Published online May 14, 2008. doi: 10.3748/wjg.14.2810
Revised: April 9, 2008
Published online: May 14, 2008
AIM: To present an approach for selectively killing retrovirus-infected cells that combines the toxicity of Pseudomonas exotoxin (PE) and the presence of reverse transcriptase (RT) in infected cells.
METHODS: PE antisense toxin RNA has palindromic stem loops at its 5’ and 3’ ends enabling self-primed generation of cDNA in the presence of RT. The RT activity expressed in retrovirus-infected cells converts “antisense-toxin-RNA” into a lethal toxin gene exclusively in these cells.
RESULTS: Using cotransfection studies with PE-expressing RNAs and β-gal expressing reporter plasmids, we show that, in HepG2 and HepG2.2.15 hepatoma cells as well as in duck hepatitis B virus (DHBV) infected cells, HBV or DHBV-polymerase reverse transcribe a lethal cDNA copy of an antisense toxin RNA, which is composed of sequences complementary to a PE gene and eukaryotic transcription and translation signals.
CONCLUSION: This finding may have important implications as a novel therapeutic strategy aimed at the elimination of HBV infection.
- Citation: Hafkemeyer P, Brinkmann U, Brinkmann E, Pastan I, Blum HE, Baumert TF. Pseudomonas exotoxin antisense RNA selectively kills hepatitis B virus infected cells. World J Gastroenterol 2008; 14(18): 2810-2817
- URL: https://www.wjgnet.com/1007-9327/full/v14/i18/2810.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.2810
Hepatitis B virus (HBV) infection results in a broad spectrum of clinical manifestations, ranging from asymptomatic course to acute self-limited or fulminant hepatitis or chronic hepatitis that may progress to liver cirrhosis and hepatocellular carcinoma (HCC). The individual host immune response as well as viral mutations largely determines the natural course of HBV infection.
Infection with retroviruses causes among others leukemias (HTLV-1) or AIDS (HIV)[12]. Because retroviruses encode a reverse transcriptase (RT), one therapeutic strategy is inhibition of RT. RT inhibitors such as lamivudine, adefovir, entecavir and other nucleotide analogues are presently being used to treat HBV infection[3–6].
RT inhibitors are of limited efficacy, however, due to the development of viral resistance[78]. An alternative therapeutic approach for retroviral infection is to target infected cells by recombinant toxins or immunotoxins that bind to surface antigens[9–13] and kill the cells by the toxin moiety, as shown for HIV-infected cells[910]. One example is the CD4-Pseudomonas exotoxin (PE), composed of a CD4 binding domain fused to a truncated derivative of PE[1415], which is an extremely potent toxin. Because only few PE molecules are required to kill a cell, the full-length or truncated PE results in the death of eukaryotic cells[16]. The fusion of the HIV-binding domain CD4 to a truncated PE derivative proved extremely effective in HIV infection[1718].
Here, we describe a strategy for the selective killing of retrovirus-infected cells that combines the extreme toxicity of PE and the presence of RT in infected cells. The RT activity of HBV is recruited to selectively convert PE antisense toxin RNA into a lethal toxin gene exclusively in HBV infected cells. This experimental strategy was first evaluated in uninfected COS and MCF7 cells and in RT positive MM5MT(murine mammary tumor) and HUT102 (human T-cell leukemia) cells (data not shown). Transfection of sense toxin RNA kills cells independently of RT, whereas antisense toxin RNA requires reverse transcription for toxin expression. In COS monkey kidney or MCF7 human breast carcinoma cells in which RT cannot be detected, antisense toxin RNA was not toxic, because it does not express a sense toxin. However, it reduces cell viability when transfected into RT positive HUT102 or MM5MT cells, infected with a human (HTLV1) and a murine (M-MLV) retrovirus, respectively. Here, we demonstrate that HBV RT converts PE antisense RNA into a cDNA expressing PE that selectively eliminates HBV infected cells. The antisense toxin RNA is complementary to the PE gene, flanked by eukaryotic transcription and translation signals. In addition, antisense toxin RNA has palindromic stem loops at its 5’ and 3’ ends enabling self-primed generation of cDNA in the presence of RT. Antisense toxin RNA does not affect uninfected cells but is lethal for RT containing cells and may therefore represent a novel antiviral strategy.
HepG2 and HepG2.2.15 cells (American Type Culture Collection, ATCC, Manassas, VA, USA) were maintained as described[19]. HepG2.2.15 is a hepatoblastoma cell line based on Hep G2 cells transfected with a plasmid carrying the gene that confers resistance to G418 and four 5’-3’ tandem copies of the HBV genome positioned such that two dimers of the genomic DNA are 3’-3’ with respect to one another[19].
