Published online Mar 21, 2008. doi: 10.3748/wjg.14.1699
Revised: January 1, 2008
Published online: March 21, 2008
In many parts of the world hepatocellular carcinoma (HCC) is among the leading causes of cancer-related mortality but the underlying molecular pathology is still insufficiently understood. There is increasing evidence that activins, which are members of the transforming growth factor β (TGFβ) superfamily of growth and differentiation factors, could play important roles in liver carcinogenesis. Activins are disulphide-linked homo- or heterodimers formed from four different β subunits termed βA, βB, βC, and βE, respectively. Activin A, the dimer of two βA subunits, is critically involved in the regulation of cell growth, apoptosis, and tissue architecture in the liver, while the hepatic function of other activins is largely unexplored so far. Negative regulators of activin signals include antagonists in the extracellular space like the binding proteins follistatin and FLRG, and at the cell membrane antagonistic co-receptors like Cripto or BAMBI. Additionally, in the intracellular space inhibitory Smads can modulate and control activin activity. Accumulating data suggest that deregulation of activin signals contributes to pathologic conditions such as chronic inflammation, fibrosis and development of cancer. The current article reviews the alterations in components of the activin signaling pathway that have been observed in HCC and discusses their potential significance for liver tumorigenesis.
- Citation: Deli A, Kreidl E, Santifaller S, Trotter B, Seir K, Berger W, Schulte-Hermann R, Rodgarkia-Dara C, Grusch M. Activins and activin antagonists in hepatocellular carcinoma. World J Gastroenterol 2008; 14(11): 1699-1709
- URL: https://www.wjgnet.com/1007-9327/full/v14/i11/1699.htm
- DOI: https://dx.doi.org/10.3748/wjg.14.1699
Hepatocellular carcinoma (HCC) is the predominant form of primary malignancy of the liver and accounts for more than half a million deaths per year[1]. In some geographical regions it is the most prevalent form of malignancy and the most common cause of death from cancer[2] making its containment a top priority. Chronic infection with the hepatitis B or C virus (HBV, HCV), dietary exposure to the hepatocarcinogen aflatoxin B1 (AFB1), ethanol abuse, and obesity are among the main risk factors for liver cancer[3]. Despite recent advances, the molecular pathology of the disease is not well understood and the therapeutic possibilities are largely limited to surgical procedures including resection, liver transplantation, or local tumor ablation[4]. A consistent pattern of changes comparable to the sequential mutations in tumor suppressor genes and oncogenes, like the one identified in colon carcinogenesis during adenoma to carcinoma progression[56], has not been defined for HCC. Nevertheless, multiple genetic alterations including mutations of p53, inactivation of the Rb pathway, and activation of the Wnt/β-catenin pathway have been linked to HCC development and progression[37]. In addition deregulated expression of growth factors and their cognate receptors has been described in HCC for both, positive regulators of hepatocyte growth, such as insulin-like growth factor 2 (IGF-2), hepatocyte growth factor (HGF), and transforming growth factor α (TGFα), as well as for negative regulators like TGFβ[8].
In recent years activins, a subgroup of the TGFβ family of growth, differentiation, and death factors which share part of their signaling mechanisms with TGFβ, have gained attention with respect to their role in tumor development in several organs[9]. Activin subunits, their receptors and several antagonistic proteins are expressed in the normal liver. Deregulation of this balanced expression appears to contribute to hepatic dysfunctions like impaired regeneration, fibrogenesis and tumorigenesis[10]. The current knowledge about the role of activins and activin antagonists in HCC is discussed in this review.
Activins are secreted polypeptides and represent a subgroup of the TGFβ superfamily of growth and differentiation factors. Additional members of this superfamily include TGFβ1-3, bone morphogenetic proteins (BMPs), growth and differentiation factors (GDFs), myostatin, Muellerian inhibiting substance (MIS), nodal and several others[1011]. Activins are homo- or heterodimers composed of four different β subunits (βA, βB, βC, βE), each encoded by a single gene. The β subunits can either form activins by dimerization with a second β subunit, or alternatively can form inhibins by dimerizing with a single α subunit encoded in mammalian genomes[12]. Activin terminology is dependent on the dimer configuration with a single letter designating homodimers (activins A, B, C, and E) and two letters designating heterodimers according to their subunit composition (activins AB, AC, AE, BC etc.). With respect to tissue expression, transcripts of the βA and βB subunits were found to be detectable in almost all tissues analyzed with especially high expression in reproductive organs[1314]. The βC and βE subunits, in contrast, are predominantly expressed in the liver and at lower levels in a limited number of additional organs[14–18].
Activin β subunits are synthesized as precursor molecules with 350-426 amino acids and molecular weights between 38 kDa and 50 kDa[19]. The prodomains are removed in the ER and in the early Golgi by members of the protease family of subtilase-like pro-protein convertases (SPC)[20] to release mature peptides with either 115 (βB, βE) or 116 (βA, βC) amino acids. The amino acid sequences of the mature peptides are approximately 50% conserved among the four human β subunits, whereas the sequence homology in the prodomain is only about 20%. Analysis of the phylogenetic relationship of the mature human peptides groups together βA and βB on the one and βC and βE on the other hand[21].
