Published online Jun 21, 2008. doi: 10.3748/wjg.14.3754
Revised: April 19, 2008
Accepted: April 26, 2008
Published online: June 21, 2008
AIM: To evaluate the inhibitory effects of human fragile histidine triad (FHIT) gene on cell proliferation and apoptosis in human hepatocellular carcinoma line Hep3B in vitro.
METHODS: A recombinant pcDNA3.1 (+)/FHIT including the functional region of FHIT gene was constructed and transferred into human hepatocellular carcinoma cells in vitro. mRNA and protein expression of the FHIT gene in the transfected cells was detected by RT-PCR and Western blot, respectively. The effect of FHIT on proliferation was detected by MTT assay. Changes in cell cycle and apoptosis were assayed by flow cytometry. Five mice received subcutaneous transplantation of Hep3B-FHIT; 5 mice received subcutaneous transplantation of normal Hep3B and Hep3B-C as controls. The body weight of nude mice and tumor growth were measured.
RESULTS: RT-PCR and Western blot analysis showed that the expression level of FHIT-mRNA and FHIT protein was higher in Hep3B cells after infection with pcDNA3.1 (+)/FHIT. The growth of Hep3B cells treated with pcDNA3.1 (+)/FHIT was significantly inhibited. The pcDNA3.1 (+)/FHIT-transfected Hep3B cells showed a significantly higher cell rate at G0-G1 phase and increased apoptosis in comparison with controls (P < 0.05). The growth of transplanted tumor was inhibited markedly by FHIT. Tumors arising from the Hep3B-FHIT cells occurred much later than those arising from the Hep3B and Hep3B-C cells. The growth of Hep3B-FHIT cells was slow and the tumor volume was low.
CONCLUSION: Transduction of FHIT gene inhibits the growth of human hepatocellular carcinoma cells and induces cell apoptosis in vivo and in vitro.
- Citation: Xu RH, Zheng LY, He DL, Tong J, Zheng LP, Zheng WP, Meng J, Xia LP, Wang CJ, Yi JL. Effect of fragile histidine triad gene transduction on proliferation and apoptosis of human hepatocellular carcinoma cells. World J Gastroenterol 2008; 14(23): 3754-3758
- URL: https://www.wjgnet.com/1007-9327/full/v14/i23/3754.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.3754
Fragile histidine triad (FHIT) gene has been successfully cloned on chromosome 3p14.2 using an exon trapping method for gene capture[1–4]. It was reported that abnormal FHIT gene exists in majority of tumors, and is an important candidate tumor-suppressing gene[5–10]. In the hepatoma cell line Hep3B, the FHIT gene, mRNA, and protein are abnormal[11]. We constructed a recombinant pcDNA3.1 (+) /FHIT vector containing human (FHIT) gene, which was used to transfect human hepatoma Hep3B cells in vitro and in vivo to explore the effect of FHIT gene on proliferation or apoptosis of hepatocellular carcinoma cells.
E.coli DH5α and eukaryotic expression vector pcDNA3.1 (+) were routinely kept in our laboratory. PBluescript SK FHIT plasmid with a full-length FHIT cDNA was kindly given by Professor Xiao-Fan Wang, Duke University, USA. Hep3B cells were purchased from the Chinese Academy of Sciences in Shanghai.
A reverse transcription-polymerase chain reaction (RT-PCR) kit was purchased from Takara, Japan. FHIT antibody was purchased from Zymed, USA. A cell cycle assay kit was purchased from Becton Dickinson, USA.
