Published online Aug 28, 2008. doi: 10.3748/wjg.14.5059
Revised: July 14, 2008
Accepted: July 21, 2008
Published online: August 28, 2008
AIM: To investigate whether hepatitis B virus (HBV) infection activates DNA damage response and DNA repair cofactors inhibit HBV infection and replication.
METHODS: Human hepatocyte cell line HL7702 was studied. Immunoblotting was performed to test the expression of ataxia telangiectasia-mutated (ATM)-Rad3-related protein (ATR), p21 and the level of phosphorylation of Chk1, p53, H2AX, ATM in HBV-infected or non-infected-cells. Special short RNAi oligos was transfected to induce transient ATR knockdown in HL7702. ATR-ATM chemical inhibitors caffeine (CF) and theophylline (TP), or Chk1 inhibitor 7-hydroxystaurosporine (UCN01) was studied to determine whether they suppress cellular DNA damage response and MG132 inhibits proteasome.
RESULTS: The ATR checkpoint pathway, responding to single-strand breaks in DNA, was activated in response to HBV infection. ATR knockdown cells decreased the HBV DNA yields, implying that HBV infection and replication could activate and exploit the activated DNA damage response. CF/TP or UCN01 reduced the HBV DNA yield by 70% and 80%, respectively. HBV abrogated the ATR-dependent DNA damage signaling pathway by degrading p21, and introduction of the p21 protein before HBV infection reduced the HBV DNA yield. Consistent with this result, p21 accumulation after MG132 treatment also sharply decreased the HBV DNA yield.
CONCLUSION: HBV infection can be treated with therapeutic approaches targeting host cell proteins by inhibiting a cellular gene required for HBV replication or by restoring a response abrogated by HBV, thus providing a potential approach to the prevention and treatment of HBV infection.
- Citation: Zhao F, Hou NB, Song T, He X, Zheng ZR, Ma QJ, Li L, Zhang YH, Zhong H. Cellular DNA repair cofactors affecting hepatitis B virus infection and replication. World J Gastroenterol 2008; 14(32): 5059-5065
- URL: https://www.wjgnet.com/1007-9327/full/v14/i32/5059.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.5059
Eukaryotic cells employ multiple strategies of checkpoint signaling and DNA repair mechanisms to monitor and repair damaged DNA. There are two branches in the checkpoint response pathway: ataxia telangiectasia-mutated (ATM) branch and ATM-Rad3-related (ATR) branch[1,2]. Many viruses are now known to interact with DNA damage sensing and repair machinery. These viruses have evolved tactics to eliminate, circumvent, or exploit various aspects of the DNA damage response to the host cells. Strategies include activation of repair proteins or targeting of specific cellular factors for degradation or mislocalization[3-6]. It is necessary to examine the DNA damage pathway activated by viral replication for generation of antiviral drugs. In human immunodeficiency virus (HIV), it has been clearly determined that prevention of viral integration inhibits viral replication and promotes cellular apoptosis. The ATM-specific inhibitor ku55933 can inhibit HIV replication in primary T cells[7].
Although a safe and efficient vaccine is available at present, chronic hepatitis B virus (HBV) infection remains a major health problem worldwide. Interferon treatment is effective in only approximately one-third of the patients and produces considerable side effects. Long-term treatment with the second-generation of nucleoside analogue lamivudine (lam) efficiently inhibits HBV replication with frequent viral polymerase mutations. We found that HBV infection could trigger ATR-dependent DNA damage response, resulting in increased ATR and Chk1 phosphorylation levels. However, ATR checkpoint signaling is blocked downstream of the p53-dependent pathway to evade apoptosis by degrading p21. We designed a strategy to select new drug targets that inhibit a cellular gene required for HBV replication or restore a response stalled by HBV in the ATR DNA damage pathway.
The ATM and ATR kinases are targets of a known inhibitor, caffeine (CF), of DNA damage response. CF has shown to inhibit the activity of these kinases in vitro and the response to cellular DNA damage controlled by ATM and ATR. Theophylline (TP) is a product of CF metabolism in vivo and has been clinically used in treatment of asthma. It has also been demonstrated that TP exhibits CF-like effects on DNA damage response[8].
Originally developed as a protein kinase C (PKC) inhibitor, 7-hydroxystaurosporine (UCN-01) has been subsequently shown to inhibit several other cell survival/cell cycle regulatory proteins, including Chk1 and 3-phosphoinositide-dependent protein kinase-1 (PDK1)/protein kinase B (Akt). By inhibiting Chk1, UCN-01 blocks the proteasome degradation of the cdc25C phosphatase, resulting in dephosphorylation (activation) of p34cdc2. In this way, UCN-01 functions as a checkpoint abrogator that potentiates the lethal effects of several DNA-damaging agents, including ara-C18 and camptothecin[9,10].
