Published online May 14, 2008. doi: 10.3748/wjg.14.2872
Revised: February 2, 2008
Published online: May 14, 2008
AIM: To develop a simple and convenient method for extracting genomic DNA from intestinal microflora for enterobacterial repetitive intergenic consensus (ERIC)-PCR detection.
METHODS: Five methods of extracting bacterial DNA, including Tris-EDTA buffer, chelex-100, ultrapure water, 2% sodium dodecyl sulfate and 10% Triton-100 with and without sonication, were compared with the commercial fecal DNA extraction kit method, which is considered as the gold standard for DNA extraction. The comparison was based on the yield and purity of DNA and the indexes of the structure and property of micro-organisms that were reflected by ERIC-PCR.
RESULTS: The yield and purity of DNA obtained by the chelex method was similar to that obtained with the fecal DNA kit. The ERIC-PCR results obtained for the DNA extracted by the chelex method and those obtained for DNA extracted with the fecal DNA kit were basically the same.
CONCLUSION: The chelex method is recommended for ERIC-PCR experiments in view of its simplicity and cost-effectiveness; and it is suitable for extracting total DNA from intestinal micro-organisms, particularly for handling a large number of samples.
- Citation: Yang JL, Wang MS, Cheng AC, Pan KC, Li CF, Deng SX. A simple and rapid method for extracting bacterial DNA from intestinal microflora for ERIC-PCR detection. World J Gastroenterol 2008; 14(18): 2872-2876
- URL: https://www.wjgnet.com/1007-9327/full/v14/i18/2872.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.2872
The intestinal tract of animals harbors a large, active, and complex community of microbes[1]. There are at least 400-500 different microbial species, constituting a complex ecosystem[2]. They play significant roles in colonization resistance, i.e., the prevention of colonization of pathogens in the gastrointestinal tract and the growth of autochthonous opportunistic micro-organisms[3]. Intestinal bacteria that synthesize or metabolize potential carcinogens and produce anti-tumorigenic products may be related to colorectal cancer, which is the second most common cause of cancer death in the USA[4].
Probiotics are defined as living organisms which, on ingestion in certain numbers, exert health benefits beyond the inherent basic nutrition in the host[56]. In humans, probiotics are effective in preventing the onset and relapse of pouchitis[78], and in mice, they effectively prevent experimental colitis and reduce gut bacterial translocation[910].
Commensal intestinal microflora have been monitored by cultivation-based techniques and molecular detections[1112]. Enterobacterial repetitive intergenic consensus (ERIC)-PCR uses oligonucleotides targeting short repetitive sequences that are dispersed throughout various bacterial genomes[11]. Commensal intestinal microflora can be identified at the genus, species and strain levels based on the electrophoretic pattern of amplified products[13–15].
The capacity of these assays to monitor bacteria in animal intestines depends on the efficiency of PCR. One of the key factors in obtaining the PCR fingerprinting patterns from the intestinal microflora is the efficiency of the DNA extraction procedure.
Usually, two factors have to be particularly considered during the extraction procedure. The first is to maximize the DNA yield. The second is to ensure that the extracted DNA is amenable to several enzymatic treatments like PCR amplification[16]. In other words, the greatest challenge is the extraction of high-quality PCR-compatible DNA from the intestinal microflora. Several methods have been evaluated for bacterial cell wall lysis and DNA extraction using detergents, proteolytic enzymes, lysozyme, mechanical disruption, temperature changes alone or in various combinations, DNA stool mini- kit, etc. Although the DNA stool mini-kit method is convenient, rapid and highly efficient, it is not widely applied on account of its high cost[17–20].
In this study, 5 methods of extracting bacterial DNA were compared with that using the fecal DNA kit (FDK). The purpose of this study was to establish an economical, simple, and convenient method for extracting genomic DNA from the intestinal microflora for ERIC-PCR detection.
Fresh fecal samples were collected from 20 10-d-old healthy goslings. No subject had received antibiotics, probiotics, or prebiotics. Gosling care and experimental procedures were performed in compliance with the Institutional Animal Care and Use Guidelines provided by the Centers for Disease Control and Prevention. Two goslings were randomly selected and their fecal samples from rectum were collected separately. The samples were collected in sterile bags, refrige rated, and immediately taken to the laboratory.