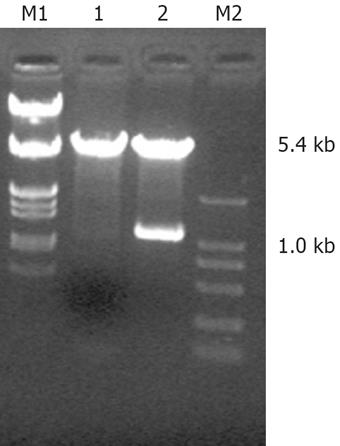
To establish an enzyme-cutting reaction, pcDNA3.1 (or PBluescript SK FHIT), BamH I and Xba I were mixed in a water bath at 37°C for 2 h. The mixture was electrophoresed, and 1.0 kb FHIT gene fragments, and a 5.4 kb linear pcDNA3.1 were retrieved separately with electrophoresis coagulation plastic boxes. For constructing the pcDNA3.1 (+) /FHIT vector, FHIT gene fragments, linear pcDNA3.1 and T4 ligase were mixed in an Eppendorf tube at 16°C for 12 h to establish a recombinant plasmid reaction. The pcDNA3.1 FHIT vector was transferred into competent bacteria DH5, and the clones were cultured and identified.
Hep3B cells were cultured at 37°C in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum (Gibco), 1.0 mmol/L sodium pyruvate, 0.1 mmol/L non-indispensable amino acid and 5% CO2, at a saturated humidity. Hep3B cells with strong growth were seeded in 6-well plates. pcDNA3.1 (+) /FHIT was transfected into the Hep3B cells at the density of 70% with Lipofectamine 2000 (liposome transfection kit) following its manufacturer’s instructions. After transfection, the cells were screened using G418 (500 mg/L) and the screening was maintained with G418 (250 mg/L). Hep3B cells transfected with FHIT gene and empty vector were named Hep3B-FHIT and Hep3B-C, respectively. Parent Hep3B cells were used as an untransfected control.
FHIT gene was amplified with primers (P1: 5' -ATGTCGTTCAGA-3', P2: 5'-CTGAAAGTACAC-3') using the Hep3B-FHIT cells screened by G418[12]. Total RNA was extracted with TRIZOL method and underwent reverse-transcription PCR. The PCR products were electrophoresed on a 1% agarose gel. One &mgr;L of the resulting cDNA was added into the PCR reaction mixture (containing 1.0 &mgr;L of forward primer (10 pmol/&mgr;L), 1.0 &mgr;L of reverse primer(10 pmol/&mgr;L), 1.0 &mgr;L of dNTPs(10 mmol/L), 0.5 &mgr;L Taq DNA polymerase). PCR was performed for 30 cycles, each amplification cycle consisting of denaturation at 94°C for 1 min, primer annealing at 44°C for 1 min, and extension at 72°C for 45 s. The PCR products were analyzed on 10 g/L agarose gel containing ethidium bromide.
The screened Hep3B-FHIT cells were lysed with a cell-lysing solution. The protein concentration in the supernatant after centrifugation was measured by Bradford assay. The supernatant was electrophoresed on a 12% SDS-polyacrylamide gel. The electrophoresed proteins were transferred to nitrocellulose membrane, blocked with calf serum, combined with FHIT antibody and horseradish peroxidase secondary antibody, developed by enhanced chemiluminescence (ECL) and photographed.
Ten thousand cells/well were seeded in 96-well plates (200 &mgr;L/well), with 6 wells for each cell group and 3 wells for control. MTT (5 g/L) was used as an incubation solution. DMSO was added to the cell culture for 4 h. The luminous absorbance was measured by ELISA (wavelength of 570 nm). The measurement was repeated once a day for 6 days.
After trypsinization, the cells were washed twice with phosphate-buffered saline (PBS), fixed with cold ethanol (70%) overnight, suspended in PBS after ethanol was purged from PBS, and stained with propidium iodide (50 &mgr;g/mL). The samples were examined by flow cytometry.
