Published online Oct 14, 2008. doi: 10.3748/wjg.14.5887
Revised: September 16, 2008
Accepted: September 23, 2008
Published online: October 14, 2008
AIM: To explore the expression of differential gene expression profiles of target cell between non-invasive submucosal and invasive advanced tumor in colon carcinoma using laser microdissection (LMD) in combination with polypeptide analysis.
METHODS: Normal colon tissue samples from 20 healthy individuals and 30 cancer tissue samples from early non-invasive colon cancer cells were obtained. The cells from these samples were used LMD independently after P27-based amplification. aRNA from advanced colon cancer cells and metastatic cancer cells of 40 cases were applied to LMD and polypeptide analysis, semiquantitative reverse transcribed polymerase chain reaction (RT-PCR) and immunohistochemical assays were used to verify the results of microarray and further identify differentially expressed genes in non-invasive early stages of colon cancer.
RESULTS: Five gene expressions were changed in colon carcinoma cells compared with that of controls. Of the five genes, three genes were downregulated and two were upregulated in invasive submucosal colon carcinoma compared with non-invasive cases. The results were confirmed at the level of aRNA and gene expression. Five genes were further identified as differentially expressed genes in the majority of cases (> 50%, 25/40) in progression of colon cancer, and their expression patterns of which were similar to tumor suppressor genes or oncogenes.
CONCLUSION: This study suggested that combined use of polypeptide analysis might identify early expression profiles of five differential genes associated with the invasion of colon cancer. These results reveal that this gene may be a marker of submucosal invasion in early colon cancer.
- Citation: Zhu JS, Guo H, Song MQ, Chen GQ, Sun Q, Zhang Q. Gene profiles between non-invasive and invasive colon cancer using laser microdissection and polypeptide analysis. World J Gastroenterol 2008; 14(38): 5887-5892
- URL: https://www.wjgnet.com/1007-9327/full/v14/i38/5887.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.5887
| Characteristic | Non-invasive tumor (n) | Invasive tumor (n) |
| Gender | ||
| Male | 19 | 19 |
| Female | 11 | 21 |
| Age | ||
| Median yr | 57.9 ± 3.24 | 58.7 ± 3.46 |
| Ranger yr | 45-80 | 43-80 |
| Cancer TNM stage | ||
| IA | 30 | 0 |
| III-IV | 0 | 40 |
| Histological type | ||
| Differentiated | 16 | 22 |
| Undifferentiated | 14 | 18 |
| GeneBank | Description | Cy3:Cy52 |
| NM_006500 | MCAM (melanoma adhesion molecule) | 10.791 |
| NM_002545 | OPCML (opioid binding protein/cell adhesion molecule-like) | 10.561 |
| NM_002933 | RNASE1 (ribonuclease, RNase A family, 1) | 6.81 |
| NM_001993 | F3 (coagulation factor III) | 6.291 |
| NM_005433 | YES1 (v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1) | 5.9161 |
| NM_016525 | UBAP (ubiquitin associated protein) | 5.876 |
| NM_001428 | ENO1 (enolase 1) | 5.6921 |
| S68616 | SLC9A1 (Na+/H+ exchanger NHE-1 isoform) | 5.484 |
| NM_003254 | TIMP1(tissue inhibitor of metalloproteinase 1) | 5.291 |
| NM_005903 | MADH5 (mothers against decapentaplegic, homolog 5) | 5.083 |
| U82828 | ATM (ataxia telangiectasia) | 0.2051 |
| NM_006343 | MERTK (c-mer proto-oncogene tyrosine kinase) | 0.19 |
| NM_002985 | SCYA5 (small inducible cytokineA5) | 0.191 |
| NM_005348 | HSPCA (heat shock 90 kDa protein 1, alpha) | 0.164 |
| NM_003968 | UBE1C (ubiquitin-activating enzyme E1C) | 0.161 |
| NM_004374 | COX6C (cytochrome c oxidase subunit VIc) | 0.131 |
| NM_002990 | SCYA22 (small inducible cytokine subfamily A, member22) | 0.129 |
| NM_005781 | ACK1 (activated p21cdc42Hs kinase) | 0.1281 |
| NM_005139 | ANXA3 (annexin A3) | 0.124 |
| AF053630 | SERPINB1 (serine proteinase inhibitor, clade B, member 1) | 0.123 |
| NM_012090 | MACF1 (microtubule-actin crosslinking factor 1) | 0.0981 |
| XM_042551 | CAMK2A (calcium/calmodulin-dependent protein kinase) | 0.0645 |
| NM_000909 | NPY1R (neuropeptide Y receptor Y1) | 0.02521 |
| NM_015230 | CENTD1 (centaurin, delta 1) | 0.024 |
| NM_004958 | FRAP1 (FK506 binding protein 12-rapamycin associated protein 1) | 0.0205 |
Colon cancer ranks second of all gastrointestinal malignant tumors, it is one of the leading causes of cancer-related deaths worldwide. In the past five years, the incidence of colon cancer is reported to be especially high in Asia. Its clinical behavior depends on the potential metastasis of the tumor; its prognosis of advanced colon cancers remains very poor. Until now, several molecules have been reported to play an important role in gastroenterological tumorigenesis and tumor metastasis, but the molecular mechanisms involved tumor development and progression still remain unclear in colon cancer[1-3].