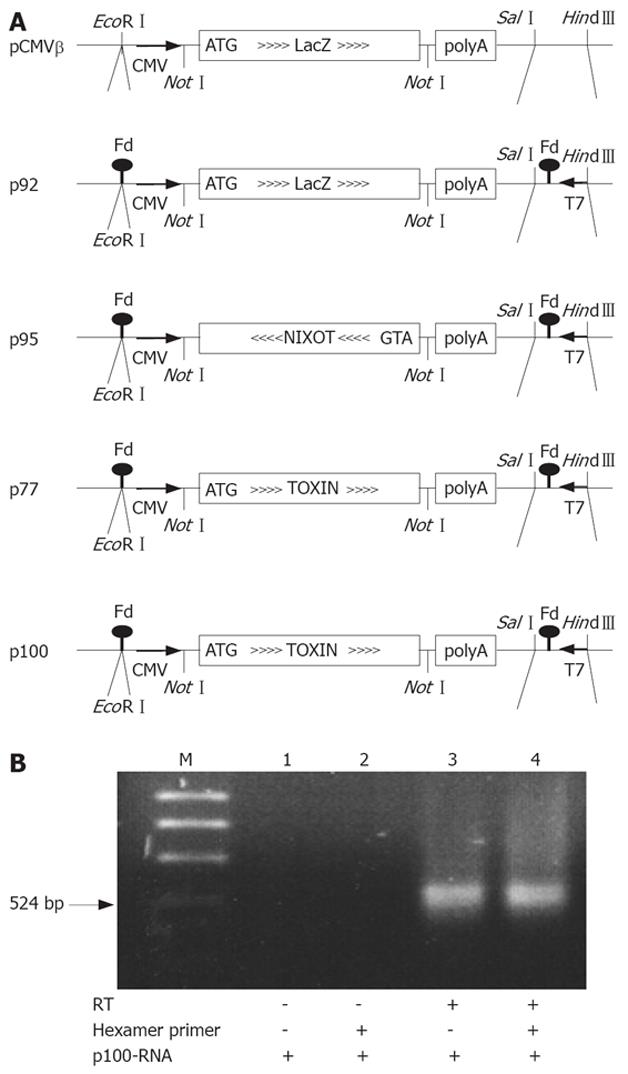
The plasmids for PE expression and production of RNA are derived from pCMVβ (Clontech, Palo Alto, CA, USA)[17] which contain the E. coli LacZ gene under the control of the CMV promoter for expression of β-galactosidase (β-Gal) in eukaryotic cells, and SV40 splice and polyadenylation signals (Figure 1A). We introduced the T7 phage gene 10 promoter, the and 5’-stem loop sequence[20] and the Fd-phage transcription terminator[21] in opposite direction to the CMV expression cassette behind the polyadenylation signal and in front of the CMV promoter of pCMVβ, respectively. This generates a T7-transcription cassette that contains, and is complementary to, the CMV-βGal expression cassette (p92). p92 has the promoter and stem loop sequence of the T7 phage gene 10, obtained by PCR with the primers 5’-GTTCTTTCCAAAGCTTGTTGAATTGAT-3’ and 5’-GGGCGGAGTCGACCCCGCTAGAGGGAAA-3’ inserted between the Sal I and HindIII restriction sites of pCMVβ and, in addition, the Fd phage transcription terminator inserted in the EcoRI site. The annealed terminator oligos, 5’-AATTACAAAATTAAAGGCTCCTTITGGAGCCTLTITTG-3’ and 5’-AATTCAAAAAAGGCTCCAAAAGGAGCCTTTAATTTTGT-3’ are EcoRI compatible at both ends but reconstitute the site after ligation in the plasmid only at the 3’-end of the terminator. The T7 transcription cassette generated by these insertions is in opposite orientation to the CMV promoter. Toxin sense or antisense RNA coding plasmids were generated by replacing LacZ of the modified pCMVβ by a truncated but enzymatically fully active PE (PE35KDEL)[22]. We also constructed a PE frameshift mutation (p77) in which the active site of the toxin (Glu553) is inactivated[23]. In p95, p100 and p77, the LacZ gene NotI-fragment of p92 is replaced by fragments containing a translation initiation signal (Kozak) and ATG start codon at the 5’-end of truncated mutated forms of PE obtained by PCR with 5’-ACCCGTCATCGCGGCCGCCACCATGGGCTGGGAACAACTGGA-3’ and 5’-TCGGGCTTTGGCGGCCGCCGAATTCTTAGAGCTCG-3’; an EcoRI site in the latter primer was later filled in by polymerase, religated and thereby removed. p9 (active toxin)[23] was used as template for PE38KDEL; the introduction of an additional (fill-in) frameshift mutation in the BamHI site located in the enzymatically active domain III of the toxin gene provided the template for p77.
The Fd-terminator stem loop at the 3’ end of the RNAs (SL1, ..ACAAAATTAAAGGCTCCTTTTGGAGCCTTTTTTG-3’) can prime the first strand synthesis, the 3’ end of which forms a stem loop (SL2, previous RNA-5’-stemloop 5’-GGGAGACCACAACGGTTTCCC..) initiating second strand synthesis. cDNA multimers might also be obtained by repeated flip-over polymerizations around the stem loops. The cDNAs would represent functional and lethal toxin expression cassettes. Depending on the orientation of the toxin coding region in the T7/CMV promoter cassette, these plasmids contain either a functional eukaryotic toxin expression cassette (p100) or no toxic activity (p77), or a cassette in which the toxin is encoded by the opposite strand (p95). In this latter plasmid p95, the CMV promoter driven expression generates nonsense RNA in eukaryotic cells. Since the T7 and CMV promoters are in opposite orientation, this situation is inverted for the RNAs from T7-promoter driven in vitro transcription: RNA produced in vitro from p100 and p77 is nonsense RNA (T7-nixoT) while RNA from p95 (T7-Toxin) encodes the toxin.