After forty-eight hours, 2 &mgr;g of pcDNA3.1 (+) and pcDNA3.1 (+)/FHIT plasmid was transfected with Lipofectamine 2000 into Hep3B cell lines, and clones were obtained by screening with G418 for 14 d. The clones were cultured. The study was approved by the Experimental Animal Committee of Tongji Medical College, Huazhong University of Science and Technology, and all animal experiments adhered to the Animal Welfare Committee Guidelines. Male athymic BALB/c nu/ nu mice (4-6 wk old) were obtained from the Institute of Materia Medica (Tongji Medical College, Wuhan, China) and housed in laminar-flow cabinets under specific pathogen-free (SPF) conditions. Fifteen male mice were randomized into Hep3B-FHIT/nude, Hep3B-C/nude and Hep3B/nude groups, 5 in each group. A suspension of Hep3B-FHIT or Hep3B-C or Hep3B cells (107 cells in 0.15 mL Hanks’ solution) was injected into the back of mice in each group. After implantation, tumor growth was detected weekly by measuring its diameter with a Vernier caliper. Tumor volume (TV) was calculated using the following formula: TV (mm3) = d2× D/2, where d is the shortest diameter and D is the longest diameter. Animals were sacrificed 7 wk after implantation, and samples were harvested. The body weight of mice, and tumor growth were measured.
The data were expressed as mean ± SD. Student’s two-sided t-test was used to compare the values of the test and control samples by software SPSS11.5. P < 0.05 was considered statistically significant.
The sequence of PBluescript SK FHIT detected in Shanghai Biological Engineering Company was identical to that of FHIT mRNA in GenBank (Lot. Number: NM-002012). After enzyme-cut, agarose gel electrophoresis showed two DNA bands of plasmid PBluescript FHIT (1.0 kb and 2.7 kb), two DNA bands of plasmid pcDNA3.1 (+)/FHIT (1.0 kb and 5.4 kb) (Figure 1); thus, pcDNA3.1 (+)/FHIT was successfully constructed.
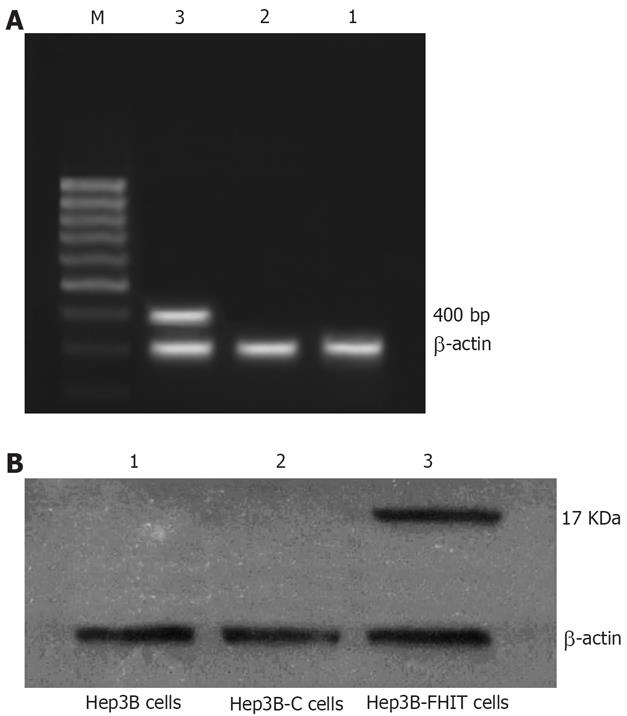
After transfected with pcDNA3.1 (+)/FHIT, RT-PCR showed a 400 bp band of Hep3B-FHIT cells. However, the empty vector-transfected, or native cells did not show any band (Figure 2A).
Western blotting demonstrated FHIT protein (17 kDa) in the cells transfected with pcDNA3.1 (+)/FHIT, but no FHIT protein in empty vector-transfected or native cells (Figure 2B).