In this research, by using the combined methods of laser microdissection (LMD), P27-based RNA amplification, and polypeptide, we evaluated differentially expressed genes between early carcinoma and lymph node metastatic patients. Moreover, we further identified four differentially expressed genes in the progression of colon cancer in another group of 15 patients by means of semiquantitative reverse transcribed polymerase chain reaction (RT-PCR), and the expression patterns of these four genes were similar to tumor suppressor genes or oncogenes.
It is now widely accepted that many malignant tumors contain heterogeneous subpopulations of cells. This heterogeneity is exhibited in a wide range of genetic, biochemical and immunologic characteristics. It is likely that specific tumor cells or colonies within the larger heterogeneous tumor specimen are the forerunners of distant metastases[4]. Therefore, many biologic differences exist between tumor cells in early carcinoma and those in metastatic lesions. Furthermore, the interaction of tumor cells within their living environment may add more differences between these two groups of cells[5]. As a result, tumor metastasis related genes can be identified by comparing the gene expression profiles between them.
LMD and polypeptide analysis are two new emerging technologies in the post-genomic era. LMD allows a specific area of tissue or group of cells to be obtained, and in this particular system, intracellular macromolecules remain intact. Since there is no heating of the sample or direct mechanical contact with the section, there is a reduced risk of contamination, sample loss or alteration of the protein sample[6,7].
The large-scale analysis of gene expression with a polypeptide allows us to evaluate the gene-expression profiles of hundreds to tens of thousands of genes in a single experiment[8]. This technology is a powerful tool for analyzing gene expression of which can be correlated with pathological phenotypes of various diseases. However, the expression profile of a specific cell type may be primarily masked or even lost because of the bulk of surrounding cells. Therefore, combined use of LMD and polypeptide analysis can provide a unique opportunity to study gene expression of subpopulations of cells in their native (in vivo) tissue environment.
Forty cases of advanced colon adenocarcinoma (TNM stage III to IV) patients with lymph node metastasis were investigated in this study. There was 21 male and 19 female patients, with an average age of 58.7 ± 3.46 years old. Histologically, 22 cases had differentiated adenocarcinoma and 18 had poorly undifferentiated adenocarcinoma. Thirty cases of non-invasive submucosal colon adenocarcinoma patients (TNM stage IA) were obtained that included 19 male and 11 female patients. Histologically, 16 cases had differentiated adenocarcinoma and 14 had poorly undifferentiated adenocarcinoma, whose ages ranged from 45 to 80 years with an average age of 57.9 ± 3.24 years old (Table 1). All patients underwent gastrectomy with regional lymph nodes dissection and informed consent was obtained from each patient. Tissue blocks of normal colon epithelium from 20 healthy individuals, early tumors, advanced colon adenocarcinoma tissue and corresponding metastatic lymph nodes were obtained within 30 min after removal from the patient. Each block was cut into 2 pieces, one for routine pathologic diagnosis, and the other for molecular analysis. The latter samples were frozen in liquid nitrogen immediately and stored at -260°C until use.