Like other members of the TGFβ family, the activin β subunits contain nine conserved cysteines in the mature peptides. The sixth is used for dimerization, whereas the other eight form intramolecular disulfide bonds which determine the three-dimensional structure of the peptides[22]. While all cysteines in the mature chain of activin βA are necessary for biosynthesis of activin A dimers or for their full biological activity, four additional cysteines in the prodomain are dispensable for dimerization and secretion. Protein folding and dimerization take place in the lumen of the ER and are catalyzed by members of the protein disulfide isomerase (PDI) family[2324]. Unlike TGFβ, which is secreted as a latent complex consisting of the TGFβ homodimer, its prodomain (also termed latency-associated propeptide, LAP), and the latent TGFβ binding protein (LTBP)[25], activins are secreted as dimers of the mature peptides and need no further processing in the extracellular space to gain bioactivity. Activin A signals are transduced via two types of single-pass transmembrane serine threonine kinase receptors, termed activin receptors typeIand type II[26]. Activin A first binds to the type II receptors which in turn recruit and phosphorylate the typeI receptors[27]. Two type II receptors for activin A (ActR-II (A) or ACVR2 (A) and ActR-IIB or ACVR2B) have been identified. The main typeIreceptor for activin A is ALK (Activin Receptor-Like kinase) 4, also designated as ActR-IB or ACVR1B, whereas activins B and AB have a preference for ALK 7 (ACVR1C) as typeIreceptor[28]. Receptors for activins containing βC or βE subunits have not been identified so far. Activin C, however, did not compete with activin A for receptor binding[29] and a chimeric activin construct in which the receptor binding sequence (amino acids 46-78) of βA was replaced by the corresponding region of βC retained type II receptor binding but was unable to recruit the typeIreceptor ALK 4[30].
Inhibins have been shown to form a complex with type II receptors via their β subunits and with betaglycan also known as TGFβ type III receptor. The α subunit, however, is unable to bind typeIreceptors and consequently activin receptor signaling is inhibited[3132]. There is in general a considerable degree of promiscuity in receptor usage by different TGFβ superfamily members. In addition to activin A, for instance, myostatin, and several BMPs were shown to signal via ActR-IIB[33].
Phosphorylated TGFβ family receptors recruit intracellular mediators called Smads, which transduce activin signals to the nucleus[26]. Smads can be divided into receptor Smads (Smads 1, 2, 3, 5 and 8), a common mediator Smad (Smad 4) and inhibitory Smads (Smads 6 and 7). Activin A receptors, as well as TGFβ receptors, recruit and phosphorylate the receptor Smads 2 and 3, whereas receptor Smads 1, 5, and 8 are recruited by BMP receptors but not activin receptors[34]. Recent evidence suggests that-similar to TGFβ-additional Smad-independent signaling pathways may contribute to activin A signaling, as for instance, RhoA, MEKK1, JNK, and p38 were found to be involved in activin-induced cytoskeleton reorganization and cell migration in keratinocytes and in promoter activation of the transcription factor Pit-1 in pituitary lactotrope cells[3536].
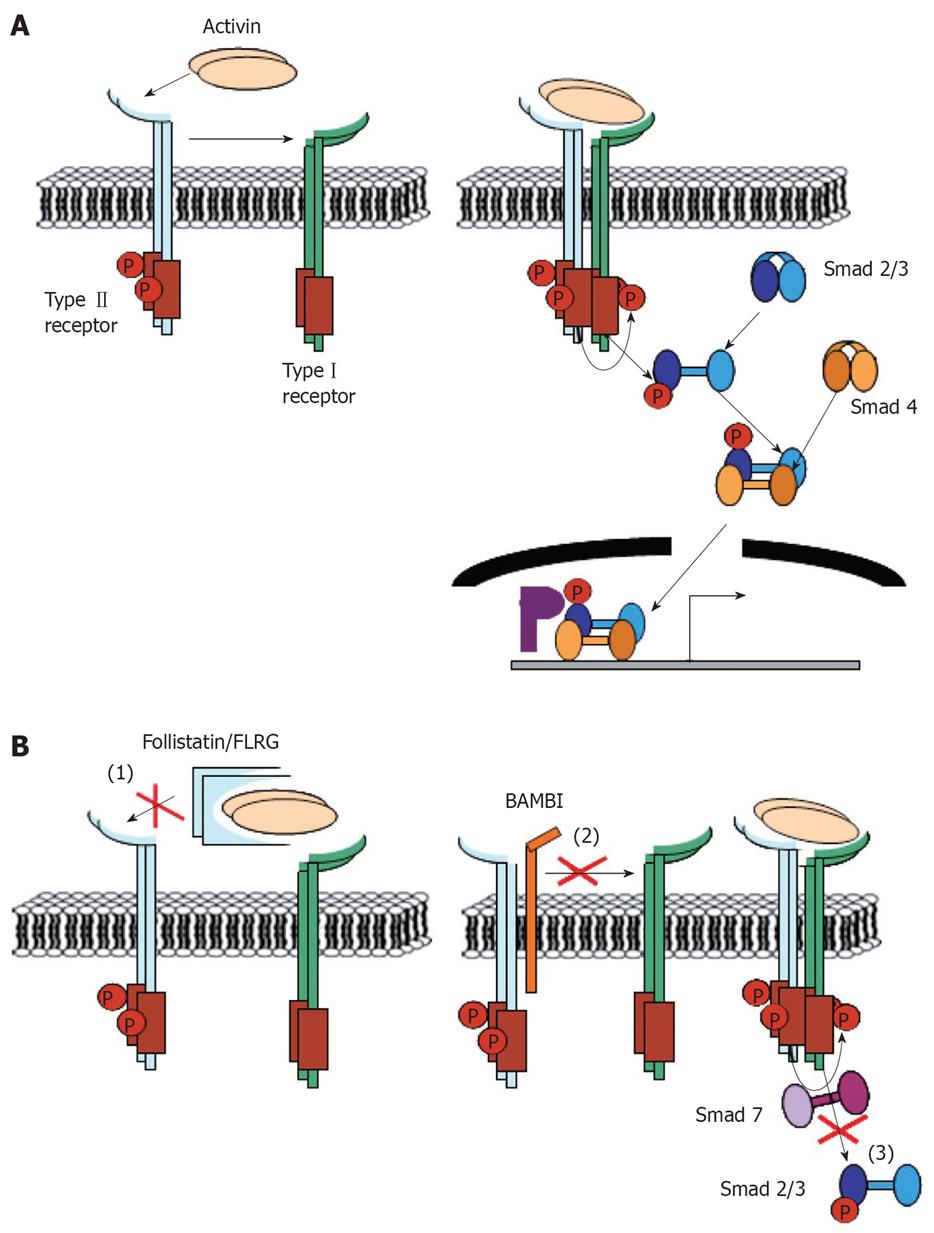
Activin signals are tightly regulated on the one hand by a spatially and temporally restricted production of activin subunits and on the other hand by the expression of several extra- as well as intracellular antagonists of activin signaling. An overview of activin-mediated signaling events and the corresponding interaction points with endogenous activin antagonists is presented in Figure 1.