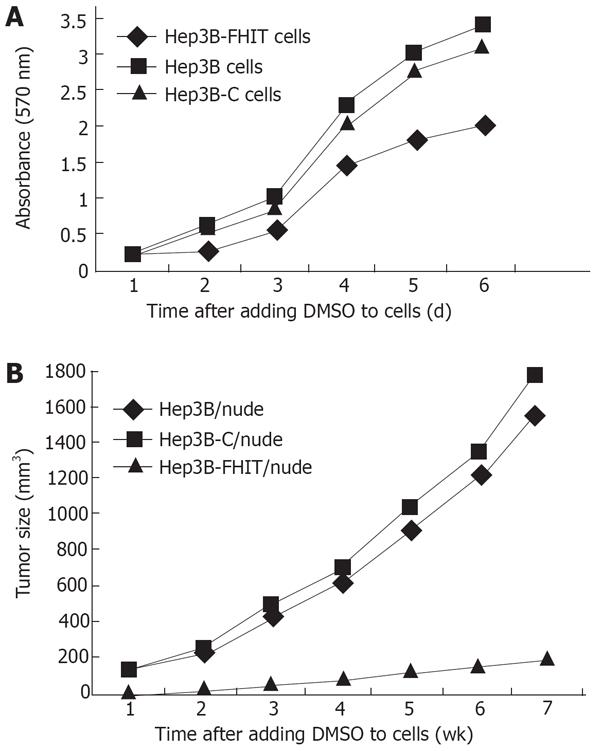
Thiazolyl tetrazolium (MTT) assay showed that the growth rate of transfected cells was significantly lower than that of native or empty vector-transfected cells, especially after the logarithm growth phase (P < 0.05, Figure 3A).
Flow cytometry analysis revealed that the number of Hep3B cells decreased in the G2/M and S phases, but increased in the G0/G1 phase. The apoptosis rate was higher for transfected cells than for empty vector-transfected or native cells (P < 0.05, Table 1).
| Cell type | G0/G1 | S | G2/M | Apoptosis rate (%) |
| Hep3B | 54.36 ± 0.78 | 17.40 ± 1.32 | 28.01 ± 1.12 | 3.78 ± 0.36 |
| Hep3B-C | 53.17 ± 0.52 | 18.23 ± 2.51 | 28.55 ± 0.55 | 3.52 ± 0.33 |
| Hep3B-FHITa | 72.23 ± 0.84 | 12.57 ± 0.42 | 15.12 ± 1.31 | 9.74 ± 0.43 |
The growth of transplanted tumor was inhibited markedly by FHIT, showing that tumors arising from the Hep3B-FHIT cells occur much later than those arising from the Hep3B and Hep3B-C cells. Hep3B-FHIT cells grew slowly and their volume was small (Table 2, Figure 3B).
| Group | Mean tumor formation time (d) | Mean tumor weight (g) |
| Hep3B/nude | 3.22 ± 0.31 | 3.66 ± 0.40 |
| Hep3B-C/nude | 3.08 ± 0.34 | 3.94 ± 0.39 |
| Hep3B-FHIT/nude | 12.17 ± 1.19 | 0.60 ± 0.04 |
| Total F value | 72.38 | 168.36 |
| Total P value | 0.00 | 0.00 |
It has been reported that FHIT is the first cancer-suppressive gene which links fragile sites to tumors. Abnormality of the FHIT gene is an early event during tumor development[13–17]. Fracture at the fragile site results in mutation and inactivation of the FHIT gene; thus leading to abnormal cell growth[18–22]. Like many other tumors, hepatoma is characterized by highly mutated FHIT gene or gene loss. Yuan et al[11] found that down-regulation of the FHIT gene is detected in 64.3% cell lines (four cell lines showing mRNA down-regulation did not express FHIT protein); allelic loss of intron 5 of the FHIT gene was detected in 29.4% hepatomas by in situ hybridization; structural alterations of chromosome 3p were identified in 61.5% of hepatocellular carcinomas; expression of FHIT protein was not detectable in 50% primary tumors with immunostaining[11].