Before sectioning, tissue blocks were embedded in Tissue Tek OCT compound medium (VWR Scientific Products, San Diego, CA, USA) in a cryostat. Then serial 8-micron-thick slices were prepared and mounted onto a foiled slide and stored at -70°C until use.
Frozen section slides were stained just before LMD and the staining process was performed on ice. Briefly, the slides were fixed in 70% ethanol for 30 s and stained with 0.1% toluidine blue (TBO) for 15 s, followed by a 5-s dehydration step in each of 75%, 95% and 100% ethanol and a final 5-min dehydration step in xylene. Once air-dried, the sections were laser microdissected with a LMD system (Leica Microsystems, Wetzlar, Germany) and the target cells were selectively collected. Next, total RNA from the interest cells was extracted independently with the RNA-lyase Micro Kit (Qiagen, Hilden, Germany), according to the manufacturer’s instructions. The integrity of the total RNA was measured by Lab-on-chip (Agilent, Palo Alto, USA).
We carried out polypeptide analysis in 30 cases with early carcinoma, 40 cases with metastatic lymph node and 20 healthy individuals. All technical services were provided by Shanghai Biochip Corporation (Shanghai, China). Among the extracted total RNA, 200 ng was amplified for array analysis, while the other 100 ng was kept unamplified for later certification. After P27-based RNA amplification, aliquots (2.5 mg) of aRNA from early carcinoma tissues and the corresponding metastatic lymph node were labeled with Cy3-dCTP and Cy5-dCTP, respectively. The labeled probes were hybridized with Human cDNA Chip version 2.0 (SBC-R-HC-100-20, Shanghai, China) which contained 13 824 genes (including 10 positive controls and 6 negative ones), and the signals were detected by Agilent Scanner (Agilent, Palo Alto, USA). We set the cutoff values for signal intensities, i.e., the signal to noise ratios of Cy3 or Cy5 must be greater than 2. Genes with Cy3:Cy5 ratios > 4 or < 0.25 in both cases were defined as commonly up- or down-regulated genes respectively.
Nonamplified total RNA (100 ng) was reverse transcribed into cDNA using Superscript II reverse transcriptase (Invitrogen, USA) for 60 min at 42°C and 15 min at 70°C. Each single-stranded cDNA was diluted for subsequent PCR amplification and the content of cDNA were semiquantitatively normalized by housekeeping gene β-actin. PCR conditions of different genes included an initial denaturation at 94°C for 3 min, and 30-35 cycles of denaturation at 94°C for 30 s, annealing for 30 s, and elongation at 72°C for 1 min. The primer sequences, annealing temperatures and cycle numbers of each gene are available on request. Amplified PCR products were visualized by electrophoresis in 1% agarose gel containing ethidium bromide.
The target cells were successfully laser microdissected in every sample. Consequently, about 6-8 × 106 cells were collected for the total RNA extraction and near 200-400 ng total RNA was obtained. The integrity of every sample was proved by Lab-on-chip.
After P27-based amplification, we evaluated the expression profiles of the carcinoma cells from early colon cancer (IA) in 30 cases in comparison with advanced colon adenocarcinoma and corresponding metastatic lymph node (TNM stage III to IV) in 40 cases, scatter plots of polypeptide analysis showed in Figure 1. Amplified aRNA from early colon carcinoma cells (Cy-3) and metastatic carcinoma cells (Cy-5) were labeled and hybridized to the polypeptide.
The analysis of the polypeptide data showed that 49 genes (including 31 of unknown function) were commonly overexpressed (Cy3:Cy5 > 4) in 22 of 30 patients (TNM IA) in early colon carcinoma, another 37 genes (including 9 of unknown function) were significantly suppressed (Cy3:Cy5 < 0.25) in the early carcinoma cells. The up-regulated genes were related to cell division, cell adhesion, cytoskeleton, and cell defense and cell metabolism. Meanwhile, the down-regulated genes included those associated with cell development, cell cycle, signal transduction, adhesion, cell defense, gene expression and cell metabolism (Table 2).