Activin A, the homodimer of two βA subunits, is by far the most extensively investigated activin. Multiple biological functions of activin A in a variety of cells and tissues have been described. Activin A has been implicated for instance in mesoderm induction[37], stem cell biology[38], reproductive biology[39], erythroid differentiation[40], systemic inflammation[41], cell death induction[42], wound healing[43], and fibrosis[44]. Knock-out mice for βA have severe defects in craniofacial development and die shortly after birth[45]. Concerning the liver, activin A potently inhibits mitogen-induced DNA synthesis and induces apoptosis in hepatocytes in vivo and in vitro[46–48]. Activin βA antisense oligonucleotides stimulated cell proliferation in the human hepatoma cell line HLF suggesting a growth inhibitory function of endogenous activin A[49]. In regenerating liver, activin βA gene expression was reduced at time points when hepatocyte replication took place and was increased at later periods when liver regeneration terminated[50]. Increased expression of βA at earlier time points after partial hepatectomy, however, has also been described[5152]. Besides the effects on DNA synthesis and cell growth, activin A also regulates restoration of liver architecture after partial hepatectomy by stimulating collagen production in hepatic stellate cells (HSC) and tubulogenesis of sinusoidal endothelial cells[5354]. Stimulation of HSC may also contribute to liver fibrosis and several investigations have found elevated levels of activin βA in fibrotic and cirrhotic rat livers[55–58]. Elevated levels of circulating activin A were found in patients suffering from chronic viral hepatitis or alcohol induced liver cirrhosis and in HCC patients[59–61]. Reduced expression of activin βA transcripts in contrast, was observed in tumor tissue from chemically-induced rat liver tumors and in 5 of 11 HCC specimens[62]. In addition to a pro-apoptotic effect on the parenchymal cells and a pro-fibrotic effect on HSC, activin A has also been linked to neoangiogenesis via stimulation of VEGF expression in human hepatoma cells[63].
Like activin βA, the βB subunit is expressed in multiple tissues and organs[1314]. Despite a considerable overlap in tissue expression and in some biological activities, important differences exist[64]. Knock-out mice for βB are viable but have defects in eyelid development and female reproduction[65]. When the coding region of the mature peptide of the βA subunit gene was replaced with the corresponding region of the βB subunit gene the developmental defects of the βA knock-out mice were only partially rescued[66]. Concerning the liver, the role of the βB subunit is not well characterized. In the normal rat liver the βB subunit was the only activin subunit undetectable by RNAse protection assay[14]. Weak positive immunoreactivity for βB was, however, detected in hepatocytes of normal rat livers and in connective tissue septa in fibrotic livers when analyzed by immunohistochemistry[55]. Activin βB mRNA was induced in stellate cells of CCl4 treated rat livers[55]. Exposure to the peroxisome proliferator di-n-butyl phthalate led to a transient surge of βB mRNA expression 6 h after treatment in rat livers[67]. With respect to biological activities, recombinant activins A and AB but not activin B inhibited EGF induced DNA synthesis in primary rat hepatocytes[68]. In normal human liver the βB transcript is readily detectable by RT-PCR (M.G. unpublished observation), but no data with regard to expression changes of the βB subunit in liver tumors compared to normal liver have been reported yet.