FHIT may modulate APnA by hydrolysis of ApnA to yield adenosine 5'-monophosphate (AMP). Due to mutation of FHIT, loss of ApnA hydrolase activity results in elevated Ap3A levels, intracellular accumulation of APnA strengthens the growth signal transduction, and blocks growth inhibition and apoptosis, thus contributing to carcinogenesis. Additionally, the activity of FHIT on mRNA cap analogs raises the possibility that failure of a decapping function might be tumorigenic[122324].
Gramantieri et al[13] examined the mRNA FHIT expression in both cancerous and matched non-cancerous tissues in 28 cases of hepatocellular carcinoma (HCC) and 10 normal livers, and abnormal FHIT transcripts were detected in 13 cases (in cancerous tissue from 11 cases and in non-cancerous tissue from 2 cases). No abnormal FHIT transcripts were found in normal livers. It was recently reported that many other factors can also induce liver carcinogenesis in rats[25–29].
Siprashvili et al[30] found that transduction of wild-type FHIT or mutative FHIT with no hydrolase activity into tumor cells lacking FHIT gene did not show any advantage to cell growth, and no difference was found in suppression of tumorigenicity, suggesting that FHIT suppresses the tumorigenicity through FHIT protein in combination with its substrate rather than through hydrolysis of APnA. Its more precise mechanism needs to be further explored.
In this study, the full-length of the FHIT cDNA eukaryotic expression vector was transfected into human hepatocellular carcinoma cell line, Hep3B, to manipulate FHIT expression, which alters the biological features of Hep3B cell line. The results indicate that successfully transfected Hep3B cells could express FHIT mRNA and FHIT protein, whereas the empty-vector transfected Hep3B or native cells did not express them. Moreover, cell proliferation and differentiation were significantly decreased at the G2/M and S phases, and cell apoptosis increased at the G0/G1 phases of Hep3B cells transfected with FHIT gene as compared with the control group, suggesting that FHIT gene plays an important role in blocking cell growth at the G1 phase, and in inducing cell apoptosis synergized by other apoptosis-inducing factors. Furthermore, transfer of FHIT gene could inhibit the growth of human hepatocellular carcinoma cells, and induce cell apoptosis in vivo.
In conclusion, FHIT mRNA, and protein are expressed in FHIT-infected Hep3B cells, thus leading to low proliferation, and high apoptosis of HCC cells. Transfection of FHIT gene into human hepatocellular carcinoma cells is a promising therapeutic approach to HCC.
Abnormal fragile histidine triad (FHIT) gene, an important candidate tumor-suppressing gene, exists in a majority of tumors. In the hepatoma cell line Hep3B, the FHIT gene, mRNA, and protein is abnormal. Therefore, we constructed a recombinant pcDNA3.1 (+)/FHIT vector containing human fragile histidine triad (FHIT) gene, which was used to transfect human hepatoma Hep3B cells in vitro and in vivo to explore the effect of FHIT gene on proliferation or apoptosis of hepatocellular carcinoma cells.
Application of gene transfer technologies in treatment of cancer has led to the development of new experimental strategies like inhibition of oncogenes, restoration of tumor-suppressor genes and enzyme/prodrug therapy (GDEPT). These strategies are being evaluated for the treatment of primary and metastatic liver cancer.
In this study, we constructed a recombinant pcDNA3.1 (+)/FHIT vector containing human FHIT gene, which can be used to transfect human hepatoma Hep3B cells (FHIT null) in vitro and in vivo. Transfection of FHIT gene could inhibit the growth of human hepatocellular carcinoma cells, and induce cell apoptosis.
Transfection of FHIT gene into human hepatocellular carcinoma cells is a promising therapeutic approach to HCC.