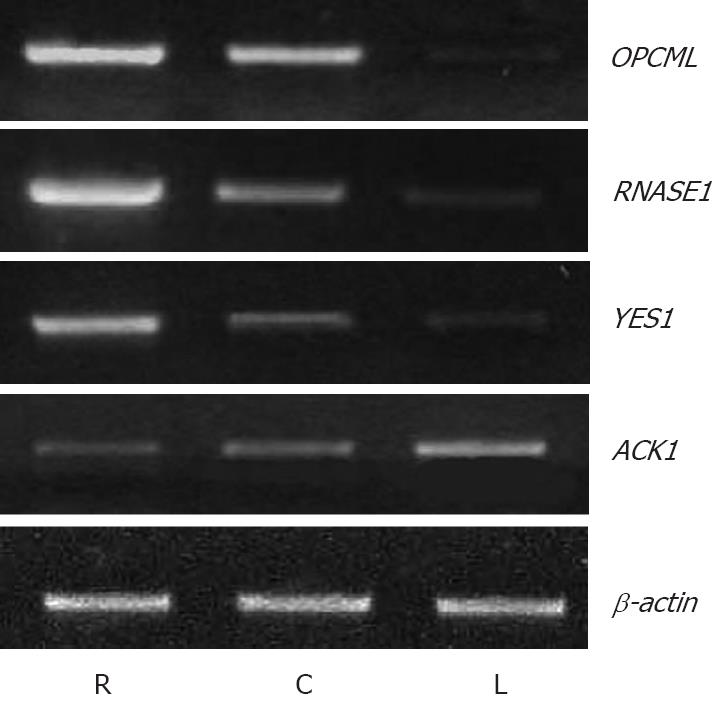
By means of semiquantitative RT-PCR, we further evaluated the expression levels of 25 selected genes, which included 10 up-regulated and 15 down-regulated genes, in microdissected colon epithelium, early colon cancer cells and metastatic cells in lymph node from other 22 cases. First, we investigated the expression levels of these genes in early colon cancer and metastatic lymph node. The results showed that 12 genes were expressed in the same pattern in > 50% (8/15) cases as revealed by polypeptide analysis.
We further measured the expression levels of these 12 genes in paired normal colon epithelium from healthy individuals and early carcinoma from the same 22 patients. We found that the expression pattern of 3 genes (OPCML, RNASE1 and YES1) was similar to tumor suppressor genes in > 50% (8/15) cases, e.g. the expression level of these genes were highest in normal colon epithelium, decreased in early carcinoma, and further decreased in metastatic lymph node. Meanwhile, the expression level of ACK1 demonstrated the opposite tendency, the pattern of which was similar to oncogene in > 50% (8/15) cases (Figure 2). The expression pattern of OPCML was also confirmed at the gene level by immunohistochemical staining.
To examine the reliability of microarray data, we confirmed our data at the level of aRNA and gene expression. First, we selected three up-regulated genes (SLC9A1, OPCML and UBAP) and three down-regulated genes (COX6C, SERPINB1 and CCL22) to measure their expression levels by semiquantitative RT-PCR. To obtain truly comparable results, we used the nonamplified total RNA (from the same batch that was used for the array hybridizations) as the template. The results were very similar to the microarray data for these genes.
As an additional means to confirm our data at the gene level, the results paralleled the differential expression pattern observed with polypeptide and RT-PCR.
Overall, the above results demonstrated that the samples obtained by means of P27-based amplification reflected the status of the original RNA in a proportional manner, and supported the reliability of our previous data[9].
Polypeptides allow an effective investigation of functional genomics. However, the existence of of the amount of surrounding cells can produce a lot of useless, noisy information because of the high sensitivity[9]. Therefore, the selection of cancer cells using LMD is of indispensable value in combination with polypeptide analysis. The LMD system used in this study integrated a UV laser of 337 nm wavelength with an upright microscope. The ultraviolet laser microbeam causes dissection by local photolysis of the supporter membrane and tissue section due to the high photon density of the microbeam rather than by local heating or coagulation. The cut sample falls down into PCR tubes placed underneath by gravity without any mechanical contact or further destruction of energy, and the integrity of the extracted aRNA is maximally kept.