Each of the six extraction methods was repeated 3 times. Each sample (20 mg) was first thawed and suspended in 5.0 mL phosphate-buffered saline (PBS; pH 7.2). Centrifugation was carried out at 100 g for 15 min at 4°C in order to remove the fecal pellets; and the obtained supernatant was centrifuged at 13 000 g at 4 °C for 10 min. The pellet was then washed 3 times by suspending it in 1.5 mL acetone. Each preparation was centrifuged at 13 000 g for 10 min at 4°C in order to remove potential PCR inhibitors in stool[21]. The supernatant was discarded, and the pellet was processed for each procedure as follows. (1) The TE boil extraction method (T method): It is a modification of the bacterial DNA extraction protocol described by Li et al[22]. The pellet was suspended in 200 &mgr;L TE buffer [10 mmol/L Tris-HCl (pH 8.0), 1 mmol/L EDTA][23], and the mixture was briefly mixed on a vortex mixer. The suspension was placed in a boiling water bath for 1 min, subjected to 3 freeze-thaw cycles alternating between -70°C for 3 min and 100°C for 2 min, and then centrifuged at 10 000 g for 5 min. A 100 &mgr;L aliquot of the supernatant was transferred to a sterile tube and stored at -20°C until PCR testing. (2) The ultrapure water method (UW method): A 200 &mgr;L aliquot of ultrapure water was added to the pellet, and the suspension was treated as described above for the TE buffer. (3) The chelex method (C method): This method is a modification of bacterial DNA extraction protocol described by Emi Suenaga[24]. A 200 &mgr;L aliquot of Chelex-100 (5%) and 0.2 mg protease K was added to the pellet and the sample was incubated at 56°C for 30 min in a water bath. The mixture was then briefly mixed on a vortex mixer and centrifuged at 10 000 g for 5 min. A 100 &mgr;L aliquot of the supernatant was transferred to a sterile tube and stored at -20°C until PCR testing. (4) SDS method: The pellets were treated as described above for the TE buffer, except that 200 &mgr;L of the nonionic detergent mix (2% SDS containing 10% Triton X-100) was substituted for the TE buffer[18]. (5) The SDSS method: A 200 &mgr;L aliquot of 2% SDS containing 10% Triton X-100 was added to the pellet, and the mixture was briefly agitated on a vortex mixer. The suspension was sonicated for 15 min and then treated as described above for the TE buffer[18]. (6) The FDK method: The pellets were processed using the Fecal DNA Kit™ (Tianli, China) and the DNA was purified with a spin column according to the manufacturer’s instructions.
The concentration and purity of DNA were determined spectrophotometrically (BIO-RAD Smart Spec 3000; USA); for this purpose, DNA absorbance was measured at 260 nm (&mgr;g DNA/g sample; 1 A260 = 50 &mgr;g/mL DNA) and protein impurities were checked at 280 nm[25]. The concentration and purity of each DNA extraction method was statistically analyzed by Excel 2003 for Windows.
For each method tested, the presence and quality of the extracted genomic DNA from one of the triplicate samples was analyzed using a 0.5% agarose gel containing ethidium bromide at 4°C. Ten microliters of the DNA extracted by each method was added into the gel and electrophoresed for 30 min at 150 V. Gel images were acquired as tagged image file format (TIFF) files with a Gel Imaging System (Bio-Rad). Unless mentioned otherwise, the following molecular procedures were carried out with the extracted genomic DNA obtained from 1 or 2 of the triplicate samples.
For fingerprinting the bacterial population in fecal samples, the total fecal DNA was used as a template for ERIC-PCR, and the sequence of the ERIC primers were E1 (ERIC1R): 5’-ATGTAAGCTCCTGGGGATTCAC-3’ and E2 (ERIC2): 5’-AAGTAAGTGACTGGGGTGAGCG-3’ which was described by Versalovic et al[11]. The ERIC-PCRs were performed under the conditions described by Di Giovanni et al[26].
We analyzed the PCR products by agarose gel electrophoresis as described by Melanie W Syrmis[27]. We acquired gel images as TIFF files using a Gel Imaging System (Bio-Rad).
To interpret the ERIC-PCR fingerprint profiles, the Quantity One software package (Applied Bio-Rad) was used with pairwise similarity coefficient (Cs)[28]. Fingerprints were assigned to a different type when any band differences were observed. Variations in band intensity were not considered as band differences. The fingerprints were considered similar when the Cs was more than 90%, different when the Cs was 0, and identical when the Cs was 100%[28].