In this paper, the authors evaluated the effect of human FHIT gene on cell proliferation and apoptosis in hepatocellular carcinoma in vitro and in vivo. It is a very interesting paper, and the study is well designed. It provides a potential therapeutic target for hepatocellular carcinoma.
| 1. | Ohta M, Inoue H, Cotticelli MG, Kastury K, Baffa R, Palazzo J, Siprashvili Z, Mori M, McCue P, Druck T. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587-597. |
| 2. | Sozzi G, Veronese ML, Negrini M, Baffa R, Cotticelli MG, Inoue H, Tornielli S, Pilotti S, De Gregorio L, Pastorino U. The FHIT gene 3p14.2 is abnormal in lung cancer. Cell. 1996;85:17-26. |
| 3. | Negrini M, Monaco C, Vorechovsky I, Ohta M, Druck T, Baffa R, Huebner K, Croce CM. The FHIT gene at 3p14.2 is abnormal in breast carcinomas. Cancer Res. 1996;56:3173-3179. |
| 4. | Virgilio L, Shuster M, Gollin SM, Veronese ML, Ohta M, Huebner K, Croce CM. FHIT gene alterations in head and neck squamous cell carcinomas. Proc Natl Acad Sci USA. 1996;93:9770-9775. |
| 5. | Sozzi G, Pastorino U, Moiraghi L, Tagliabue E, Pezzella F, Ghirelli C, Tornielli S, Sard L, Huebner K, Pierotti MA. Loss of FHIT function in lung cancer and preinvasive bronchial lesions. Cancer Res. 1998;58:5032-5037. |
| 6. | Campiglio M, Pekarsky Y, Menard S, Tagliabue E, Pilotti S, Croce CM. FHIT loss of function in human primary breast cancer correlates with advanced stage of the disease. Cancer Res. 1999;59:3866-3869. |
| 7. | Sorio C, Baron A, Orlandini S, Zamboni G, Pederzoli P, Huebner K, Scarpa A. The FHIT gene is expressed in pancreatic ductular cells and is altered in pancreatic cancers. Cancer Res. 1999;59:1308-1314. |
| 8. | Birrer MJ, Hendricks D, Farley J, Sundborg MJ, Bonome T, Walts MJ, Geradts J. Abnormal Fhit expression in malignant and premalignant lesions of the cervix. Cancer Res. 1999;59:5270-5274. |
| 9. | Thiagalingam S, Lisitsyn NA, Hamaguchi M, Wigler MH, Willson JK, Markowitz SD, Leach FS, Kinzler KW, Vogelstein B. Evaluation of the FHIT gene in colorectal cancers. Cancer Res. 1996;56:2936-2939. |
| 10. | Huebner K, Croce CM. Cancer and the FRA3B/FHIT fragile locus: it's a HIT. Br J Cancer. 2003;88:1501-1506. |
| 11. | Yuan BZ, Keck-Waggoner C, Zimonjic DB, Thorgeirsson SS, Popescu NC. Alterations of the FHIT gene in human hepatocellular carcinoma. Cancer Res. 2000;60:1049-1053. |
| 12. | Barnes LD, Garrison PN, Siprashvili Z, Guranowski A, Robinson AK, Ingram SW, Croce CM, Ohta M, Huebner K. Fhit, a putative tumor suppressor in humans, is a dinucleoside 5',5"'-P1,P3-triphosphate hydrolase. Biochemistry. 1996;35:11529-11535. |
| 13. | Gramantieri L, Chieco P, Di Tomaso M, Masi L, Piscaglia F, Brillanti S, Gaiani S, Valgimigli M, Mazziotti A, Bolondi L. Aberrant fragile histidine triad gene transcripts in primary hepatocellular carcinoma and liver cirrhosis. Clin Cancer Res. 1999;5:3468-3475. |
| 14. | Zochbauer-Muller S, Wistuba II, Minna JD, Gazdar AF. Fragile histidine triad (FHIT) gene abnormalities in lung cancer. Clin Lung Cancer. 2000;2:141-145. |