Plasmid pCH-391wt contains the complete HBV2 (ayw) genome (infectious in chimpanzees) under control of a CMV promoter. The CMV promoter allows transcription of an authentic pregenomic RNA. Plasmid pCH-391mt is derived from pCH-391wt. pCH-391mt contains a G2024C mutation that inactivates the endogenous polymerase because the active polymerase motif YMDD in pCH-391mt is mutated to YMDH[24]. In pCH-391mt a new Nsi I restriction site is created. pCH-391mt was cloned by restriction digest of pCH-391wt with XhoI and BsrGI, removing the wild-type XhoI/BsrGI fragment and insertion of a PCR fragment containing the YMDH motif using primers HBV-pol G2024C Rev (5’-CTCAAGATGCTGTACAGACTTGGCCCCCAATACCACATCATGCATATAACTG-3’) and HBV-pol 1400 For (5’-GTCAATCTTCTCGAGGATTGGGGACCCT-3’) and pCH-391wt as template for PCR amplification. The mutation was verified by sequencing as well as NsiI digestion.
Templates for in vitro transcription were linearized with EcoRI to eliminate read-through transcripts. Transcription was performed using a large-scale RNA synthesis kit (RiboMAX, Promega, Mannheim, Germany) without Cap analog. RNA was incubated for 1 h at 37°C with 10 units RNase free DNaseI (Promega, Mannheim, Germany) to remove template DNA, extracted with phenol, phenol/chloroform and chloroform, precipitated with ethanol, dissolved in DEPC-treated water and stored at -70°C. For reverse transcription 1 &mgr;g p100 RNA (r100) was incubated for 30 min in 20 &mgr;L RT buffer (Advantage RT-PCR-Kit, Clontech, Palo Alto, CA, USA) with or without random hexamer primers and M-MLV RT and subsequently treated with 1 &mgr;g RNAse A for 1 h at 37°C. One &mgr;L of the reaction mixture was used to PCR amplify [25 × (95°C, 1 min; 60°C, 2 min; 72°C, 3 min)] a PE gene fragment with the primers 5’-ATGGTCTCCAGGCGCCCGCCTTCCTC-3’ and 5’-GCTATGTGTTCGTCGGCTACCACGGC-3’. Ethidium bromide stained nondenaturing 2% agarose gel-electrophoresis was performed to identify the PE cDNA PCR fragments.
Transfections were performed using the calcium phosphate transfection kit (GIBCO, Heidelberg, Germany). 1.5 × 105 cells were plated 24 h prior to transfections. Transfections were performed with 7.5 &mgr;g β-Gal expressing plasmid p92 and 7.5 &mgr;g toxin expressing plasmid. β-Gal activities obtained after cotransfection with the β-Gal expressing plasmid p92 (7.5 &mgr;g) and p92 plasmid without insert (7.5 &mgr;g) were set to 100%. Activities resulting from coexpression of toxin plasmids (p77, p95, p100) and β-Gal expressing plasmid p92 are expressed as relative levels. RT dependent toxicity was determined by β-Gal activity staining.
In RNA transfection experiments, 7.5 &mgr;g β-Gal expression plasmid p92 and 7.5 &mgr;g of r95, r100, r77 or r92-RNA obtained by in vitro transcription were transfected. In experiments determining the sensitivity of HepG2 cells to sense toxin and antisense RNA a further plasmid (7.5 &mgr;g) expressing either wild-type (pCH-391wt) or mutant HBV polymerase (pCH-391mt) was transfected. β-Gal activity was assessed using the β-Gal activity staining set (Invitrogen, Groningen, Netherlands). To assess the sensitivity of Hep G2 cells to sense and antisense toxin RNA in the presence and absence of the RT inhibitor adefovir, Hep G2 cells were transfected with 7.5 &mgr;g β-Gal expressing plasmid p92 and 7.5 g of r95, r100, r77 or r92-RNA. After transfection of plasmid p92 and the respective RNA adefovir was added to a final concentration of 10 &mgr;mol/L. In addition, a plasmid expressing wild-type or mutant HBV polymerase was transfected (7.5 &mgr;g).
To demonstrate that HBV polymerase reverse transcribed p100 RNA in HepG2 cells only that were transfected with the HBV polymerase encoding plasmid DNA pCH-391wt, genomic DNA was extracted and purified using the QIA-amp-DNA mini Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol, following transfection with β-Gal expression plasmid p92 and r100 PE expressing RNA. Genomic DNA was stored at -70°C. For reverse transcription 1 &mgr;L of the genomic DNA was used to PCR amplify [25 × (94°C, 1 min; 58°C, 2 min; 72°C, 3 min)] a gene fragment with the primers PE-FOR (5’-GCTATGTGTTCGTCGGC-3’) and 4648 REV (5’-ATTAATGTGAGTTAGCTCACTCAT-3’).