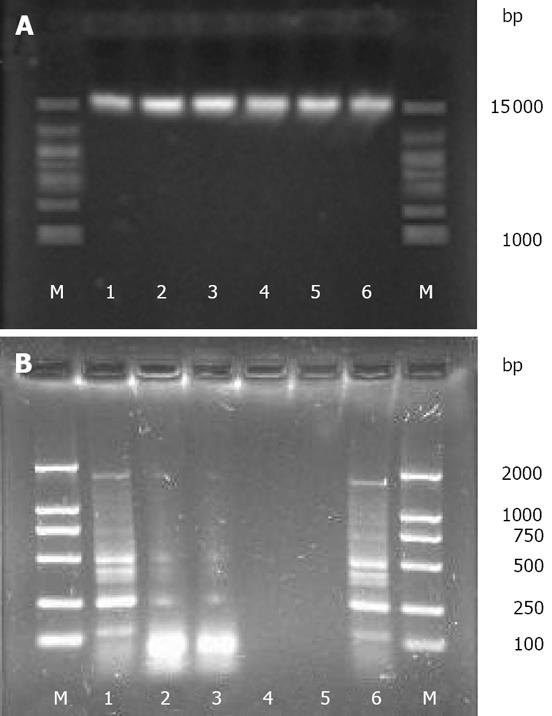
The yield, purity, and quality of genomic DNA obtained from the six extraction methods are shown in Table 1. The quality of the extracted DNA was evaluated by the A260/280 ratio, and values close to 1.8 indicate a good DNA extract with little protein contamination[25]. Although high-molecular-weight DNA (approximately 15 kb) was acquired by all the six methods (Figure 1A), discrepancies in the yield and purity of DNA were observed among different methods. For instance, the SDSS and SDS methods yielded the maximum amount of DNA (mean, 1820.12 &mgr;g/g and 1710.95 &mgr;g/g, respectively), but the lowest A260/280 ratio (mean, 1.49 and 1.54, respectively), thus implying the lowest purity of DNA. The T and UW methods yielded the least amount of DNA (mean, 601.52 &mgr;g/g and 586.03 &mgr;g/g, respectively) and lower purity of DNA (mean A260/280 ratio, 1.58 and 1.60, respectively). The C and FDK methods yielded similar amounts of DNA (mean, 1301.52 &mgr;g/g and 1326.07 &mgr;g/g, respectively) and similar A260/280 ratios (mean, 1.84 and 1.82, respectively). The purity of the DNA obtained by the two methods was the highest.
| Samples | T- | C- | UW- | SDS- | SDSS- | FDK- |
| method | method | method | method | method | method | |
| Yield | 601.52 | 1301.52 | 586.03 | 1710.95 | 1820.12 | 1326.07 |
| Purity (A260/280) | 1.58 | 1.84 | 1.6 | 1.54 | 1.49 | 1.82 |
The amplifications of ERIC-PCR were confirmed by agarose gel electrophoresis. Except for the SSDS and SDS methods, the DNA extracted by the remaining methods was effectively amplified (Figure 1B).
The ERIC-PCR fingerprint profiles of the genomic DNA of intestinal bacterium extracted from the same sample by the six methods showed that the results of the C and FDK methods were consistent, and the Cs value for these methods was 92.1%. However, the Cs values by other methods were lower than 30%, and the Cs by SSDS and SDS methods was 0 (Table 2).
| Samples | T- | C- | UW- | SDS- | SDSS- | FDK- |
| method | method | method | method | method | method | |
| C-method | 25 | - | 25 | 0 | 0 | 92.1 |
| UW-method | 100 | 25 | - | 0 | 0 | 25 |
| SDS-method | 0 | 0 | 0 | - | 100 | 0 |
| SDSS-method | 0 | 0 | 0 | 100 | - | 0 |
| FDK-method | 25 | 92.1 | 25 | 25 | 25 | - |
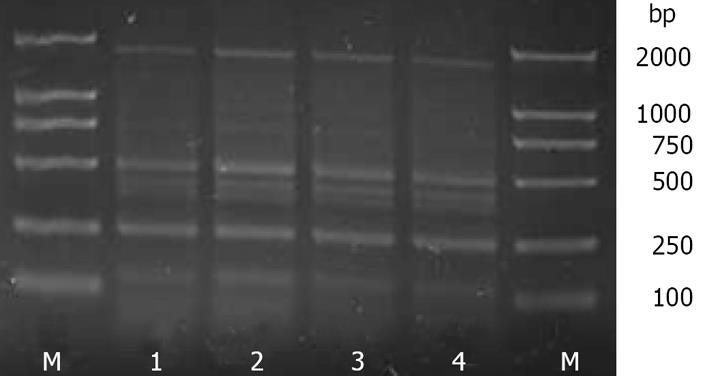
The ERIC-PCR fingerprint profiles of the intestinal bacterial genomic DNA extracted from the same sample by the C and FDK methods showed that the C method (1) had a 100% Cs value when compared with the C method (2), and the FDK method (1) had a 100% Cs value when compared with FDK method (2) (Figure 2 and Table 3). These results indicated that the stability and reproducibility of the C and FDK methods were equally good.