| 15. | Ishii H, Ozawa K, Furukawa Y. Alteration of the fragile histidine triad gene early in carcinogenesis: an update. J Exp Ther Oncol. 2003;3:291-296. |
| 16. | Guler G, Uner A, Guler N, Han SY, Iliopoulos D, McCue P, Huebner K. Concordant loss of fragile gene expression early in breast cancer development. Pathol Int. 2005;55:471-478. |
| 17. | Weiske J, Albring KF, Huber O. The tumor suppressor Fhit acts as a repressor of beta-catenin transcriptional activity. Proc Natl Acad Sci USA. 2007;104:20344-20349. |
| 18. | Ishii H, Furukawa Y. Alterations of common chromosome fragile sites in hematopoietic malignancies. Int J Hematol. 2004;79:238-242. |
| 19. | Lee SH, Kim HY, Kim TJ, Park HK, Kim WH, Woo KM, Cho MH. Aberrant splicing of FHIT transcripts in human gastric cancer cell lines. Res Commun Mol Pathol Pharmacol. 2002;112:39-49. |
| 20. | McAvoy S, Ganapathiraju SC, Ducharme-Smith AL, Pritchett JR, Kosari F, Perez DS, Zhu Y, James CD, Smith DI. Non-random inactivation of large common fragile site genes in different cancers. Cytogenet Genome Res. 2007;118:260-269. |
| 21. | Smith DI, McAvoy S, Zhu Y, Perez DS. Large common fragile site genes and cancer. Semin Cancer Biol. 2007;17:31-41. |
| 22. | Iliopoulos D, Guler G, Han SY, Druck T, Ottey M, McCorkell KA, Huebner K. Roles of FHIT and WWOX fragile genes in cancer. Cancer Lett. 2006;232:27-36. |
| 23. | Zawacka-Pankau J, Podhajska AJ. Expression and simple, one-step purification of fragile histidine triad (Fhit) tumor suppressor mutant forms in Escherichia coli and their interaction with protoporphyrin IX. Biotechnol Lett. 2007;29:877-883. |
| 24. | Huang K, Frey PA. Engineering human Fhit, a diadenosine triphosphate hydrolase, into an efficient dinucleoside polyphosphate synthase. J Am Chem Soc. 2004;126:9548-9549. |
| 25. | Tsujiuchi T, Sasaki Y, Oka Y, Konishi Y, Tsutsumi M. Fhit gene alterations in hepatocarcinogenesis induced by a choline-deficient L-amino acid-defined diet in rats. Mol Carcinog. 2003;36:147-152. |
| 26. | Golebiowski F, Kowara R, Pawelczyk T. Distribution of Fhit protein in rat tissues and its intracellular localization. Mol Cell Biochem. 2001;226:49-55. |
| 27. | Tsujiuchi T, Sasaki Y, Kubozoe T, Tsutsumi M, Konishi Y, Nakae D. Alterations of the Fhit gene in hepatocellular carcinomas induced by N-nitrosodiethylamine in rats. Mol Carcinog. 2002;34:19-24. |
| 28. | Asensio AC, Rodriguez-Ferrer CR, Oaknin S, Rotllan P. Biochemical and immunochemical characterisation of human diadenosine triphosphatase provides evidence for its identification with the tumour suppressor Fhit protein. Biochimie. 2006;88:461-471. |
| 29. | Han SY, Iliopoulos D, Druck T, Guler G, Grubbs CJ, Pereira M, Zhang Z, You M, Lubet RA, Fong LY. CpG methylation in the Fhit regulatory region: relation to Fhit expression in murine tumors. Oncogene. 2004;23:3990-3998. |
| 30. | Siprashvili Z, Sozzi G, Barnes LD, McCue P, Robinson AK, Eryomin V, Sard L, Tagliabue E, Greco A, Fusetti L. Replacement of Fhit in cancer cells suppresses tumorigenicity. Proc Natl Acad Sci USA. 1997;94:13771-13776. |