Metastasis of cancer is a highly selective sequential step, which favors the survival of a subpopulation of metastatic cells preexisting within the early tumor mass to produce clinically relevant metastases, the successfully metastatic cells must exhibit a complex phenotype that is regulated by transient or permanent changes in different genes at the DNA and/or aRNA level(s). This was also proved in colon cancer by work of other researchers[10].
Previously, our former study had discovered differential gene expression profiles between primary tumor and metastatic lesions in gastric cancer patients using LMD and cDNA microarray[11]. The results clearly demonstrated that an analysis of the gene-expression profile could be performed by using LMD, P27-based RNA amplification and polypeptide. Moreover, it was confirmed at the level of aRNA and gene. Although the majority of these genes have been implicated in various aspects of tumor biology, few have been demonstrated to be associated with colon cancer.
Among the above genes, some may be differentially expressed because of different living environments[12-15]. Therefore, to further confirm and screen the results of polypeptide analysis, we measured the expression levels of 25 selected genes in another group using semiquantitative RT-PCR. These target cells were collected by LMD, and the normal colon epithelium were included. As a result, we identified four genes, the expression level of were not only different between early carcinoma and metastatic lymph node (same as the results of polypeptide), but also different between normal mucosa and early tumor. These results suggested that these four genes could play a role in the tumorigenesis and metastasis of colon cancer .The expression pattern of three genes (OPCML, RNASE1 and YES1) was similar to tumor suppressor genes[16], e.g. the expression level of these genes were highest in normal colon epithelium, decreased in early carcinoma, and further decreased in metastatic lymph node. OPCML (opioid binding gene/cell adhesion molecule-like) encodes a member of the IgLON subfamily in the immunoglobulin gene superfamily and acted as a GPI-anchored gene[17,18]. Sellar et al had found that OPCML has tumor-suppressor function in epithelial ovarian cancer, which was in accordance with our findings in gastric cancer[19-22]. Interestingly, YES1 is the cellular homolog of a virus oncogene and was considered to be associated with esophageal tumorigenesis[23-25], which was on the contrary to our results. In respect to the gene RNASE1, it encoded a member of the pancreatic-type of secretory ribonucleases, a subset of the ribonuclease A superfamily, and it had not been shown to have any relationship with human cancers previously[26].
On the contrary, the expression pattern of gene ACK1 demonstrated the opposite tendency to the genes mentioned above, which was similar to oncogene[27]. ACK1 encodes a tyrosine kinase that binds Cdc42Hs in its GTP-bound form and inhibited both the intrinsic and GTPase-activating gene (GAP)-stimulated GTPase activity of Cdc42Hs[28]. It was directly linked to a tyrosine phosphorylation signal transduction pathway. Up to now, its effect on tumor progression had not been reported.
Tumor suppressor genes and oncogenes were important target molecules in the clinical diagnosis and therapy of tumors. We reported the different expression of these four genes during the progression of colon cancer. In conclusion, using carefully controlled conditions, analysis of gene expression profile by means of LMD, P27-based amplification, and polypeptide has been provided.
Useful information for clarifying the mechanism underlying development and metastasis of colon cancers[29,30]. These results not only reveal the differentially expressed genes in progression of colon cancer, but also provide information that may prove useful for identifying novel diagnostic and therapeutic targets.
Colon cancer is the second leading cause of cancer death in the worldwide. Its clinical behavior depends on the potential metastasis of the tumor. However, up to now, the difference of genes had rarely been reported between early colon adenocarcinoma (TNM stage I) patients and advanced adenocarcinoma (TNM stage III-IV) cases with lymph node metastasis.
In this study, the authors revealed the gene profiles in non-invasive and invasive colon cancer using laser microdissection (LMD) and polypeptide analysis. Using these techniques they have identified early expression profiles of five differential genes associated with invasive colon cancer.
These studies have shown that certain genes are involved in the development of colon cancer and that the gene expression of non-invasive early colon adenocarcinoma was obviously different from that of invasive cases. This offers a new potential molecular mechanism of invasive colon cancer using LMD and polypeptide analysis. Our major finding in the study is that five genes are consequently differently expressed between early and advanced colorectal cancer (CRC) cases. Three of these resembled the expression pattern of tumor suppression genes (OPCML, RNASE1 and YES1).