Proteasome, a multicatalytic enzyme complex, controls the regulation (by means of degradation) of many proteins involved in cell cycle arrest and apoptosis. The proteasome inhibitor MG132 can restore wild-type (WT) p53 levels reduced by E6 and sensitize HPV-positive cervical cancer cell lines to apoptotic stimuli such as rhTRAIL. It was also reported that MG13 could inhibit HSV replication by preventing HSV-1-induced NF-κB activation[11,12].
In this paper, we report that HBV infection activates and exploits DNA damage response to replication stress. We investigated whether inhibition of DNA damage response by CF, TP and UCN01 or restoration of p21 expression by p21 transfection or proteasome inhibition leads to suppression of HBV replication. We set up a chronic HBV infection model by culturing HL7702 liver cells with HBV-positive serum without washing off input virus as conventional. HBV DNA titers inside the infected cells represent the final viral amount including the infected DNA not degraded and the newly synthesized HBV DNA. In this way, we could study the efficacy of DNA damage response inhibitors on HBV infection and replication. In addition, since DNA damage response is an acute response occurring quickly after virus infection, we assume that early intervention of the DNA damage pathway would function more efficiently, and thus can be used in clinical practice as a HBV infection therapy during its early infectious stage or fulminant HBV infection.
CF, TP, UCN01 and MG132 were obtained from Sigma (St. Louis, MO). The stock concentration was 100 mmol/L. CF and UCN01 were dissolved in water, whereas TP was dissolved in 0.1 mol/L NaOH, and MG132 was dissolved in DMSO.
The human hepatocyte cell line HL7702, isolated from a HBV sera-negative individual, was obtained from Shanghai Biochemistry Institute. HL7702 cells were cultured in RPMI-1640 with 10% heat-inactivated fetal bovine serum (FBS). Serum samples from HBV carriers obtained for infection test were analyzed. The number of serum HBV viral particles was 7 × 109 copies/mL, as quantified by FQ-PCR. The sera were stored at -80°C until use.
HBV infection was monitored by culturing 105 HL7702 cells in a 6-cm plate in 3 mL RPMI-1640 containing 106 HBV virus particles. Infected cells were simultaneously treated with different types of drugs (CF, TP and UCN01) at various concentrations. The cells were washed thoroughly 8 times to remove excessive viral inputs and drugs before harvesting. Serum from healthy individuals was used as a negative control. All procedures were performed under level P2 biosafety conditions to minimize the possibility of cross-contamination.
siRNA duplex composed of 21-bp sense and antisense oligonucleotides of ATR was synthesized by Beijing AuGCT Biotechnology Co. Ltd (China). The sequence of siRNA oligos used in this study was (only the sense strand is shown) siATR: 5'-AACCUCCGUGAUGUUGCUUGATT-3'. HL7702 cells were transfected with 50 nmol/L siRNA using lipofectamine 2000 (Promega Corp., Madison, WI, USA) according to the manufacturer’s protocol. At 24 h after transfection, cells were subjected to HBV infection and then the HBV DNA load was tested at indicated time points post-infection.
For FQ-PCR analyses, virus DNA was extracted from the culture medium using an alkaline lysis method. Briefly, HBV DNA was measured using a FQ-PCR diagnostic kit from Da-An Gene Corporation. FQ-PCR was performed using a Lightcycler™ Roche FQ-PCR system with the following amplification cycle: an initial denaturation at 93°C for 2 min, followed by 40 cycles of annealing at 93°C for 5 s, at 57°C for 45 s, and a final extension at 37°C for 10 s.
Cell extracts were lysed in ice-cold Tris buffer (50 mmol/L, pH 7.5) containing 5 mmol/L EDTA, 300 mmol/L NaCl, 0.1% Igepal, 0.5 mmol/L NaF, 0.5 mmol/L Na3VO4, 0.5 mmol/L PMSF, and antiprotease mixture (Roche Molecular Biochemicals) for 30 min and centrifuged at 13 000 ×g for 10 min. The supernatant protein concentration was determined with the Bradford procedure (BioRad). The proteins were resolved on a 7%-15% SDS-PAGE gel and transferred onto nitrocellulose membranes. Blots were blocked in TBST containing 5% nonfat dried milk and incubated with the primary antibodies. Antibodies against p21, ATR (Santa Cruz) and tubulin (Sigma) were incubated at room temperature for 1 h, while antibodies against ATM phosphoserine 1981 (ATMp), Chk1 phosphoserine 345 (Chk1p), and p53 phosphoserine 15 (p53p) (cell signaling) were incubated at 4°C overnight. Secondary antibodies were from Jackson Laboratories. Horseradish peroxidase-based detection was performed using a chemiluminescence reagent (Amersham Biosciences), according to the manufacturer’s instructions.