DHBV was obtained from the serum of 2-wk to 3-wk-old Pekin ducks congenitally DHBV-infected. One day after hatching, Pekin ducks were infected by intravenous injection of 100 L DHBV DNA positive serum (109 virions/mL). Ten days later the ducks were sacrificed. Hepatocytes were obtained by liver perfusion. After the ducks were anesthesized with pentobarbital sodium, the livers were perfused via the portal vein with 200 mL of 0.5 mmol/L EGTA [ethyleneglycol-bis (β-aminoethyl ether)-N,N,N’,N’-tetraacetic acid] in Swimms 77 medium (GIBCO Heidelberg, Germany) that was buffered with 20 mmol/L HEPES (N-2-hydroxyethylpiperazine-N’-2-ethanesulfonic acid) followed by 200 mL of 0.5 mg of collagenase type 1 (Sigma, Munich, Germany) per mL, 2.5 mmol/L MgCl2 in Swimms 77 medium. To maintain the perfusion at 37°C, all solutions were kept at 39°C, and the livers were removed and cells dispersed in Williams’ medium (GIBCO Heidelberg, Germany) containing 5% fetal bovine serum (FBS), 20 mmol/L HEPES (pH 7.4), 300 mg of penicillin per liter, 1.5 mg of glucose per liter, 10-5 mol/L hydrocortisone-hemisuccinate (Sigma, Munich, Germany), and nystatin (10 U/mL). Cells were filtered through gauze and centrifuged at 50 ×g for 4 min. The cell pellet was washed three times with Williams’ medium supplemented 20 mmol/L HEPES (pH 7.4), 5 mmol/L glutamine, 0.066 mmol/L insulin, 10 mmol/L dexamethasone, 100 &mgr;g/mL penicillin, 100 &mgr;g/mL streptomycin and 1.5% dimethyl sulfoxide containing 5% FBS. Cells were counted in a hemacytometer, and 60 mm dishes were seeded with 1.5 × 105 cells per dish in Williams’ medium containing the above mentioned supplements. Cultures were incubated at 37°C and 5% CO2 in a humidified incubator.
Transfections were performed using the calcium phosphate transfection kit (GIBCO, Heidelberg, Germany). DNA and RNA transfections were performed identical to “Transfection of HepG2 and HepG2.2.15 cells with toxin expression plasmids”.
To study the ability of a RT to reverse transcribe an antisense toxin RNA, we first constructed a series of PE expression plasmids allowing primer-independent synthesis of cDNA by RT. RNA was obtained by in vitro transcription from the T7 promoter. The 5’ end of the RNA can form 5-end stem loop of T7 gene 10 promoter transcripts[20]. The 3’ end consists of the Fd-terminator[21] followed by a residual EcoRI sequence (used to linearize the plasmid templates prior to in vitro transcription) that can participate in a 3’ stem loop. Primer independent cDNA synthesis was used to verify that these secondary structures at the 5’- and 3’- ends of the RNA allow self-primed, i.e., primer-independent synthesis of cDNA by RT. Figure 1B shows the conversion of these RNAs to cDNA when incubated with dNTPs and RT either with or without primers. The self-primed in vitro cDNA synthesis is as effective as random-primed cDNA synthesis under otherwise identical conditions.
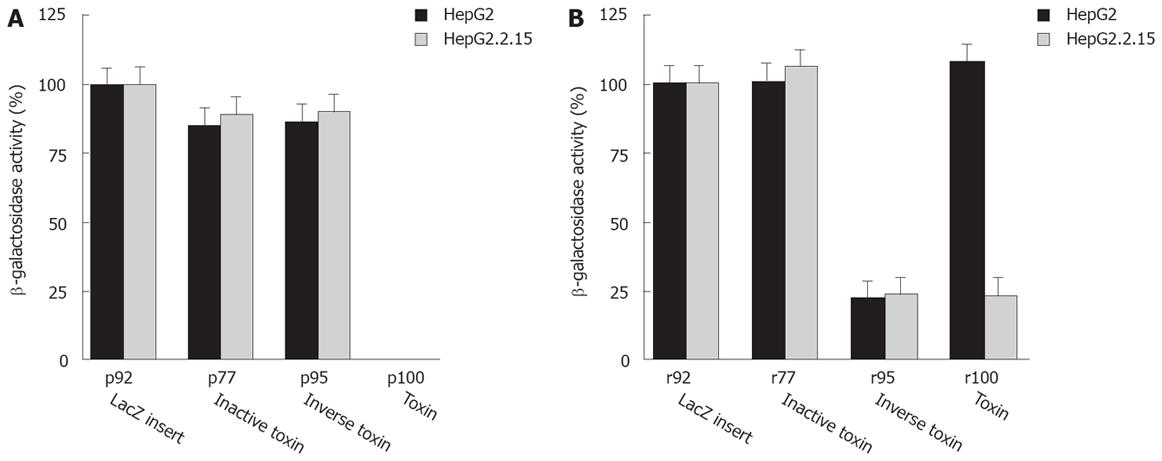
Expression of PE or truncated active PE derivatives in animal cells leads to cell death resulting in lack of viable PE expressing transfectants[16]. The analysis of cell viability is based upon transfection of toxin expression plasmids, sense or antisense toxin RNA, and a LacZ reporter plasmid (p92) followed by colorimetric quantitation of the LacZ gene product β-galactosidase (β-Gal). The β-Gal activity correlates with the number of transfectants as well as cell viability, because β-Gal protein is synthesized in viable cells only. Thus, reduced β-Gal activity reflects cell death induced by intracellular PE expression. Our data indicate that plasmids expressing an active toxin (p100) kill cells independent from RT (Figure 2A). By contrast, a plasmid expressing a mutated inactive toxin gene (p77) is not toxic. p95 is not toxic, because the PE cassette has an inverse orientation to the CMV promoter resulting in synthesis of nonsense RNA. We conclude that transfection and expression of a fully active or only partially active sense PE gene kills target cells. By contrast, a PE gene inactivated by a frameshift mutation or inversion of the cassette is not toxic.