These results revealed that expression profiles of differential genes were associated with the progression of colon cancer, and these insights might play a role in identifying novel diagnostic and therapeutic markers in colon cancer.
This is an interesting manuscript investigating the expression profile differences of early and advanced CRC at aRNA level using laser microdissection and polypeptide analysis. The major finding of the study was that five genes were consequently differently expressed between early and advanced CRC cases. Three of these resembled the expression pattern of tumor suppression genes (OPCML, RNASE1 and YES1) while one further gene showed an expression pattern similar to oncogenes.
Peer reviewer: Peter L Lakatos, MD, PhD, Assistant Professor, 1st Department of Medicine, Semmelweis University, Koranyi S 2A, Budapest H1083, Hungary
S- Editor Li DL L- Editor Alpini GD E- Editor Yin DH
| 1. | Zhu JS, Shen B, Chen JL, Chen GQ, Yu XH, Yu HF, Zhu ZM. Molecular action mechanisms of NM-3 on human gastric cancer SGC-7901 cells in vivo or in vitro. World J Gastroenterol. 2003;9:2366-2369. |
| 2. | Zhu JS, Jiang XH, Cheng WX. Studies on peritoneal chemotherapy treatment of gastric cancer ascites and correlative effects. Hepato-Gastroenterol. 2000;15:B108. |
| 3. | Wang ZN, Xu HM, Jiang L, Zhou X, Lu C, Zhang X. Expression of survivin mRNA in peritoneal lavage fluid from patients with gastric carcinoma. Chin Med J (Engl). 2004;117:1210-1217. |
| 4. | Portera CA Jr, Berman RS, Ellis LM. Molecular determinants of colon cancer metastasis. Surg Oncol. 1998;7:183-195. |
| 5. | Fidler IJ. Critical determinants of metastasis. Semin Cancer Biol. 2002;12:89-96. |
| 6. | Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang Z, Goldstein SR, Weiss RA, Liotta LA. Laser capture microdissection. Science. 1996;274:998-1001. |
| 7. | Böhm M, Wieland I, Schütze K, Rübben H. Microbeam MOMeNT: non-contact laser microdissection of membrane-mounted native tissue. Am J Pathol. 1997;151:63-67. |
| 8. | Liotta L, Petricoin E. Molecular profiling of human cancer. Nat Rev Genet. 2000;1:48-56. |
| 9. | Wang L, Zhu JS, Song MQ, Chen GQ, Chen JL. Comparison of gene expression profiles between primary tumor and metastatic lesions in gastric cancer patients using laser microdissection and cDNA microarray. World J Gastroenterol. 2006;12:6949-6954. |
| 10. | Versteeg HH, Spek CA, Peppelenbosch MP, Richel DJ. Tissue factor and cancer metastasis: the role of intracellular and extracellular signaling pathways. Mol Med. 2004;10:6-11. |
| 11. | Kakinuma T, Hwang ST. Chemokines, chemokine receptors, and cancer metastasis. J Leukoc Biol. 2006;79:639-651. |
| 12. | Cui Y, Ying Y, van Hasselt A, Ng KM, Yu J, Zhang Q, Jin J, Liu D, Rhim JS, Rha SY. OPCML is a broad tumor suppressor for multiple carcinomas and lymphomas with frequently epigenetic inactivation. PLoS ONE. 2008;3:e2990. |
| 13. | Ye F, Zhang SF, Xie X, Lu WG. OPCML gene promoter methylation and gene expression in tumor and stroma cells of invasive cervical carcinoma. Cancer Invest. 2008;26:569-574. |
| 14. | Hayashida T, Ueda M, Aiura K, Tada H, Onizuka M, Seno M, Yamada H, Kitajima M. Anti-angiogenic effect of an insertional fusion protein of human basic fibroblast growth factor and ribonuclease-1. Protein Eng Des Sel. 2005;18:321-327. |