| Samples | C-method (1) | C-method (2) | FDK-method (1) | FDK-method (2) |
| C-method (2) | 100.0 | 92.1 | 92.1 | |
| FDK-method (1) | 92.1 | 92.1 | 100.0 | |
| FDK-method (2) | 92.1 | 92.1 | 100.0 |
Although extensive researches with ERIC-PCR have been conducted on the bacterial microflora in the intestine, the effect of the method selected for DNA extraction on the analysis of a bacterial community has barely been considered. This study is important as DNA extraction is one of the most commonly used procedures in genetics, molecular biology, and biochemistry.
Some important factors that should be considered when choosing a DNA extraction method are the time required to complete the extraction, the cost of extraction, and the safety of the chemical reagents employed. Moreover, DNA fragmentation should be avoided during the extraction.
From the extraction methods already published for various bacteria, we chose and compared six methods for extracting DNA from the intestinal microflora: the T, C, UW, SDSS, SDS, and commercial FDK methods.
The aim of an extraction procedure is to obtain a high quality and high yield of DNA from the samples. The extracted DNA should contain the least amount of proteins, RNA, or any other PCR inhibitors such as bile salt and cholerythrin, which are present in the feces[2829]. Removing those inhibitors is one of the key factors for a successful PCR. In this study, we used acetone to remove the inhibitors from the stool samples, as reported by Zhong Hua et al[21]. Our ERIC-PCR results was found successful in removal of these inhibitors.
DNA absorbance was measured at 260 nm (A260) to evaluate the quantity of the extracted DNA, and the ratio of the absorbance at 260 nm to that at 280 nm (A260/280) was used to evaluate the DNA quality. This method was employed previously by other researchers to compare different DNA extraction methods[3031].
In this study, the results of the concentration and the purity for each method were correlated. The SDSS and SDS methods seemed to yield a greater quantity of DNA, but their A260/280 ratio indicated a high protein contamination. On the other hand, the concentrations of the DNA obtained using the C and FDK methods were lower, but the A260/280 ratio showed a high purity of the DNA obtained. When applied to engorged individuals, the C method resulted in 100% successful DNA amplification that was similar to the FDK method. In our study, the C method provided the best results with a Cs close to 92.1% when compared with the FDK method.
Bacterial lysis is the key to obtain bacterial DNA. Although the SDS and SDSS methods can provide the highest DNA yield, the SDS residue inhibits the PCR process. This result is consistent with that reported by Khan[18] and Wade[17]. The excessive SDS above 0.01% has been shown to inhibit PCR by denaturing the Taq polymerase.
Many reports have described DNA extraction from tissues of other organisms using chelex-100. The results vary by different methods[3233]. The use of chelex-100 has been recommended for DNA extraction in some papers, but other reports have not regarded it as being optimum because of its lowest efficiency for DNA amplification[32]. However, we found that the modified chelex-100 protocol was the best method to extract DNA from stool. Although there are methodological differences between our study and others[3233], the conclusion is the same that this method is simple and efficient for PCR.
Theoretically, column-purified DNA should be the cleanest, containing the least PCR-inhibitory substances. One purpose of our study was to identify a method for rapid DNA extraction that did not compromise PCR sensitivity. We used the FDK extraction method as the gold standard for PCR purification. Our results showed that for PCR, column purification was unnecessary for DNA extracted from the intestinal microflora. Compared to FDK that requires 2 h, DNA extraction with chelex-100 can be completed within less than 1 h, and it does not involve 3-4 transfers of samples to new tubes.