Next, we analyzed whether PE sense RNA synthesized in vitro can be transfected and expressed in target cells resulting in cytotoxicity. The T7 in vitro transcription cassette of p95 is in the same orientation as the CMV eukaryotic expression cassette encoding the toxin. Indeed, RNA derived from p95 is cytotoxic even though transfected DNA plasmid itself is not toxic. Cell viability after transfection of r95 toxin RNA is reduced in both HepG2 and HepG2.2.15 cells (Figure 2B compare to p100 DNA in Figure 2A) as well as in DHBV infected cells (Figure 3). These data show that transfected PE RNA is expressed in cells, resulting in cytotoxicity. By comparison transfection of r95 (inverse orientation of PE cassette) is less toxic than transfection of p100.
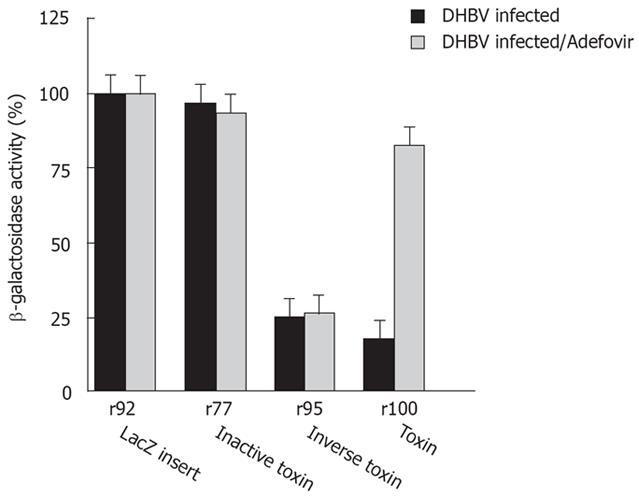
Transfection of sense RNA kills HBV expressing target cells as well as normal cells in a non-specific manner. Using PE antisense RNA we analyzed the effect of HBV-mediated reverse transcription on cytotoxicity. PE antisense RNA was synthesized by in vitro transcription of the plasmids p77 and p100. lt contains stem loop sequences at both ends, a toxin gene, poly A signals and CMV promoter sequences, all in inverse orientation to the toxin reading frame and transcription/translation signals (Figure 1A). Antisense RNA should be nontoxic to cells because, similarly to p95 the toxin gene has an inverse orientation. However, if antisense toxin RNA is reverse transcribed in RT positive cells, similar to M-MLV RT in vitro (Figure 1B), an active toxin should be synthesized resulting in cell death of RT positive cells only. Indeed, transfection of antisense RNA into HepG2.2.15 cells or DHBV infected hepatocytes expressing HBV or DHBV polymerase (HBV or DHBV-RT) results in cell death (Figure 2B, Figure 4) while HepG2 cells expressing no HBV or DHBV polymerase are not affected. In contrast to transfection of sense toxin RNA r95 (or toxin genes, Figure 2A), that kills cells independent from the presence of RT, antisense toxin RNA r100 derived in vitro from p100 is selectively toxic in HBV or DHBV positive cells. We therefore conclude that HBV and DHBV RT reverse transcribes PE antisense RNA, resulting in cytotoxicity.
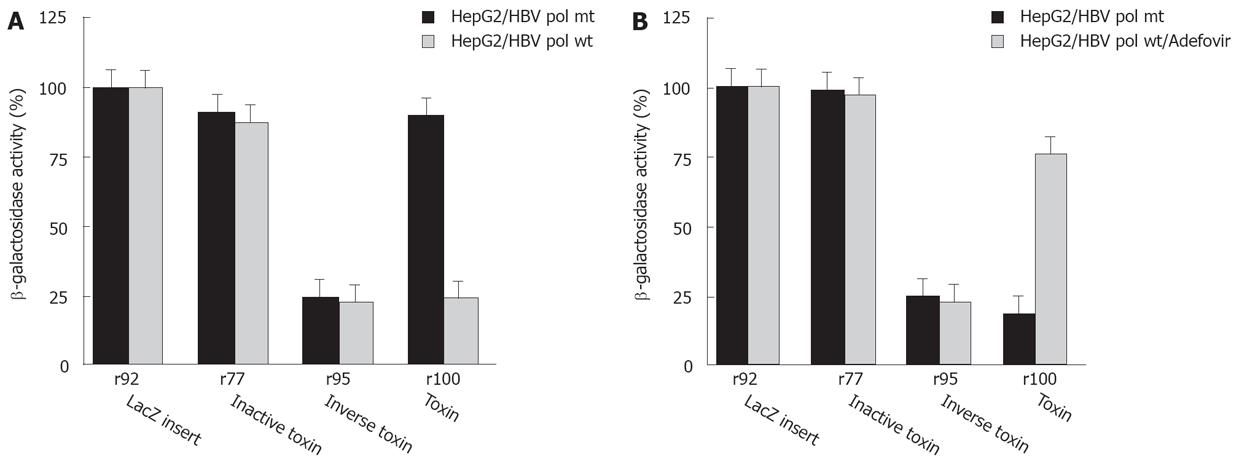
To further analyze the selective cytotoxicity to HBV infected cells, HepG2 cells were transfected with the HBV polymerase construct pCH-391wt expressing wild-type HBV polymerase or pCH-391mt encoding an inactive HBV polymerase with a mutation in the YMDD-motif (Figure 5A). A similar effect on cell viability was observed after transfection of HepG2 cells with r100 RNA and pCH-391wt or of HepG2.2.15 cells with r100 RNA (Figure 2B). Again, r95 RNA kills HepG2 cells independent from the cotransfection of pCH-391wt or pCH-391mt (Figure 5A). By contrast, markedly reduced cytotoxicity was observed after HepG2 cells were transfected with PE antisense RNA r100 and pCH-391mt. This finding indicates that cell death caused by toxin antisense RNA r100 depends on the presence of active HBV polymerase.