The activin βC subunit was cloned from liver cDNA and demonstrated to be predominantly expressed in hepatocytes by Northern blot analysis and RNAse protection assays[14185269]. By immunohistochemistry significant activin βC expression has been detected in cells from additional organs including the prostate, ovary, testes, and pituitary gland[1570]. Formation of homodimeric activin C as well as heterodimeric activins AC, BC, CE, as well as inhibin C has been demonstrated by ectopic expression of the respective subunits in different cell models[147071]. After partial hepatectomy a transient down-regulation of activin βC expression was observed by several studies[50527273]. A decrease of activin βC expression has also been observed in HepG2 and Hep3B hepatoma cells versus normal liver tissue[74] and in rat hepatocytes during primary culture with and without EGF treatment[52]. In contrast, increased activin βC expression was reported in the rat liver during the development of CCl4 induced cirrhosis[5675] and in response to treatment with the peroxisome proliferator bi-n-butyl phthalate[67]. The functions of the activin βC subunit are controversial. Activin βC knock-out mice developed normally and liver regeneration after partial hepatectomy proceeded similarly in knock-out animals and wild-type littermates[76]. Ectopic expression of activin βC induced apoptosis in human (HepG2, Hep3B) and rat (H4IIEC3) hepatoma cells and delayed liver regeneration in mice[7477]. In AML12 cells, an immortalized mouse hepatocyte cell line in contrast, and in primary rat hepatocytes, activin βC increased DNA synthesis[29]. Adenovirus-mediated expression of activin βC accelerated liver regeneration after partial hepatectomy in rats[78]. A specific association of activin βC immunoreactivity with mitotic hepatocytes was observed in regenerating liver after partial hepatectomy[50]. It was shown that activin C does not activate activin A-responsive promoters, and it was suggested that the βC subunit regulates the levels of bioactive activin A via the formation of signaling-incompetent activin AC heterodimers in PC3 human prostate cancer cells[7980]. Data regarding the expression of the βC subunit in human liver tumors are not available yet.
Similar to activin βC, the βE subunit is predominantly expressed in hepatocytes but has also been detected in human heart, testis, peripheral blood leucocytes, placenta, and skeletal muscle[14162181]. Formation of homodimeric activin E as well as heterodimeric activins AE and CE has been demonstrated after ectopic co-expression of the respective subunits[1416]. Activin βE mRNA expression was transiently up-regulated after partial hepatectomy or portal vein branch ligation[7376] and in response to lipopolysaccharide treatment[81]. Increased βE expression has also been observed in hepatic fibrosis induced by CCl4[75]. Recently, induction of βE expression has been described as a marker for phospholipidosis in HepG2 hepatoma cells[82]. Similar to βC, βE subunit knock-out mice and double knock-outs lacking both βC and βE expression developed normally and had no defects in liver function[76]. When ectopically expressed in HepG2 or Hep3B hepatoma cells or in the murine hepatocyte cell line AML12, activin βE reduced cell number and increased apoptosis rates[7483]. Transient overexpression of βE by non-viral gene transfer in the mouse liver inhibited regenerative DNA synthesis[77]. These observations suggest that activin E may have a growth-limiting function similar to activin A, however, the two subunits show a reciprocal pattern with respect to diurnal variations of expression[10]. In line with a growth-limiting function of activin E, transgenic mice overexpressing βE in the pancreas showed reduced proliferation of pancreatic exocrine cells[84]. Regarding liver cancer, reduced expression of the βE subunit was found in human HCC specimens as well as in N-nitroso morpholine-induced rat liver tumors[6285]. Interestingly, activin βE expression was found to be regulated by the tumor suppressor gene RASSF1A[86], a gene frequently inactivated by promoter hypermethylation in HCC[8788].
The inhibin α subunit is part of inhibins but not activins and in many biological systems activins and inhibins have antagonistic effects[89]. Historically activins received their name from the fact that they activated follicle stimulating hormone (FSH) secretion from the pituitary, whereas the previously described inhibins represented the long sought-after gonadal feed-back inhibitor of pituitary FSH secretion[12]. Knock-out mice for the inhibin α subunit developed gonadal sex-cord stromal tumors suggesting a tumor suppressive function of the inhibin α subunit[90]. In several human tumor types including some types of ovarian carcinoma and adrenal tumors, in contrast, overexpression of inhibin α has been demonstrated, and inhibins have been used as serum markers for early detection of ovarian germ cell tumors and monitoring of recurrence[9]. With regard to liver cell growth, treatment with inhibin A per se had no effect on DNA synthesis of HepG2 hepatoma cells but antagonized the inhibitory effect of activin A[91]. In normal and fibrotic rat liver absence of inhibin α subunit immunoreactivity has been reported[55]. Immunostaining for inhibin α has been used to distinguish adrenal cortical tumors, which are positive in about 70% of cases, from HCC and renal cell carcinoma, which are mostly negative[9293].