In conclusion, of the 5 extraction methods evaluated, the chelex method is technically simpler, less expensive, and more rapid than the FDK method; it is the best method for extracting genomic DNA from the intestinal microflora. Although the extraction with TE buffer and ultrapure water is extremely easy and inexpensive, the DNA yield is the lowest. The SDS and SDSS methods consistently inhibit PCR, therefore, they cannot be recommended for DNA extraction from the intestinal microflora.
The term probiotic is a relatively new term that implies “for life”. Currently, it is used to refer to intestinal bacteria that have beneficial effects on humans and animals. Probiotics can play an important role in immunological, digestive, and respiratory functions and could have a significant effect in alleviating infectious diseases in children and other high-risk populations. Thus, knowledge about probiotics could contribute to improving human health. It is necessary to understand the bacterial flora in the gastrointestinal tract, particularly at the species level of bacterium. The DNA-based techniques often represent the most appropriate approach in this regard.
To date, the chelex method has mainly been used for medicolegal investigations. Since it is a rapid, simple, and inexpensive technique for extracting genomic DNA from intestinal microflora, it will be an ideal method to study the microflora in the gastrointestinal tract.
In previous studies on DNA extraction from the intestinal microflora, researchers used the traditional DNA extraction process that involved proteinase K digestion by phenol chloroform extraction or the FDK method. However, the chemical products employed in these experiments were either toxic or expensive. Through the present study, we provide a significantly improved method for the rapid, safe, simple and economical extraction of DNA from the intestinal microflora.
This study details a new, simple protocol for extracting DNA from the intestinal bacterial microflora and may ultimately provide new insights in the field of probiotics.
In this study, through the development of new methods for extracting bacterial DNA from the intestinal microflora, the authors found that the chelex method is a simpler, more rapid, and a less expensive method than the FDK method. Moreover, the results presented herein may lead to the development of a novel convenient approach for monitoring the intestinal microflora by molecular detection methods.
| 1. | Steinhoff U. Who controls the crowd? New findings and old questions about the intestinal microflora. Immunol Lett. 2005;99:12-16. |
| 2. | Moore WE, Cato EP, Holdeman LV. Some current concepts in intestinal bacteriology. Am J Clin Nutr. 1978;31:S33-S42. |
| 3. | Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157-1170. |
| 4. | Nechvatal JM, Ram JL, Basson MD, Namprachan P, Niec SR, Badsha KZ, Matherly LH, Majumdar AP, Kato I. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Methods. 2008;72:124-132. |
| 5. | Guarner F, Schaafsma GJ. Probiotics. Int J Food Microbiol. 1998;39:237-238. |
| 6. | Food and Agriculture Organization of the United Nations/ World Health Organization. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. World Health Organization online, 2001-10-01, cited 2007-11-08. Available from: URL: http://www.who.int/foodsafety/publications/fs_management/en/probiotics.pdf. |
| 7. | Mimura T, Rizzello F, Helwig U, Poggioli G, Schreiber S, Talbot IC, Nicholls RJ, Gionchetti P, Campieri M, Kamm MA. Once daily high dose probiotic therapy (VSL#3) for maintaining remission in recurrent or refractory pouchitis. Gut. 2004;53:108-114. |
| 8. | Gionchetti P, Rizzello F, Helwig U, Venturi A, Lammers KM, Brigidi P, Vitali B, Poggioli G, Miglioli M, Campieri M. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology. 2003;124:1202-1209. |
| 9. | Shen TY, Qin HL, Gao ZG, Fan XB, Hang XM, Jiang YQ. Influences of enteral nutrition combined with probiotics on gut microflora and barrier function of rats with abdominal infection. World J Gastroenterol. 2006;12:4352-4358. |
| 10. | Foligne B, Nutten S, Grangette C, Dennin V, Goudercourt D, Poiret S, Dewulf J, Brassart D, Mercenier A, Pot B. Correlation between in vitro and in vivo immunomodulatory properties of lactic acid bacteria. World J Gastroenterol. 2007;13:236-243. |
