, thus indicating the the absence of other genotypes. The results of genotype distribution are shown in Table 3 and Figure 4.
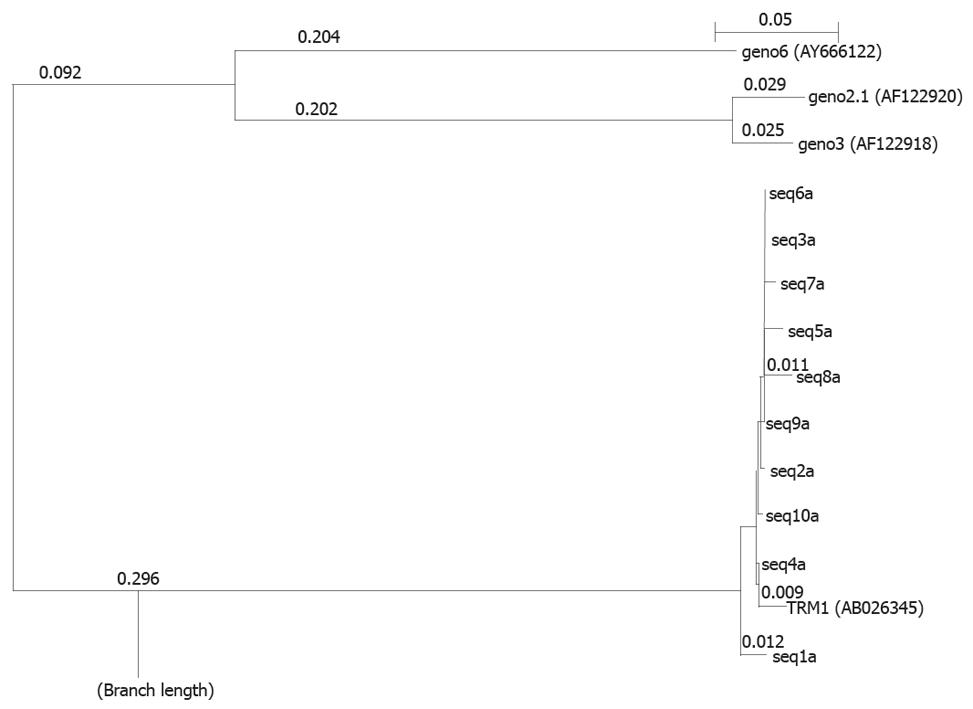
Genotypes of TTV in these cases were also confirmed by phylogenetic analysis of DNA sequences obtained from above. The results are shown in Figure 5. This analysis shows the maximum alignment of the sample sequence with TRM1 isolate of genotype 1 indicating a major presence of TRM1 isolate genotypes 1 and supporting the results of RFLP analysis. From these results, it is clear that genotype 1 is predominantly prevalent in all these cases with TTV infection.
TTV was assumed to be one of the possible agents causing non A-G hepatitis. Its characterization and significance in causing hepatitis became more interesting with the findings by Okamoto et al[27], showing level of TTV-DNA in liver tissue to be 10-100 times higher than those in serum. A similar report by Nishizawa et al[1], who reported the appearance of TTV-DNA in the sera of patients with post-transfusion hepatitis of unknown etiology and displayed a close correlation with ALT levels, triggered an additional interest in this virus for its potential relationship with hepatitis. In India, viral hepatitis is a common disease in all parts of the country, with an established endemicity of all known hepatitis viruses. Previous studies from India have reported the existence of a group of viruses other than A-G hepatitis viruses, causing liver diseases[28]. This remains an enigma to investigate and characterize the responsible agent for non A-G hepatitis in this country. TTV attracted our attention to study this virus for its prevalence and molecular form in Indian populations to understand its possible role in causing hepatitis as well as other blood transmitted diseases.