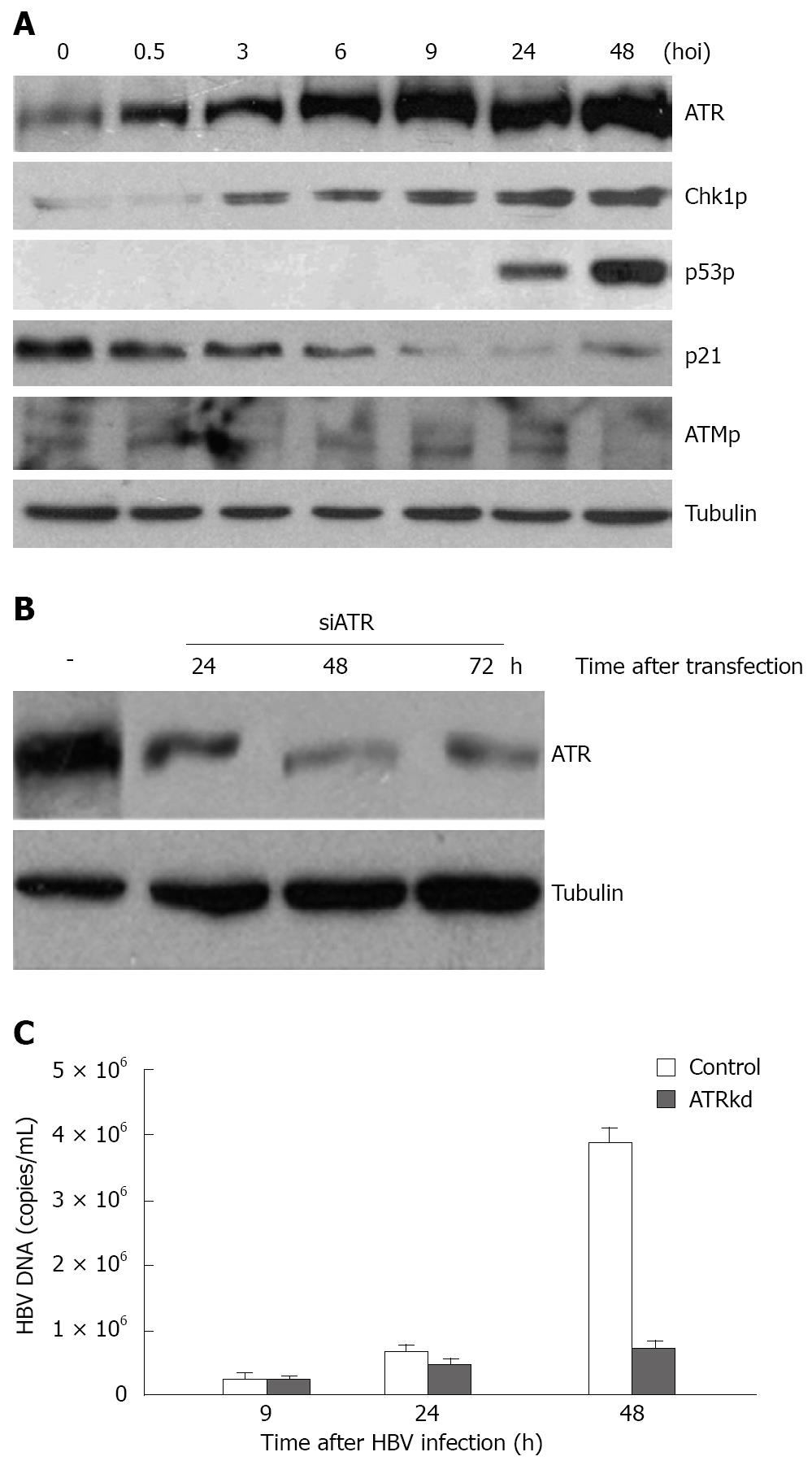
To identify whether a cellular DNA damage response was induced upon HBV infection, we set up a HBV infection and replication model by culturing normal HL7702 liver cells with HBV-positive serum[13]. A monolayer (105) of the human HL7702 liver cells in 6-cm plates was infected with 106 HBV-DNA copies/mL serum at 37°C in an atmosphere containing 50 mL/L CO2. HBV infection induced an increase in the steady state levels of the ATR protein and in the phosphorylation levels of its downstream substrates Chk1 and p53 (Figure 1A). Chk1 phosphorylation at Ser-345 was evidently increased at the start of infection with a further increase from 6 to 48 h. p53 phosphorylation at Ser-15 was elevated beginning at 24 hpi and increased considerably at 48 hpi.