To further support this conclusion, HepG2 cells were transfected with antisense toxin RNA r100 and wild-type HBV polymerase pCH-391wt in the presence of adefovir - a potent inhibitor of HBV polymerase. Inhibition of HBV polymerase indeed results in reduced cytotoxicity (Figure 5B). A similar result was achieved when adefovir was added to DHBV infetced cells after transfection with antisense toxin RNA r100 (Figure 4). Taken together these findings demonstrate that the toxicity of r100 is dependent on HBV RT.
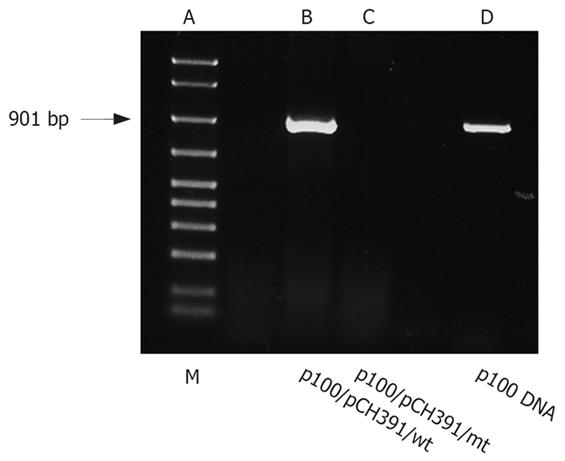
To demonstrate that r100 is reverse transcribed in HBV-infected cells and to visualize the resulting p100 cDNA, genomic DNA was extracted from HepG2 cells transfected with either wild-type or mutant HBV polymerase expression plasmid (pCH-391wt, pCH-391mt). RNA was transcribed from linearized p100 DNA. RT-PCR (Figure 3) demonstrates that r100 was reverse transcribed from p100 only in cells cotransfected with pCH-391wt, resulting in the amplification of a 901 bp DNA fragment. In cells cotransfected with pCH-391mt p100 DNA could not be amplified.
HBV infection is a major cause of chronic hepatitis, liver cirrhosis and HCC worldwide. HCC develops 30-50 years after HBV infection and is a leading cause of death, especially in sub-Sahara Africa and South East Asia where up to 20% of the population are HBV infected.
The HBV genome replicates via reverse transcription of a RNA intermediate. This property has been used in a model system of HBV infection based on HBV transfected cell lines to study the selective cytotoxicity of a PE antisense toxin RNA.
Despite the availability of a safe and efficient vaccine, chronic HBV infection remains a major health problem. The problems of non-responding individuals and emergence of vaccine escape mutants are largely still unsolved. New treatment options with nucleotide analogues initially designed for HIV infection have become available. Lamivudine efficiently inhibits HBV infection but drug resistant mutations are frequent. Adefovir is an alternative for the treatment of chronic HBV infection. Different from lamivudine, viral resistance to adefovir is used lesser frequent. Viral variants may influence the course of disease and require special attention during antiviral therapy[8]. Antiviral agents such as lamivudine, adefovir, entecavir and others work directly inhibit HBV viral polymerase and thereby suppress viral replication[25]. The viral cccDNA template in the cell nucleus, however, is not affected by antiviral therapy. Therefore, HBV infection persists, because the turnover rate of hepatocytes is rather slow. Therefore, cessation of treatment or the development of breakthrough mutations promptly results in eradication of replication. HBV polymerase is highly error-prone and lacks proofreading activity, as do other RT such as HIV-RT. Thus, viral quasispecies will develop during persistent HBV infection[26]. In this setting, the use of nucleotide chain terminators such as lamivudine, adefovir, entecavir and others carry the risk of selecting drug resistant viral mutants by suppressing the replication of the major wild-type population. These agents allow noncompetitive drug-resistant quasispecies to replicate and become the dominant viral population.
To overcome the problem of drug resistance we explored a novel experimental strategy approach to eliminate HBV infected cells. PE has the advantage as a candidate gene for gene therapy, because only very few molecules are necessary to induce cell death[16]. It is widely used as an experimental therapeutic approach as part of immunotoxins for a variety of malignant diseases. A major requirement for in vivo gene therapy is to target the therapeutic gene expression to tissues of interest by selecting potent therapeutic genes. The mechanism of action of plant or bacterial toxins is internalization into an endosome, and translocation to the cytosol followed by enzyme induced modification or destruction of components critical for the cellular translation machinery resulting in lethal inhibition of protein synthesis or induction of programmed cell death[2728]. An example for a retroviral disease is HIV infection where infected cells were selectively killed by an immunotoxin, consisting of a truncated PE joined to the variable region of a broadly neutralizing antibody (3B3) recognizing the viral envelope glycoprotein (env)[17].