The prevalence of TTV infections in India has not been well documented. Only a few reports are available[19,29] that demonstrate the presence of TTV in patients population randomly selected from various studies. In this study, we found the presence of TTV-DNA in 22 to 35% of patients with different liver diseases. In CRF, on the contrary, its prevalence was relatively higher, reaching the level of 58.5%. A low prevalence of TTV in liver diseases, comparable to that reported in healthy persons (27%) supports our earlier preliminary report demonstrating a little role of TTV in liver diseases[19]. TTV infection has been found to be common in humans with prevalence which may exceed 90%. Its prevalence in healthy populations in India is lower than those previously reported for Turkish (51.6%), Japanish (92.0%) and Polish (78%) blood donors[30].
Our findings indicate that TTV prevalence in all tested groups was comparable to that among healthy controls, with the exception of a higher value in chronic renal failure patients, thus indicating that TTV infection in both the healthy population and liver disease patients is not very frequent in the northern part of India. Moreover, presence of TTV in liver diseases does not necessarily indicate its role in the etiology of liver damage. This is supported by the low level of ALT detected in all liver disease groups carrying TTV infection. Several other studies have similarly suggested that TTV does not seem to cause disease, simply acting as a by-stander virus. High prevalence in CRF patients may be attributed to repeated blood transfusion or procedural transmission in these patients, due to the fact that TTV is frequently transmitted via parenteral routes.
The results of the sequence analysis in these cases has shown a point mutation at a fixed position of TTV genome, corresponding to ORF2. A nucleotide sequence heterogeneity is not very frequent, but is pre-mature to conclude from these data about the impact of the genomic variation on its pathogenicity or change in etiological potency. It was interesting to find here that the heterogeneity in nucleotide sequence was at the same position in all amplicons studied. This suggests the likelihood of a single isotype circulating in all of the patients studied.
In this study, the genotyping of TTV was based on a RFLP procedure involving a set of four restriction enzymes targeting the N22 nucleotide region. The results indicate that most isolates assessed in our study belong to genotype 1, with only a minor contribution of genotype 2. Since there are very few reports available from our country, it is not possible to support or contradict other observations and therefore this report should be taken as accepted at this stage. While comparing TTV genotypes circulating in India with those reported from other countries, there are reports available showing evidence for the existence of 4 major genotypes, G1, G2, G3 and G4 with most belonging to G1 and G2. There are three major genotypes, i.e. G1, G2, G3, that are prevalent worldwide. Different countries have reported different genotypes in their population. G2 was reported from Western Anatolia, where appears to be very common[31]. Similarly, G3 & G4 genotypes were reported from Turkey, whereas G3 is detected primarily from Europe[32]. Asian countries have reported predominantly G4 genotype[24]. All these findings indicate diversity in the prevalence of TTV genotypes in different countries of the world. The reason(s) why G1 is predominant in India is difficult to explain and needs further investigations. The predominance of G1 was also supported by phylogenetic tree analysis of the amplicons studied in our patients’ population.
In conclusion, our results indicate a low prevalence of TTV infections in North India. This also demon-strates that the prevailing genotype in North India is G1 with minor prevalence of G2 genotype. The presence of strains belonging to G1 reveals the limited genetic diversity in TTV genome circulating in this country.
Our results indicate that Torque teno virus (TTV) infection is moderately present in North India. Analysis of different genotypes of TTV demonstrate that G1 is the predominant genotype prevailing in North India. The presence of strains belonging mostly to G1 reveals that there is a limited genetic diversity in the TTV genome circulating in this part of country.
There are only preliminary informations available on the status of TTV in India. There is a need to study and understand the endemicity, epidemiology and etiological potential of TTV infection in various diseases. The present study was planned to determine the prevalence of TTV in North India and which was the prevailing genotype(s).
The prevalence of TTV infection was observed in various categories of liver and renal diseases. In particular, the rate of TTV infection in liver diseases was comparable to that of a normal healthy populations, although it was significantly higher in chronic renal failure patients. In all these cases genotype 1 (G1) was the predominant genotype with a minor contribution of genotype 2 (G2).
The results of prevalence and genotyping of TTV infection in Indian patients’ will be helpful in understanding the role of TTV in causing various diseases.
Authors have studied TTV infection and found that TTV infection has minor role in causation of liver diseases. Its significant prevalence in renal diseases needs further investigations.