In contrast to ATR and its target, the phosphorylated form of ATM at Ser-1981 did not effectively increase upon infection. Furthermore, the phosphorylation of its downstream substrate Chk2 at Thr-68 began to decrease from 3 hpi (data not shown). p53 transcriptional target p21cip1/waf1-, a cyclin-dependent kinase inhibitory protein, decreased substantially with time after infection, suggesting that p53-dependent downstream signaling can be blocked during HBV infection despite the appearance of phosphorylated p53. Taken together, these results indicate that HBV infection elicited activation of the ATR DNA damage checkpoint pathway that responds to replication stress rather than the ATM DNA damage checkpoint pathway.
To investigate the effect of the activated DNA damage response on HBV infection and replication, HBV infection was performed in ATR knockdown cells. ATR knockdown was conducted with short RNAi oligos transfection using lipofectamine 2000. RNAi technique diminished total ATR protein from 24 h to 72 h after RNA oligos transfection (Figure 1B). Based on this, HBV-positive serum was added to ATR knockdown cells 24 h after the transfection, cells were then harvested for HBV DNA titer assay at 9, 24 and 48 h, respectively, after infection. HBV DNA load in ATR knockdown cells was reduced to about 20% of control (Figure 1C), indicating that HBV replication is dependent on the presence of ATR protein.
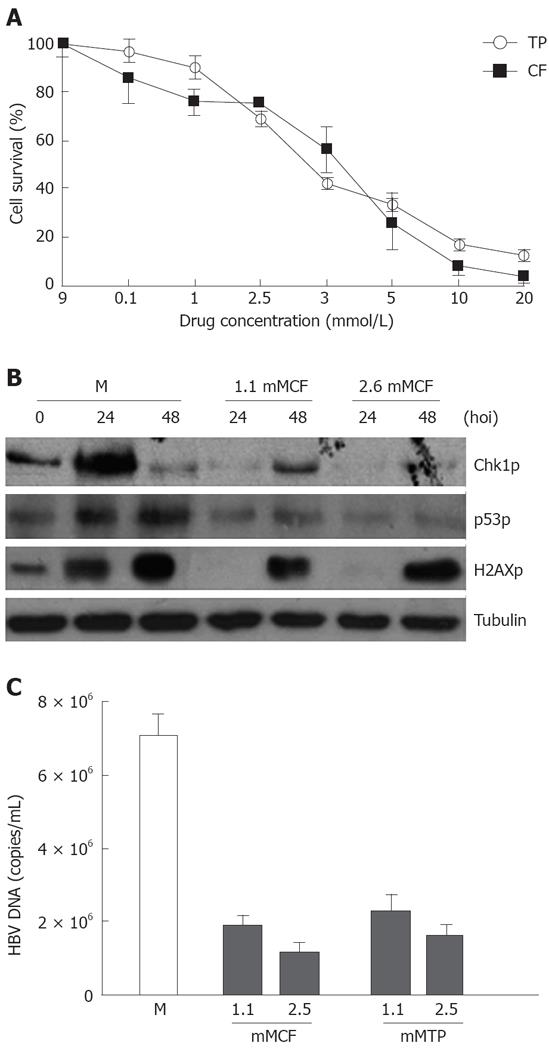
To evaluate the influence of the ATR and ATM kinase inhibitor CF and its methylxanthines on HBV replication, the infected cells were simultaneously treated with different types of drugs (CF and TP) at various concentrations. Trypan blue staining was performed and viable cells were counted at different time points after drug addition. The percentage of cells survived was determined by the ratio of the number of treated cells divided by the number of untreated cells. The DNA extracted from the infected cells was subjected to FQ-PCR analysis, and the degree of amplification of the viral genome over the time course of infection was determined. We observed a 70% decrease in HBV DNA yields in cells treated with 1.1 mmol/L CF at 48 h during HBV infection (Figure 2C). However, such a dose did not affect cell proliferation (Figure 2A) but abrogated the phosphorylation of target proteins with ATR kinase activity such as Chk1 at Ser-345, H2AX at Ser-139, and p53 at Ser-15 (Figure 2B).