Follistatin is a secreted, monomeric glycoprotein lacking homology to the TGFβ superfamily. The biological activities described for follistatin, however, seem to depend entirely on its interaction with activins and other members of the TGFβ family. Follistatin is expressed in most of the organs, that also express activin[1394], and it binds mature secreted activin A with very high affinity (Kd 50-680 pmol/L)[95–97]. Complex formation with follistatin completely abolished receptor binding of activin A, thus blocking activin signaling[9698]. Two follistatin molecules embrace one activin dimer and bury one-third of its residues and its receptor binding sites[99]. Three major forms of secreted follistatin exist, resulting from alternative splicing and protein processing of a single follistatin gene and containing 288, 303 and 315 amino acids, respectively[95]. All forms of follistatin contain three homologous follistatin domains[100] of which the first two, but not the third, are necessary for activin A binding[97101]. Follistatin 288 binds to heparan sulfates, whereas this binding is blocked by an acidic tail in follistatin 315[95]. In addition to binding activins A, B, AB, and E, follistatin was also shown to bind and antagonize myostatin as well as BMPs 2, 4, 6 and 7[16102–105]. Follistatin administration by intraportal infusion or adenovirus-mediated overexpression caused DNA synthesis and liver growth in normal rat livers presumably by antagonizing tonic inhibition of liver growth by activin A[106107]. Following partial hepatectomy follistatin expression was up-regulated after 24-48 h, the time period in which hepatocyte replication was increased[50]. Under similar conditions administration of follistatin accelerated liver regeneration but led to impaired restoration of normal tissue architecture and compromised liver function[108–110]. Administration of exogenous follistatin in CCl4 treated rats attenuated the formation of liver fibrosis[111]. These results likely reflect the ability of follistatin to antagonize both growth-inhibitory and pro-fibrotic activities of activin A.
In human liver cancer and also in animal models follistatin expression was increased in about 60% of tumor tissues. Increased follistatin levels were also found in the blood of patients with liver cirrhosis and HCC[6062112]. Administration of follistatin stimulated DNA synthesis in preneoplastic rat hepatocytes in an ex vivo system, whereas hepatoma cell lines were unresponsive to exogenous follistatin possibly due to autocrine production of follistatin or other activin antagonists[62112–114].
Follistatin-related protein, encoded by follistatin-related gene (FLRG), also designated as follistatin-like 3 (FSTL-3) has a high similarity to follistatin and shares its ability to bind TGFβ family proteins, but contains only two instead of three follistatin domains[115]. Several other proteins, containing 1-10 follistatin domains, like the extracellular matrix-associated proteins SPARC and agrin, on the other hand were not able to bind TGFβ family members[100116]. The FLRG gene was originally identified as a target of chromosomal rearrangement in leukemia[117]. The highest tissue expression of FLRG was found in placenta, whereas highest follistatin expression was found in ovary, testis, and pituitary[115118]. In HepG2 hepatoma cells, expression of both FLRG and follistatin was induced in response to activin A treatment suggesting that they participate in a feedback loop to restrict activin A signals[119]. FLRG mRNA is up-regulated in rat livers in response to a necrogenic dose of CCl4 (M.G. unpublished observation) but otherwise the role of FLRG in liver regeneration has not been characterized. Elevated expression of FLRG was found in chemically induced rat liver tumors and H4IIE rat hepatoma cells but not in human liver tumor specimens[62] indicating species-specific differences with respect to FLRG regulation or differences between liver tumors of different etiologies.
The type II activin receptors ActR-II (A) and ActR-IIB and the typeIactivin receptors ALK4 and ALK7 are expressed in multiple cell types and tissues including the liver. Adenovirus-mediated overexpression of a dominant-negative type II activin receptor caused DNA synthesis and liver growth in normal rat livers[120]. During liver regeneration after partial hepatectomy, no change of ActRII was observed while ActRIIB was transiently decreased[50]. During CCl4 induced rat liver cirrhosis, ActRIIA was reduced after 5 wk but returned to control levels after 10 wk[56]. Ectopic overexpression of ActR-IB (ALK4) and ActR-IIB or of ALK7 induced apoptosis in hepatoma cells[121122]. In HCC tissue specimens, expression of activin receptors (ActR-I, ActR-IB, ActR-II, and ActR-IIB) was demonstrated by immunohistochemistry[63]. Inactivating mutations of activin receptors have been found in microsatellite instable colon cancer, pancreatic cancer and prostate cancer, but have not been investigated in HCC so far[123–126].
Several membrane-associated proteins exist which regulate activin-induced receptor activation. Cripto/TDGF1 is a member of the EGF-CFC (epidermal growth factor-Cripto/frl/cryptic) family of growth factor-like molecules. This secreted protein can attach to the outer cell membrane via a glycosylphosphatidylinositol anchor and functions as a co-receptor for nodal signaling during embryogenesis. Cripto has been found overexpressed in high percentages of several human malignancies including breast, pancreas, lung, colon and bladder cancer[127]. Cripto inhibits ligand receptor interactions of activins and TGFβ[128–130] and this has been suggested to contribute to its pro-tumorigenic activity. However, an additional activin receptor-independent signaling pathway for Cripto involving Glypican-1 and c-Src has also been described[127]. Expression of a shorter Cripto variant was observed in colon cancer including liver metastases, as well as in colon cancer and hepatoma cell lines[131132]. Expression of this short variant is driven by Wnt signaling which is frequently constitutively activated in colon cancer and HCC. Based on these findings, a more extensive investigation on the role of Cripto in HCC is certainly warranted.