In our experiments PE antisense toxin RNA is less toxic to RT positive cells than toxin DNA (compare Figures 4 and 5: killing of HepG2 cells by p100 DNA versus r100 RNA). Possibly, intracellular reverse transcription of toxin antisense RNA is inefficient and the transfected RNA is degraded rather than expressed. Despite these limitations, our data clearly demonstrate that, in principle, it is possible to reverse transcribe a nontoxic RNA in RT positive cells to a toxic cDNA expressing active PE, resulting in cell death.
The elimination of retrovirus infected cells in vivo by antisense toxin RNA requires effective delivery to target cells. Thus, RNA instability and low transfection efficacy limit its usefulness and make even the analysis of antisense toxin RNA in vitro difficult. It might be possible to deliver such RNA, more effectively using liposomes or recombinant viruses[2930]. Cells with retrotransposon activity could be affected by this approach when retroposon activity would be able to reverse transcribe the antisense-toxin RNA template.
Ultimately, retroviruses which naturally contain and stabilize RNA could be engineered to carry antisense toxin RNA and thus serve as a Trojan horse for the treatment of HBV infection.
Hepatitis B virus (HBV) is the prototypic member of the Hepadnaviridae, a DNA-containing virus that replicates by reverse transcription of a pregenomic intermediate. Initiation of reverse transcription is thought to be strictly template specific. Here we present data showing that HBV-reverse transcription of the non-viral template coding for Pseudomonas exotoxin (PE) is possible. The RT-activity expressed in hepadnavirus-infected cells is recruited to convert an “antisense-toxin-RNA“ into a lethal toxin exclusively in HBV-infected cells. We also present this approach for selectively killing retrovirus infected cells that combines the toxicity of Pseudomonas exotoxin (PE) and the presence of RT in infected cells.
Current therapies for chronic HBV infection are limited in their effect on viral gene expression and replication. Experimental therapies for HBV-infection are RNA interference (RNAi), ribozyme technology, target-dependent ribozymes, and gene therapeutic approaches using a variety of vectors to deliver therapeutic genes into HBV-infected cells. Pseudomonas exotoxin A kills cells either by direct inhibition of protein synthesis, or by concomitant induction of apoptosis.
Here it is shown for the first time that HBV-and DHBV infected cells can be selectively killed by Pseudomonas exotoxin. The explanation for RT dependent toxicity is likely conversion of antisense-toxin-RNA to a toxin expression module by self-primed reverse transcription in the cell. This approach has been previously used by targeting HIV- infected cells by recombinant toxins that bind to HIV-surface antigens and kill infected cells by the toxin moiety. Moreover it is shown that HBV-polymerase can replicate a non-viral template. HBV polymerase can be active in trans outside the HBV nucleocapsid.
The data demonstrate that, in principle, it is possible to generate a lethal cDNA copy of a per se nontoxic RNA in RT-containing infected cells. This principle is not limited to toxin genes but can probably be applied to other genes, e.g. inhibitors or ribozymes. The elimination of retrovirus infected cells in vivo by antisense-toxin-RNA requires effective delivery to target cells. It might be possible to deliver such RNA more effectively with liposomes or recombinant viruses. Ultimately, retroviruses which naturally contain and stabilize RNA could be loaded with antisense-toxin-RNA to serve as a “Trojan horse“ in the battle against AIDS.
Cells with retrotransposon activity could be affected by this approach when retroposon activity would be able to reverse transcribe the antisense-toxin RNA template.