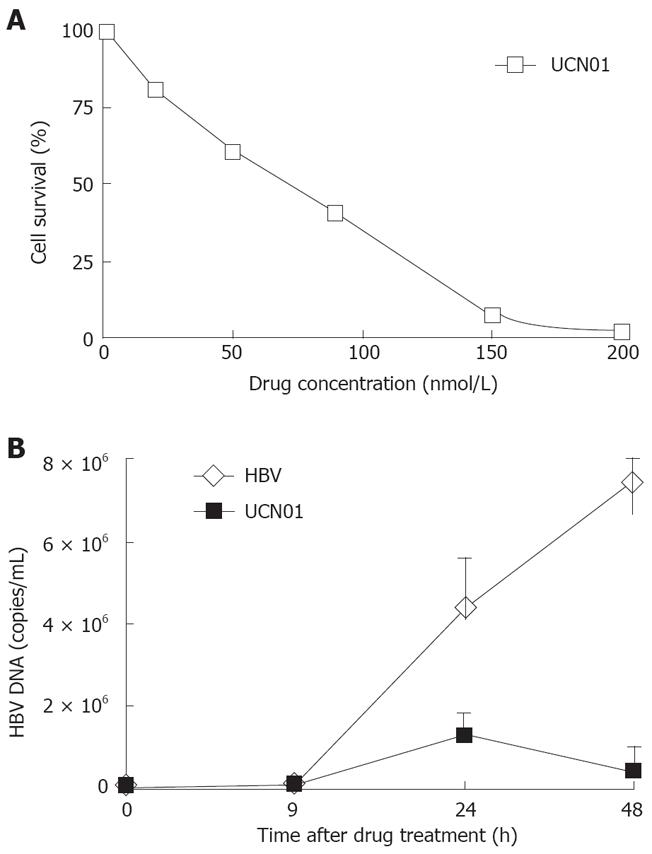
Chk1 phosphorylation was greatly increased during HBV infection. We therefore investigated whether Chk1 inhibitors would have an effect on HBV replication. Infected cells were simultaneously treated with various concentrations of UCN01. The percentage of survived cells was calculated as described above. The DNA extracted from infected cells was subjected to FQ-PCR analysis. We observed a 90% decrease in HBV DNA yields in cells treated with 50 nmol/L UCN01 (Figure 3B), which did not significantly affect the cell survival was not significantly affected in 48 h (Figure 3A).
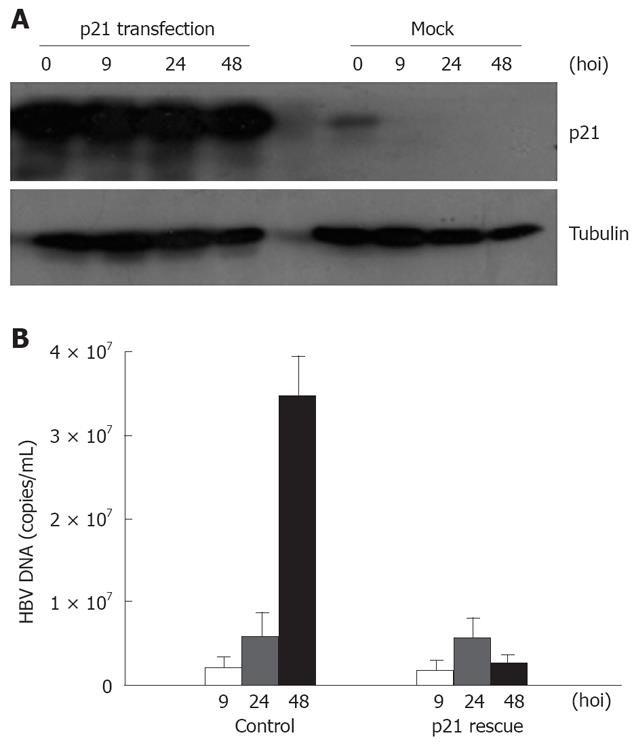
In contrast to increased levels of sustained ATR and Chk1 (ATR substrate) phosphorylation during HBV infection, HBV abrogated p53-dependent cell cycle checkpoint signaling by degrading p21 (Figure 1A). We thus introduced p21 into host cells to investigate its effect on HBV replication. WT p21 cDNA was transfected into HL7702 cells, which were infected with HBV-positive serum 24 h later, harvested at 9, 24 and 48 h respectively after HBV infection, and subjected to FQ-PCR and immunoblotting. p21 expression decreased with time after infection, and myc-tagged p21 expression was higher in the transfected host cells than in the mock-transfected cells (Figure 4A). We then investigated the effects of p21 transfection on HBV infection and replication. Figure 4B showed that the yield of HBV DNA in the p21-transfected cells was only approximately 8% of that in the cells infected only with HBV, indicating that recovery of p21 destroyed by HBV infection has a detrimental effect on replication of the virus.