BAMBI (bone morphogenetic protein and activin membrane-bound inhibitor) also known as nma (non metastatic gene A) is a pseudoreceptor related to the typeI receptors of the TGFβ family. It lacks an intracellular kinase domain and inhibits activin A, TGFβ, and BMP signaling by stably associating with TGFβ family receptors[133]. A recent study links LPS/Toll-like receptor 4-induced downregulation of BAMBI in hepatic stellate cells to hepatic fibrosis[134]. In contrast, elevated BAMBI expression driven by the Wnt/β-catenin pathway was found in HCC and CRC specimens[135].
ARIPS 1 and 2 (activin receptor-interacting proteins) are PDZ (PSD-95/Discs-large/ZO-1) protein-protein interaction domain-containing proteins that were described to interact with type II activin receptors and inhibit or augment activin signaling, depending on the isoforms expressed[136–138]. ARIP 2 was recently shown to be induced by activin A in the mouse hepatoma cell line Hepa1-6 and to decrease activin-mediated collagen IV expression, suggesting that it participates in a negative feedback regulation of activin-induced liver fibrosis[139]. Data with regard to a role of ARIPS in HCC or other tumor types are missing so far.
Downstream from activin receptors, signals are transduced by receptor Smad 2 and Smad 3 and the common mediator Smad 4, the same set of Smad proteins also used by TGFβ receptors. Mutations of Smad proteins are frequent in pancreatic and colorectal cancer and have also been detected in HCC[140–142]. Smads 6 and 7 associate with TGFβ family receptors but are not phosphorylated and thus inhibit signal transduction[143144]. Smad7 has been demonstrated to inhibit activin signaling and to protect hepatocytes from activin A-induced growth inhibition[145]. Increased expression of Smad7 has been observed in HCC tissue compared to adjacent tissue[146] and in advanced HCC compared to early HCC or dysplastic nodules[147]. No mutations of either Smad 6 or Smad 7 were found in 52 HCC samples[148].
Smurf-type ubiquitin E3 ligases, Smad anchor for receptor activation (SARA), and transcriptional co-activators and co-repressors such as CBP, p300, c-Ski, and SnoN, control Smad activation by TGFβ-family receptors or shuttling of activated Smads into the nucleus as well as transcriptional activity of Smad-containing complexes[42]. Their role in the link between activin signals and liver carcinogenesis has yet to be defined.
In summary, increasing evidence suggests that deregulation of activin signals frequently occurs in and contributes to HCC development and progression. An overview of alterations in activin subunits and activin antagonists described in liver tumors and hepatoma cells is presented in Table 1.
| Activin subunits and activin antagonists | Proposed function in activin signaling | Alterations observed in HCC and hepatoma cells |
| Activin βA subunit | Activates activin receptors | Increased activin A in circulation of HCC patients[6061] |
| Decreased expression in rat and human liver tumors[62] | ||
| Loss of expression in hepatoma cells[74114] | ||
| Activin βE subunit | Induces apoptosis by as yet undefined mechanisms | Decreased expression in rat and human liver tumors[6285] |
| Follistatin | Binds activins and blocks their interaction with receptors | Increased in circulation of HCC patients[60] |
| Increased expression in human mouse and rat liver tumors[62112] | ||
| Expressed in hepatoma cells[112114] | ||
| FLRG | Binds activins and blocks their interaction with receptors | Increased in rat but decreased in human liver tumors[62] |
| BAMBI | Binds TGFβ-family type II receptors and blocks typeIreceptor activation | Increased in HCC and colon cancer[135] |
| Cripto | Blocks interaction of activins (and TGFβ) with their receptors | Overexpressed in hepatoma cells[132] |
| Smad 7 | Inhibits activation of Smads by activin (and TGFβ) receptors | Increased expression in HCC[146147] |
Activin signaling is complex. At least three features of the activin signaling cascade contribute to this complexity. First, four activin β and one inhibin α subunit can give rise to multiple homo and heterodimers with different receptor binding capabilities. Secondly, a number of different extracellular activin-binding and receptor-interacting proteins can modulate ligand receptor interactions not only of activins but also of TGFβ, BMPs and GDFs. Thirdly, there is a considerable degree of promiscuity with respect to usage of receptors and intracellular signaling molecules between different members of the TGFβ superfamily[149]. For instance, activins and TGFβ use different typeIand type II receptors but rely on the same Smad proteins for intracellular propagation of their signals. This makes it a difficult task to dissect their specific contribution to biological activities, especially in tissues such as the liver, where both activins and TGFβ are expressed. In addition, BAMBI, Cripto and Smad7 have all been shown to interfere with signal transduction of activins as well as of TGFβ.
TGF-β1 has a well recognized dual role in carcino-genesis[150]. It acts as a tumor suppressor in early stages of hepatocarcinogenesis by inducing apoptosis and eliminating precursor lesions[151152]. At a later stage, however, liver tumor cells often become resistant to its proapoptotic effect, and produce large amounts of TGFβ themselves[153]. From the available data on both loss of expression in tumor cells and apoptosis induction[74114154], one would postulate that activin A, and possibly activin E, may have a similar tumor suppressive function in the liver as TGFβ. Whether also activins may shift to a pro-tumorigenic function during tumor progression is little explored. For activin A, a contribution to liver fibrosis, enhanced expression of the angiogenic factor VEGF in hepatoma cells, and stimulation of growth and invasiveness of esophageal squamous cell carcinoma cells has been demonstrated[5863155].