| 11. | Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823-6831. |
| 12. | Finegold SM, Attebery HR, Sutter VL. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am J Clin Nutr. 1974;27:1456-1469. |
| 13. | Rodriguez-Barradas MC, Hamill RJ, Houston ED, Georghiou PR, Clarridge JE, Regnery RL, Koehler JE. Genomic fingerprinting of Bartonella species by repetitive element PCR for distinguishing species and isolates. J Clin Microbiol. 1995;33:1089-1093. |
| 14. | Sampaio JL, Viana-Niero C, de Freitas D, Hofling-Lima AL, Leao SC. Enterobacterial repetitive intergenic consensus PCR is a useful tool for typing Mycobacterium chelonae and Mycobacterium abscessus isolates. Diagn Microbiol Infect Dis. 2006;55:107-118. |
| 15. | de Bruijn FJ. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180-2187. |
| 16. | Spaniolas S, Tsachaki M, Bennett MJ and Tucker GA. Evaluation of DNA extraction methods from green and roasted coffee beans. Food Control. 2008;3:257-262. |
| 17. | Aldous WK, Pounder JI, Cloud JL, Woods GL. Comparison of six methods of extracting Mycobacterium tuberculosis DNA from processed sputum for testing by quantitative real-time PCR. J Clin Microbiol. 2005;43:2471-2473. |
| 18. | Khan IU, Yadav JS. Development of a single-tube, cell lysis-based, genus-specific PCR method for rapid identification of mycobacteria: optimization of cell lysis, PCR primers and conditions, and restriction pattern analysis. J Clin Microbiol. 2004;42:453-457. |
| 19. | Rantakokko-Jalava K, Jalava J. Optimal DNA isolation method for detection of bacteria in clinical specimens by broad-range PCR. J Clin Microbiol. 2002;40:4211-4217. |
| 20. | Buck GE, O'Hara LC, Summersgill JT. Rapid, simple method for treating clinical specimens containing Mycobacterium tuberculosis to remove DNA for polymerase chain reaction. J Clin Microbiol. 1992;30:1331-1334. |
| 21. | Zhong H, Lai XL, Wei RP, Liu ZL. An improved protocol for DNA extraction from the faeces of giant panda. Acta Zool Sin. 2003;49:670-674. |
| 22. | Li M, Gong J, Cottrill M, Yu H, de Lange C, Burton J, Topp E. Evaluation of QIAamp DNA Stool Mini Kit for ecological studies of gut microbiota. J Microbiol Methods. 2003;54:13-20. |
| 23. | Wang RF, Cao WW, Cerniglia CE. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242-1247. |
| 24. | Suenaga E, Nakamura H. Evaluation of three methods for effective extraction of DNA from human hair. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;820:137-141. |
| 25. | Samuel M, Lu M, Pachuk CJ, Satishchandran C. A spectrophotometric method to quantify linear DNA. Anal Biochem. 2003;313:301-306. |
| 26. | Di Giovanni GD, Watrud LS, Seidler RJ, Widmer F. Compari-son of Parental and Transgenic Alfalfa Rhizosphere Bacterial Communities Using Biolog GN Metabolic Fingerprinting and Enterobacterial Repetitive Intergenic Consensus Sequence-PCR (ERIC-PCR). Microb Ecol. 1999;37:129-139. |
| 27. | Syrmis MW, O'Carroll MR, Sloots TP, Coulter C, Wainwright CE, Bell SC, Nissen MD. Rapid genotyping of Pseudomonas aeruginosa isolates harboured by adult and paediatric patients with cystic fibrosis using repetitive-element-based PCR assays. J Med Microbiol. 2004;53:1089-1096. |
| 28. | Sidransky D, Tokino T, Hamilton SR, Kinzler KW, Levin B, Frost P, Vogelstein B. Identification of ras oncogene mutations in the stool of patients with curable colorectal tumors. Science. 1992;256:102-105. |
| 29. | Deuter R, Pietsch S, Hertel S, Muller O. A method for preparation of fecal DNA suitable for PCR. Nucleic Acids Res. 1995;23:3800-3801. |
| 30. | Tan H, Wang J, Zhao ZK. Purification and refolding optimization of recombinant bovine enterokinase light chain overexpressed in Escherichia coli. Protein Expr Purif. 2007;56:40-47. |
| 31. | Ki JS, Chang KB, Roh HJ, Lee BY, Yoon JY, Jang GY. Direct DNA isolation from solid biological sources without pretreatments with proteinase-K and/or homogenization through automated DNA extraction. J Biosci Bioeng. 2007;103:242-246. |
| 32. | Desloire S, Valiente Moro C, Chauve C, Zenner L. Comparison of four methods of extracting DNA from D. gallinae (Acari: Dermanyssidae). Vet Res. 2006;37:725-732. |
| 33. | Aranishi F, Okimoto T. A simple and reliable method for DNA extraction from bivalve mantle. J Appl Genet. 2006;47:251-254. |