Peer reviewer: Dr. Adam G Testro, Department of Gastroenterology and Liver Transplantation, Austin Health Institution, Heidelberg 3032, Australia
S- Editor Zhong XY L- Editor Negro F E- Editor Yin DH
| 1. | Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, Mayumi M. A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun. 1997;241:92-97. |
| 2. | Miyata H, Tsunoda H, Kazi A, Yamada A, Khan MA, Murakami J, Kamahora T, Shiraki K, Hino S. Identification of a novel GC-rich 113-nucleotide region to complete the circular, single-stranded DNA genome of TT virus, the first human circovirus. J Virol. 1999;73:3582-3586. |
| 3. | Mushahwar IK, Erker JC, Muerhoff AS, Leary TP, Simons JN, Birkenmeyer LG, Chalmers ML, Pilot-Matias TJ, Dexai SM. Molecular and biophysical characterization of TT virus: evidence for a new virus family infecting humans. Proc Natl Acad Sci USA. 1999;96:3177-3182. |
| 4. | Okamoto H, Nishizawa T, Ukita M, Takahashi M, Fukuda M, Iizuka H, Miyakawa Y, Mayumi M. The entire nucleotide sequence of a TT virus isolate from the United States (TUS01): comparison with reported isolates and phylogenetic analysis. Virology. 1999;259:437-448. |
| 5. | Saláková M, Nemecek V, König J, Tachezy R. Age-specific prevalence, transmission and phylogeny of TT virus in the Czech Republic. BMC Infect Dis. 2004;4:56. |
| 6. | Peng YH, Nishizawa T, Takahashi M, Ishikawa T, Yoshikawa A, Okamoto H. Analysis of the entire genomes of thirteen TT virus variants classifiable into the fourth and fifth genetic groups, isolated from viremic infants. Arch Virol. 2002;147:21-41. |
| 7. | Irving WL, Ball JK, Berridge S, Curran R, Grabowska AM, Jameson CL, Neal KR, Ryder SD, Thomson BJ. TT virus infection in patients with hepatitis C: frequency, persistence, and sequence heterogeneity. J Infect Dis. 1999;180:27-34. |
| 8. | Maggi F, Pistello M, Vatteroni M, Presciuttini S, Marchi S, Isola P, Fornai C, Fagnani S, Andreoli E, Antonelli G. Dynamics of persistent TT virus infection, as determined in patients treated with alpha interferon for concomitant hepatitis C virus infection. J Virol. 2001;75:11999-12004. |
| 9. | Okamoto H, Takahashi M, Kato N, Fukuda M, Tawara A, Fukuda S, Tanaka T, Miyakawa Y, Mayumi M. Sequestration of TT virus of restricted genotypes in peripheral blood mononuclear cells. J Virol. 2000;74:10236-10239. |
| 10. | Hino S. TTV, a new human virus with single stranded circular DNA genome. Rev Med Virol. 2002;12:151-158. |
| 11. | Handa A, Dickstein B, Young NS, Brown KE. Prevalence of the newly described human circovirus, TTV, in United States blood donors. Transfusion. 2000;40:245-251. |
| 12. | Ott C, Duret L, Chemin I, Trepo C, Mandrand B, Komurian-Pradel F. Use of a TT virus ORF1 recombinant protein to detect anti-TT virus antibodies in human sera. J Gen Virol. 2000;81:2949-2958. |
| 13. | Qiu J, Kakkola L, Cheng F, Ye C, Soderlund-Venermo M, Hedman K, Pintel DJ. Human circovirus TT virus genotype 6 expresses six proteins following transfection of a full-length clone. J Virol. 2005;79:6505-6510. |
| 14. | Okamoto H, Mayumi M. TT virus: virological and genomic characteristics and disease associations. J Gastroenterol. 2001;36:519-529. |
| 15. | Okamoto H, Ukita M, Nishizawa T, Kishimoto J, Hoshi Y, Mizuo H, Tanaka T, Miyakawa Y, Mayumi M. Circular double-stranded forms of TT virus DNA in the liver. J Virol. 2000;74:5161-5167. |