Despite all the complexity, however, a general theme in HCC and in other tumor types seems to be the elevated expression of activin antagonistic proteins in the tumor cells, as observed for follistatin, BAMBI, Cripto, and Smad7[62127135146147]. These may serve to block the growth inhibitory and pro-apoptotic activity of activin A on hepatocytes. Similar observations have been made in additional tumor types, for instance for Cripto in multiple epithelial tumors, BAMBI in colon carcinoma, follistatin in melanoma and FLRG in breast cancer[127135156157].
Consequently, a targeted inhibition of activin antagonists might restore sensitivity to activin-induced growth inhibition and apoptosis, and may thus represent a feasible strategy to inhibit tumor growth. In line with this hypothesis, it has recently been shown that siRNA-mediated silencing of FLRG inhibited breast tumor cell growth in vitro, and that monoclonal antibodies to Cripto inhibited growth of testicular and colon cancer cells in xenograft models[128156]. Future studies will have to clarify whether such approaches may offer new therapeutic opportunities for combating liver cancer.
| 1. | Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74-108. |
| 2. | Chen JG, Zhu J, Parkin DM, Zhang YH, Lu JH, Zhu YR, Chen TY. Trends in the incidence of cancer in Qidong, China, 1978-2002. Int J Cancer. 2006;119:1447-1454. |
| 3. | El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557-2576. |
| 4. | Befeler AS, Di Bisceglie AM. Hepatocellular carcinoma: diagnosis and treatment. Gastroenterology. 2002;122:1609-1619. |
| 5. | Chung DC. The genetic basis of colorectal cancer: insights into critical pathways of tumorigenesis. Gastroenterology. 2000;119:854-865. |
| 7. | Teufel A, Staib F, Kanzler S, Weinmann A, Schulze-Bergkamen H, Galle PR. Genetics of hepatocellular carcinoma. World J Gastroenterol. 2007;13:2271-2282. |
| 8. | Breuhahn K, Longerich T, Schirmacher P. Dysregulation of growth factor signaling in human hepatocellular carcinoma. Oncogene. 2006;25:3787-3800. |
| 9. | Risbridger GP, Schmitt JF, Robertson DM. Activins and inhibins in endocrine and other tumors. Endocr Rev. 2001;22:836-858. |
| 10. | Rodgarkia-Dara C, Vejda S, Erlach N, Losert A, Bursch W, Berger W, Schulte-Hermann R, Grusch M. The activin axis in liver biology and disease. Mutat Res. 2006;613:123-137. |
| 11. | Chang H, Brown CW, Matzuk MM. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr Rev. 2002;23:787-823. |
| 12. | Ling N, Ying SY, Ueno N, Shimasaki S, Esch F, Hotta M, Guillemin R. Pituitary FSH is released by a heterodimer of the beta-subunits from the two forms of inhibin. Nature. 1986;321:779-782. |
| 13. | Tuuri T, Eramaa M, Hilden K, Ritvos O. The tissue distribution of activin beta A- and beta B-subunit and follistatin messenger ribonucleic acids suggests multiple sites of action for the activin-follistatin system during human development. J Clin Endocrinol Metab. 1994;78:1521-1524. |
| 14. | Vejda S, Cranfield M, Peter B, Mellor SL, Groome N, Schulte-Hermann R, Rossmanith W. Expression and dimerization of the rat activin subunits betaC and betaE: evidence for the ormation of novel activin dimers. J Mol Endocrinol. 2002;28:137-148. |
| 15. | Gold EJ, O’Bryan MK, Mellor SL, Cranfield M, Risbridger GP, Groome NP, Fleming JS. Cell-specific expression of betaC-activin in the rat reproductive tract, adrenal and liver. Mol Cell Endocrinol. 2004;222:61-69. |
| 16. | Hashimoto O, Tsuchida K, Ushiro Y, Hosoi Y, Hoshi N, Sugino H, Hasegawa Y. cDNA cloning and expression of human activin betaE subunit. Mol Cell Endocrinol. 2002;194:117-122. |
| 17. | Fang J, Wang SQ, Smiley E, Bonadio J. Genes coding for mouse activin beta C and beta E are closely linked and exhibit a liver-specific expression pattern in adult tissues. Biochem Biophys Res Commun. 1997;231:655-661. |
| 18. | Schmitt J, Hotten G, Jenkins NA, Gilbert DJ, Copeland NG, Pohl J, Schrewe H. Structure, chromosomal localization, and expression analysis of the mouse inhibin/activin beta C (Inhbc) gene. Genomics. 1996;32:358-366. |
| 19. | Grusch M, Rodgarkia-Dara C, Bursch W, Schulte-Hermann R. Activins and the liver-Transforming Growth Factor-β in Cancer Therapy. New York: Humana Press 2007; 1-20. |
| 20. | Salvas A, Benjannet S, Reudelhuber TL, Chretien M, Seidah NG. Evidence for proprotein convertase activity in the endoplasmic reticulum/early Golgi. FEBS Lett. 2005;579:5621-5625. |