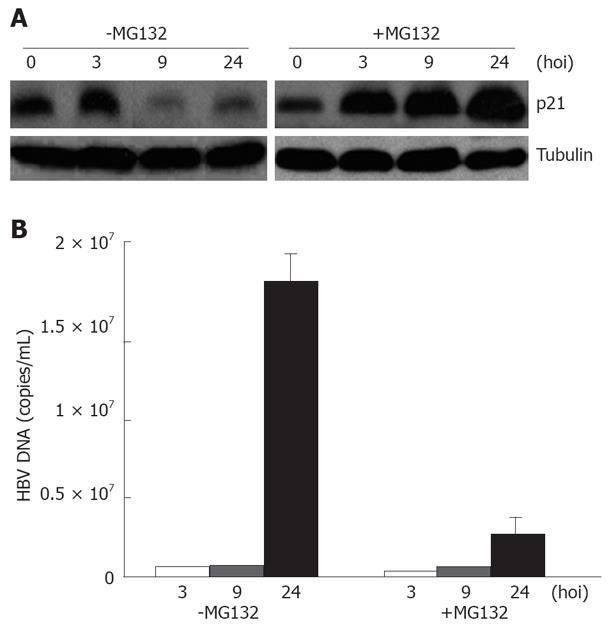
Previous studies have established that HIV-1 infection can be enhanced by treatment of infected cells with proteasome chemical inhibitors[7,8]. It is likely that such inhibitors protect the viral core from degradation in target cells. We have proved that introduction of p21 into infected cells leads to decreased HBV replication. Therefore, we tested whether the proteasome inhibitor MG132 would have an effect on HBV replication in the presence of p21 accumulation. To determine p21 levels and the effect of proteasome inhibitors on cell susceptibility to HBV infection, we pretreated cultures of HL7702 cells with MG132 (10 μM) for 2 h and then monitored HBV infection. HBV-infected cells were harvested at 0, 9, 24, 48 h after extensive washes 8 times with PBS, and then subjected to immunoblotting and FQ-PCR. The level of p21 expression decreased with time after infection, the addition of MG132 resulted in a substantial increase in the p21 expression levels (Figure 5A). To investigate whether treatment with MG132 can also effectively inhibit virus replication, we analyzed the effect of MG132 on HBV replication. The yield of HBV in the presence of proteasome inhibitors was only approximately 10% of that in control cells 48 h after HBV infection (Figure 5B), indicating that proteasome inhibitors can reduce HBV replication by up-regulating p21.
Several viruses, including DNA virus herpes simplex virus (HSV)[14,15], Kaposi’s sarcoma-associated herpesvirus[16], human cytomegalovirus (HCMV)[17] Epstein-Barr virus (EBV)[18], papillomavirus[19,20], simian virus 40 (SV40)[21] and retrovirus HIV[22-24], trigger cellular signaling cascades that are characteristic of a DNA damage response. Infection and replication of HBV have been achieved in human hepatoma cell lines and primary fetal human hepatocytes using transfected HBV genomes or HBV serum. In this study, HL7702 cells were inoculated with HBV serum consistently (106 particles per 105 cells), mimicking the HBV infection process. Serum from healthy individuals was used as the non-infected control. By using FQ-PCR and southern blotting, we demonstrated that human hepatocytes could maintain HBV infection in vitro and support the replication of HBV DNA (data not shown), suggesting that HBV infection activates the DNA damage response to replication stress accompanying increased ATR and Chk1 (ATR substrate) phosphorylation. Furthermore, HBV abrogated the checkpoint signaling by degrading p21. The results of this study show that the ATM and ATR inhibitor CF and its methylxanthine TP, the Chk1 inhibitor UCN01, the proteasome inhibitor MG132 and p21 up-regulation could suppress HBV replication. These findings suggest that some DNA damage responsive proteins are implicated in modulating HBV infections and therefore can be used in treatment of drug-resistant viral strains. In addition, targeting cellular proteins with a low mutation rate may not lead to the rapid emergence of HBV strains that are resistant to inhibitors of these proteins.
The effect of CF is mediated by inhibiting its cellular target, the ATR kinase involved in retroviral integration. The precise role of ATR in HBV replication is unclear. The integration of HBV DNA into chromosomes has been reported[13]. Interestingly, treatment with TP has an inhibitory effect on HBV replication even at a concentration as low as 0.1 mmol/L, which is too low to impact ATM/ATR kinase activity. Therefore, the antioxidant function of TP or CF may have a great inhibitory effect on HBV infection process, and further experiments should be done on this issue.
Chk1 is activated following HBV infection and the Chk1 inhibitor UCN01 can inhibit HBV replication. however, the precise mechanism underlying this inhibition remains to be determined. The resultant S-G2 phase-like cellular condition appears to favor replication of viral DNA.