| 1. | Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415-7419. |
| 2. | Gallo RC. Human retroviruses: a decade of discovery and link with human disease. J Infect Dis. 1991;164:235-243. |
| 3. | Zoulim F. Hepatitis B virus resistance to entecavir in nucleoside naive patients: Does it exist? Hepatology. 2006;44:1404-1407. |
| 4. | Matthews G. The management of HIV and hepatitis B coinfection. Curr Opin Infect Dis. 2007;20:16-21. |
| 5. | Pawlotsky JM, Dusheiko G, Hatzakis A, Lau D, Lau G, Liang TJ, Locarnini S, Martin P, Richman DD, Zoulim F. Virologic monitoring of hepatitis B virus therapy in clinical trials and practice: recommendations for a standardized approach. Gastroenterology. 2008;134:405-415. |
| 6. | Leemans WF, Ter Borg MJ, de Man RA. Review article: Success and failure of nucleoside and nucleotide analogues in chronic hepatitis B. Aliment Pharmacol Ther. 2007;26 Suppl 2:171-182. |
| 7. | Tan J, Degertekin B, Wong SN, Husain M, Oberhelman K, Lok AS. Tenofovir monotherapy is effective in hepatitis B patients with antiviral treatment failure to adefovir in the absence of adefovir-resistant mutations. J Hepatol. 2008;48:391-398. |
| 8. | Baumert TF, Thimme R, von Weizsacker F. Pathogenesis of hepatitis B virus infection. World J Gastroenterol. 2007;13:82-90. |
| 9. | Chaudhary VK, Mizukami T, Fuerst TR, FitzGerald DJ, Moss B, Pastan I, Berger EA. Selective killing of HIV-infected cells by recombinant human CD4-Pseudomonas exotoxin hybrid protein. Nature. 1988;335:369-372. |
| 10. | Ashorn P, Moss B, Weinstein JN, Chaudhary VK, FitzGerald DJ, Pastan I, Berger EA. Elimination of infectious human immunodeficiency virus from human T-cell cultures by synergistic action of CD4-Pseudomonas exotoxin and reverse transcriptase inhibitors. Proc Natl Acad Sci USA. 1990;87:8889-8893. |
| 11. | Pincus SH, Cole RL, Hersh EM, Lake D, Masuho Y, Durda PJ, McClure J. In vitro efficacy of anti-HIV immunotoxins targeted by various antibodies to the envelope protein. J Immunol. 1991;146:4315-4324. |
| 12. | Bell KD, Ramilo O, Vitetta ES. Combined use of an immunotoxin and cyclosporine to prevent both activated and quiescent peripheral blood T cells from producing type 1 human immunodeficiency virus. Proc Natl Acad Sci USA. 1993;90:1411-1415. |
| 13. | Erice A, Balfour HH Jr, Myers DE, Leske VL, Sannerud KJ, Kuebelbeck V, Irvin JD, Uckun FM. Anti-human immunodeficiency virus type 1 activity of an anti-CD4 immunoconjugate containing pokeweed antiviral protein. Antimicrob Agents Chemother. 1993;37:835-838. |
| 14. | Hwang J, Fitzgerald DJ, Adhya S, Pastan I. Functional domains of Pseudomonas exotoxin identified by deletion analysis of the gene expressed in E. coli. Cell. 1987;48:129-136. |
| 15. | Carroll SF, Collier RJ. Active site of Pseudomonas aeruginosa exotoxin A. Glutamic acid 553 is photolabeled by NAD and shows functional homology with glutamic acid 148 of diphtheria toxin. J Biol Chem. 1987;262:8707-8711. |
| 16. | Wels W, Baldrich M, Chakraborty T, Gross R, Goebel W. Expression of bacterial cytotoxin genes in mammalian target cells. Mol Microbiol. 1992;6:2651-2659. |
| 17. | McHugh L, Hu S, Lee BK, Santora K, Kennedy PE, Berger EA, Pastan I, Hamer DH. Increased affinity and stability of an anti-HIV-1 envelope immunotoxin by structure-based mutagenesis. J Biol Chem. 2002;277:34383-34390. |
| 18. | Kennedy PE, Bera TK, Wang QC, Gallo M, Wagner W, Lewis MG, Berger EA, Pastan I. Anti-HIV-1 immunotoxin 3B3(Fv)-PE38: enhanced potency against clinical isolates in human PBMCs and macrophages, and negligible hepatotoxicity in macaques. J Leukoc Biol. 2006;80:1175-1182. |
| 19. | Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005-1009. |
| 20. | Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477-535. |
| 21. | Gentz R, Langner A, Chang AC, Cohen SN, Bujard H. Cloning and analysis of strong promoters is made possible by the downstream placement of a RNA termination signal. Proc Natl Acad Sci USA. 1981;78:4936-4940. |
| 22. | Theuer CP, Kreitman RJ, FitzGerald DJ, Pastan I. Immunotoxins made with a recombinant form of Pseudomonas exotoxin A that do not require proteolysis for activity. Cancer Res. 1993;53:340-347. |
| 23. | Brinkmann U, Pai LH, FitzGerald DJ, Willingham M, Pastan I. B3(Fv)-PE38KDEL, a single-chain immunotoxin that causes complete regression of a human carcinoma in mice. Proc Natl Acad Sci USA. 1991;88:8616-8620. |
| 24. | Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product: domain structure and RNase H activity. J Virol. 1990;64:613-620. |
| 25. | Koeck J, Baumert TF, Delaney WE, Blum HE, Weizsaecker F. Inhibitory effect of adefovir and lamivudine on the initiation of hepatitis B virus infection in primary tupaia hepatocytes. Hepatology. 2003;38:1410-1408. |
| 26. | Stuyver LJ, Locarnini SA, Lok A, Richman DD, Carman WF, Dienstag JL, Schinazi RF. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology. 2001;33:751-757. |
| 27. | Kreitman RJ, Bailon P, Chaudhary VK, FitzGerald DJ, Pastan I. Recombinant immunotoxins containing anti-Tac(Fv) and derivatives of Pseudomonas exotoxin produce complete regression in mice of an interleukin-2 receptor-expressing human carcinoma. Blood. 1994;83:426-434. |
| 28. | Hafkemeyer P, Brinkmann U, Gottesman MM, Pastan I. Apoptosis induced by Pseudomonas exotoxin: a sensitive and rapid marker for gene delivery in vivo. Hum Gene Ther. 1999;10:923-934. |
| 29. | Zhu N, Liggitt D, Liu Y, Debs R. Systemic gene expression after intravenous DNA delivery into adult mice. Science. 1993;261:209-211. |
| 30. | Danos O, Mulligan RC. Safe and efficient generation of recombinant retroviruses with amphotropic and ecotropic host ranges. Proc Natl Acad Sci USA. 1988;85:6460-6464. |