| 16. | Kondili LA, Pisani G, Beneduce F, Morace G, Gentili G, Ballati G, Rapicetta M. Prevalence of TT virus in healthy children and thalassemic pediatric and young adult patients. J Pediatr Gastroenterol Nutr. 2001;33:629-632. |
| 17. | Niel C, Diniz-Mendes L, Devalle S. Rolling-circle amplification of Torque teno virus (TTV) complete genomes from human and swine sera and identification of a novel swine TTV genogroup. J Gen Virol. 2005;86:1343-1347. |
| 18. | Pistello M, Morrica A, Maggi F, Vatteroni ML, Freer G, Fornai C, Casula F, Marchi S, Ciccorossi P, Rovero P. TT virus levels in the plasma of infected individuals with different hepatic and extrahepatic pathology. J Med Virol. 2001;63:189-195. |
| 19. | Irshad M, Sharma Y, Dhar I, Singh J, Joshi YK. Transfusion-transmitted virus in association with hepatitis A-E viral infections in various forms of liver diseases in India. World J Gastroenterol. 2006;12:2432-2436. |
| 20. | Tandon BN, Joshi YK, Tandon M. Acute liver failure. Experience with 145 patients. J Clin Gastroenterol. 1986;8:664-668. |
| 21. | Acute and chronic hepatitis revisited. Review by an international group. Lancet. 1977;2:914-919. |
| 22. | Agarwal SK, Dash SC, Irshad M, Raju S, Singh R, Pandey RM. Prevalence of chronic renal failure in adults in Delhi, India. Nephrol Dial Transplant. 2005;20:1638-1642. |
| 23. | Biagini P, Gallian P, Attoui H, Cantaloube JF, de Micco P, de Lamballerie X. Determination and phylogenetic analysis of partial sequences from TT virus isolates. J Gen Virol. 1999;80:419-424. |
| 24. | Tanaka Y, Mizokami M, Orito E, Ohno T, Nakano T, Kato T, Kato H, Mukaide M, Park YM, Kim BS. New genotypes of TT virus (TTV) and a genotyping assay based on restriction fragment length polymorphism. FEBS Lett. 1998;437:201-206. |
| 25. | Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876-4882. |
| 26. | Felsenstein J. PHYLIP (Phylogeny Inference Package) version 3.5c. Distributed by the author. Department of Genetics, University of Washington, Seattle. 1993;. |
| 27. | Okamoto H, Nishizawa T, Kato N, Ukita M, Ikeda H, Iizuka H, Miyakawa Y, Mayumi M. Molecular cloning and characterization of a novel DNA virus (TTV) associated with posttransfusion hepatitis of unknown etiology. Hepatol Res. 1998;10:1-16. |
| 28. | Irshad M, Chaudhuri PS, Joshi YK. Superoxide dismutase and total anti-oxidant levels in various forms of liver diseases. Hepatol Res. 2002;23:178-184. |
| 29. | Chattopadhyay S, Das BC, Gupta RK, Kar P. Presence of TT virus infection in chronic hepatitis patients from a hospital in New Delhi, India. Indian J Med Res. 2005;122:29-33. |
| 30. | Kalkan A, Ozdarendeli A, Bulut Y, Saral Y, Ozden M, Kelestimur N, Toraman ZA. Prevalence and genotypic distribution of hepatitis GB-C/HG and TT viruses in blood donors, mentally retarded children and four groups of patients in eastern Anatolia, Turkey. Jpn J Infect Dis. 2005;58:222-227. |
| 31. | Erensoy S, Sayiner AA, Turkoglu S, Canatan D, Akarca US, Sertoz R, Ozacar T, Batur Y, Badur S, Bilgic A. TT virus infection and genotype distribution in blood donors and a group of patients from Turkey. Infection. 2002;30:299-302. |
| 32. | Erensoy S, Zeytinoglu A, Goksel S, Ozacar T, Ozkahya M, Ok E, Tuumlrkoglu S, Bilgic A. GB virus C/hepatitis G virus infection among renal transplant recipients in Izmir, Turkey: Molecular analysis of phylogenetic groups. Int J Infect Dis. 2002;6:242-243. |