In cervical carcinogenesis, the p53 suppressor pathway is disrupted by HPV E6-mediated proteasome degradation. MG132 could restore WT p53 levels and thus may sensitize papillomavirus (PV) positive human cervical cancer cells to apoptosis[11,25]. Several studies have recently demonstrated that proteasome is a suitable neoplastic target with a clinical potential[26-28]. We report here that MG132 can be used to suppress HBV replication by up-regulating p21. As a latency virus, HBV abrogates the checkpoint signaling controlled by ATR to prevent triggering of apoptotic signals. The mechanism underlying the regulation of apoptosis by HBV is via both p53-dependent pathways. The p53-dependent cell cycle checkpoint pathway involves p21-mediated inactivation of cdk2/cyclinE. HBV abrogates p53-dependent checkpoint activation by degrading p21. Our results demonstrate that MG132 could up-regulate p21 and thus suppress HBV replication, possibly due to the partial recovery of the p53-dependent checkpoint signaling pathway.
In summary, CF and its related methylxanthines can suppress replication of HIV as previously reported[8]. However, since DNA damage response is acute, DNA repair cofactors against HBV infection and replication should be used in early stage HBV infection. Further studies are necessary to determine the mechanism by which DNA damage response inhibitors down-regulate HBV infection and replication.
Eukaryotic cells employ multiple strategies of checkpoint signaling and DNA repair mechanisms to monitor and repair damaged DNA. There are two branches in the checkpoint response pathway: ataxia telangiectasia-mutated (ATM) branch and ATM-Rad3-related (ATR) branch. Many viruses are now known to interact with DNA damage and repair machinery. These viruses have evolved tactics to eliminate, circumvent, or exploit various aspects of DNA damage response to the host cells. Strategies include activation of repair proteins or targeting of specific cellular factors for degradation or mislocalization.
In human immunodeficiency virus (HIV), prevention of viral integration inhibits viral replication and promotes cellular apoptosis. Thus, the ATM-specific inhibitor ku55933 can inhibit HIV replication in primary T cells.
In this paper, we first report that (hepatic B virus) HBV infection activates and exploits DNA damage response to replication stress. We investigated whether the inhibition of DNA damage response by CF, TP and UCN01 or the restoration of p21 expression by p21 transfection or proteasome inhibition would lead to suppression of HBV replication. We set up a chronic HBV infection model by culturing HL7702 liver cells with HBV-positive serum without washing off input virus as conventional. HBV DNA titers inside the infected cells represent the final viral amount including the infected DNA not degraded and the newly synthesized HBV DNA. In this way, we can study the efficacy of DNA damage response inhibitors on HBV infection and replication.
Since DNA damage response is an acute response occurring quickly after virus infection, we assume that early intervention of the DNA damage pathway would function more efficiently and thus can be used in clinical practice as a HBV infection therapy during its early infectious stage or fulminant HBV infection.
This is an interesting article that reports data on the effect of DNA repair cofactors on HBV replication and infection. The study is well designed. The data or findings are reliable.
Peer reviewer: Dr. Carla Brady, Duke University Medical Center, DUMC Box 3913, Durham 27705, United States
S- Editor Zhong XY L- Editor Wang XL E- Editor Lin YP
| 1. | Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat Rev Cancer. 2003;3:155-168. |
| 2. | Helt CE, Cliby WA, Keng PC, Bambara RA, O'Reilly MA. Ataxia telangiectasia mutated (ATM) and ATM and Rad3-related protein exhibit selective target specificities in response to different forms of DNA damage. J Biol Chem. 2005;280:1186-1192. |
| 3. | Weitzman MD, Carson CT, Schwartz RA, Lilley CE. Interactions of viruses with the cellular DNA repair machinery. DNA Repair (Amst). 2004;3:1165-1173. |
| 4. | Everett RD. Interactions between DNA viruses, ND10 and the DNA damage response. Cell Microbiol. 2006;8:365-374. |