| 15. | Mahajan NP, Whang YE, Mohler JL, Earp HS. Activated tyrosine kinase Ack1 promotes prostate tumorigenesis: role of Ack1 in polyubiquitination of tumor suppressor Wwox. Cancer Res. 2005;65:10514-10523. |
| 16. | Lynch G, Kohler S, Leser J, Beil M, Garcia-Marin LJ, Lutz MP. The tyrosine kinase Yes regulates actin structure and secretion during pancreatic acinar cell damage in rats. Pflugers Arch. 2004;447:445-451. |
| 17. | Ntougkos E, Rush R, Scott D, Frankenberg T, Gabra H, Smyth JF, Sellar GC. The IgLON family in epithelial ovarian cancer: expression profiles and clinicopathologic correlates. Clin Cancer Res. 2005;11:5764-5768. |
| 18. | Reed JE, Dunn JR, du Plessis DG, Shaw EJ, Reeves P, Gee AL, Warnke PC, Sellar GC, Moss DJ, Walker C. Expression of cellular adhesion molecule 'OPCML' is down-regulated in gliomas and other brain tumours. Neuropathol Appl Neurobiol. 2007;33:77-85. |
| 19. | Sellar GC, Watt KP, Rabiasz GJ, Stronach EA, Li L, Miller EP, Massie CE, Miller J, Contreras-Moreira B, Scott D. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat Genet. 2003;34:337-343. |
| 20. | Guo W, Shang F, Liu Q, Urim L, West-Mays J, Taylor A. Differential regulation of components of the ubiquitin-proteasome pathway during lens cell differentiation. Invest Ophthalmol Vis Sci. 2004;45:1194-1201. |
| 21. | Wei W, Ayad NG, Wan Y, Zhang GJ, Kirschner MW, Kaelin WG Jr. Degradation of the SCF component Skp2 in cell-cycle phase G1 by the anaphase-promoting complex. Nature. 2004;428:194-198. |
| 22. | Kudo Y, Kitajima S, Sato S, Ogawa I, Miyauchi M, Takata T. Transfection of p27 (Kip1) threonine residue 187 mutant type gene, which is not influenced by ubiquitin-mediated degradation, induces cell cycle arrest in oral squamous cell carcinoma cells. Oncology. 2002;63:398-404. |
| 23. | Defatta RJ, De Benedetti A. Translational upregulation of yes accompanies eIF4E-mediated oncogenic transformation. Int J Oncol. 2003;23:1709-1713. |
| 24. | Lawson C, Goupil S, Leclerc P. Increased activity of the human sperm tyrosine kinase SRC by the cAMP-dependent pathway in the presence of calcium. Biol Reprod. 2008;79:657-666. |
| 25. | Barraclough J, Hodgkinson C, Hogg A, Dive C, Welman A. Increases in c-Yes expression level and activity promote motility but not proliferation of human colorectal carcinoma cells. Neoplasia. 2007;9:745-754. |
| 26. | Psarras K, Ueda M, Tanabe M, Kitajima M, Aiso S, Komatsu S, Seno M. Targeting activated lymphocytes with an entirely human immunotoxin analogue: human pancreatic RNase1-human IL-2 fusion. Cytokine. 2000;12:786-790. |
| 27. | Mahajan NP, Liu Y, Majumder S, Warren MR, Parker CE, Mohler JL, Earp HS, Whang YE. Activated Cdc42-associated kinase Ack1 promotes prostate cancer progression via androgen receptor tyrosine phosphorylation. Proc Natl Acad Sci USA. 2007;104:8438-8443. |
| 28. | Modzelewska K, Newman LP, Desai R, Keely PJ. Ack1 mediates Cdc42-dependent cell migration and signaling to p130Cas. J Biol Chem. 2006;281:37527-37535. |
| 29. | Castiglione F, Taddei A, Buccoliero AM, Garbini F, Gheri CF, Freschi G, Bechi P, Degl'Innocenti DR, Taddei GL. TNM staging and T-cell receptor gamma expression in colon adenocarcinoma. Correlation with disease progression? Tumori. 2008;94:384-388. |
| 30. | Iddings D, Bilchik A. The biologic significance of micrometastatic disease and sentinel lymph node technology on colorectal cancer. J Surg Oncol. 2007;96:671-677. |