| 5. | Lilley CE, Schwartz RA, Weitzman MD. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 2007;15:119-126. |
| 6. | Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc Natl Acad Sci USA. 2005;102:5844-5849. |
| 7. | Lau A, Swinbank KM, Ahmed PS, Taylor DL, Jackson SP, Smith GC, O'Connor MJ. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat Cell Biol. 2005;7:493-500. |
| 8. | Nunnari G, Argyris E, Fang J, Mehlman KE, Pomerantz RJ, Daniel R. Inhibition of HIV-1 replication by caffeine and caffeine-related methylxanthines. Virology. 2005;335:177-184. |
| 9. | Xiao Z, Xue J, Semizarov D, Sowin TJ, Rosenberg SH, Zhang H. Novel indication for cancer therapy: Chk1 inhibition sensitizes tumor cells to antimitotics. Int J Cancer. 2005;115:528-538. |
| 10. | Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14:1448-1459. |
| 11. | Hougardy BM, Maduro JH, van der Zee AG, de Groot DJ, van den Heuvel FA, de Vries EG, de Jong S. Proteasome inhibitor MG132 sensitizes HPV-positive human cervical cancer cells to rhTRAIL-induced apoptosis. Int J Cancer. 2006;118:1892-900. |
| 12. | Qi M, Aiken C. Selective restriction of Nef-defective human immunodeficiency virus type 1 by a proteasome-dependent mechanism. J Virol. 2007;81:1534-1536. |
| 13. | Lin M, Chen Q, Yang LY, Li WY, Cao XB, Wu JR, Peng YP, Chen MR. Hepatitis B virus infection and replication in primarily cultured human fetal hepatocytes. World J Gastroenterol. 2007;13:1027-1031. |
| 14. | Wilkinson DE, Weller SK. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J Cell Sci. 2006;119:2695-2703. |
| 15. | Liang X, Pickering MT, Cho NH, Chang H, Volkert MR, Kowalik TF, Jung JU. Deregulation of DNA damage signal transduction by herpesvirus latency-associated M2. J Virol. 2006;80:5862-5874. |
| 16. | Shin YC, Nakamura H, Liang X, Feng P, Chang H, Kowalik TF, Jung JU. Inhibition of the ATM/p53 signal transduction pathway by Kaposi's sarcoma-associated herpesvirus interferon regulatory factor 1. J Virol. 2006;80:2257-2266. |
| 17. | Gaspar M, Shenk T. Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc Natl Acad Sci USA. 2006;103:2821-2826. |
| 18. | Kudoh A, Fujita M, Zhang L, Shirata N, Daikoku T, Sugaya Y, Isomura H, Nishiyama Y, Tsurumi T. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J Biol Chem. 2005;280:8156-8163. |
| 19. | Kumar A, Zhao Y, Meng G, Zeng M, Srinivasan S, Delmolino LM, Gao Q, Dimri G, Weber GF, Wazer DE. Human papillomavirus oncoprotein E6 inactivates the transcriptional coactivator human ADA3. Mol Cell Biol. 2002;22:5801-5812. |
| 20. | Horner SM, DeFilippis RA, Manuelidis L, DiMaio D. Repression of the human papillomavirus E6 gene initiates p53-dependent, telomerase-independent senescence and apoptosis in HeLa cervical carcinoma cells. J Virol. 2004;78:4063-4073. |
| 21. | Shi Y, Dodson GE, Shaikh S, Rundell K, Tibbetts RS. Ataxia-telangiectasia-mutated (ATM) is a T-antigen kinase that controls SV40 viral replication in vivo. J Biol Chem. 2005;280:40195-40200. |
| 22. | Zimmerman ES, Sherman MP, Blackett JL, Neidleman JA, Kreis C, Mundt P, Williams SA, Warmerdam M, Kahn J, Hecht FM. Human immunodeficiency virus type 1 Vpr induces DNA replication stress in vitro and in vivo. J Virol. 2006;80:10407-10418. |
| 23. | Daniel R, Kao G, Taganov K, Greger JG, Favorova O, Merkel G, Yen TJ, Katz RA, Skalka AM. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. Proc Natl Acad Sci USA. 2003;100:4778-4783. |
| 24. | Roshal M, Kim B, Zhu Y, Nghiem P, Planelles V. Activation of the ATR-mediated DNA damage response by the HIV-1 viral protein R. J Biol Chem. 2003;278:25879-25886. |
| 25. | Aguilar-Lemarroy A, Gariglio P, Whitaker NJ, Eichhorst ST, zur Hausen H, Krammer PH, Rosl F. Restoration of p53 expression sensitizes human papillomavirus type 16 immortalized human keratinocytes to CD95-mediated apoptosis. Oncogene. 2002;21:165-175. |
| 26. | Poulaki V, Mitsiades CS, Kotoula V, Negri J, McMillin D, Miller JW, Mitsiades N. The proteasome inhibitor bortezomib induces apoptosis in human retinoblastoma cell lines in vitro. Invest Ophthalmol Vis Sci. 2007;48:4706-4719. |
| 27. | Spano JP, Bay JO, Blay JY, Rixe O. Proteasome inhibition: a new approach for the treatment of malignancies. Bull Cancer. 2005;92:E61-E66, 945-952. |