| 21. | Fang J, Yin W, Smiley E, Wang SQ, Bonadio J. Molecular cloning of the mouse activin beta E subunit gene. Biochem Biophys Res Commun. 1996;228:669-674. |
| 22. | Mason AJ. Functional analysis of the cysteine residues of activin A. Mol Endocrinol. 1994;8:325-332. |
| 23. | Freedman RB, Hirst TR, Tuite MF. Protein disulphide isomerase: building bridges in protein folding. Trends Biochem Sci. 1994;19:331-336. |
| 24. | Ellgaard L, Ruddock LW. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28-32. |
| 25. | Todorovic V, Jurukovski V, Chen Y, Fontana L, Dabovic B, Rifkin DB. Latent TGF-beta binding proteins. Int J Biochem Cell Biol. 2005;37:38-41. |
| 26. | Abe Y, Minegishi T, Leung PC. Activin receptor signaling. Growth Factors. 2004;22:105-110. |
| 27. | Attisano L, Wrana JL, Montalvo E, Massague J. Activation of signalling by the activin receptor complex. Mol Cell Biol. 1996;16:1066-1073. |
| 28. | Tsuchida K, Nakatani M, Yamakawa N, Hashimoto O, Hasegawa Y, Sugino H. Activin isoforms signal through type I receptor serine/threonine kinase ALK7. Mol Cell Endocrinol. 2004;220:59-65. |
| 29. | Wada W, Maeshima A, Zhang YQ, Hasegawa Y, Kuwano H, Kojima I. Assessment of the function of the betaC-subunit of activin in cultured hepatocytes. Am J Physiol Endocrinol Metab. 2004;287:E247-E254. |
| 30. | Muenster U, Harrison CA, Donaldson C, Vale W, Fischer WH. An activin-A/C chimera exhibits activin and myostatin antagonistic properties. J Biol Chem. 2005;280:36626-36632. |
| 31. | Cook RW, Thompson TB, Jardetzky TS, Woodruff TK. Molecular biology of inhibin action. Semin Reprod Med. 2004;22:269-276. |
| 32. | Lewis KA, Gray PC, Blount AL, MacConell LA, Wiater E, Bilezikjian LM, Vale W. Betaglycan binds inhibin and can mediate functional antagonism of activin signalling. Nature. 2000;404:411-414. |
| 33. | Rebbapragada A, Benchabane H, Wrana JL, Celeste AJ, Attisano L. Myostatin signals through a transforming growth factor beta-like signaling pathway to block adipogenesis. Mol Cell Biol. 2003;23:7230-7242. |
| 34. | Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465-471. |
| 35. | de Guise C, Lacerte A, Rafiei S, Reynaud R, Roy M, Brue T, Lebrun JJ. Activin inhibits the human Pit-1 gene promoter through the p38 kinase pathway in a Smad-independent manner. Endocrinology. 2006;147:4351-4362. |
| 36. | Zhang L, Deng M, Parthasarathy R, Wang L, Mongan M, Molkentin JD, Zheng Y, Xia Y. MEKK1 transduces activin signals in keratinocytes to induce actin stress fiber formation and migration. Mol Cell Biol. 2005;25:60-65. |
| 37. | McDowell N, Gurdon JB. Activin as a morphogen in Xenopus mesoderm induction. Semin Cell Dev Biol. 1999;10:311-317. |
| 38. | Beattie GM, Lopez AD, Bucay N, Hinton A, Firpo MT, King CC, Hayek A. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells. 2005;23:489-495. |
| 39. | de Kretser DM, Hedger MP, Loveland KL, Phillips DJ. Inhibins, activins and follistatin in reproduction. Hum Reprod Update. 2002;8:529-541. |
| 40. | Maguer-Satta V, Bartholin L, Jeanpierre S, Ffrench M, Martel S, Magaud JP, Rimokh R. Regulation of human erythropoiesis by activin A, BMP2, and BMP4, members of the TGFbeta family. Exp Cell Res. 2003;282:110-120. |
| 41. | Jones KL, de Kretser DM, Patella S, Phillips DJ. Activin A and follistatin in systemic inflammation. Mol Cell Endocrinol. 2004;225:119-125. |
| 42. | Chen YG, Wang Q, Lin SL, Chang CD, Chuang J, Ying SY. Activin signaling and its role in regulation of cell proliferation, apoptosis, and carcinogenesis. Exp Biol Med (Maywood). 2006;231:534-544. |
| 43. | Munz B, Smola H, Engelhardt F, Bleuel K, Brauchle M, Lein I, Evans LW, Huylebroeck D, Balling R, Werner S. Overexpression of activin A in the skin of transgenic mice reveals new activities of activin in epidermal morphogenesis, dermal fibrosis and wound repair. EMBO J. 1999;18:5205-5215. |
| 44. | Werner S, Alzheimer C. Roles of activin in tissue repair, fibrosis, and inflammatory disease. Cytokine Growth Factor Rev. 2006;17:157-171. |
| 45. | Matzuk MM, Kumar TR, Vassalli A, Bickenbach JR, Roop DR, Jaenisch R, Bradley A. Functional analysis of activins during mammalian development. Nature. 1995;374:354-356. |
| 46. | Hully JR, Chang L, Schwall RH, Widmer HR, Terrell TG, Gillett NA. Induction of apoptosis in the murine liver with recombinant human activin A. Hepatology. 1994;20:854-862. |
| 47. | Schwall RH, Robbins K, Jardieu P, Chang L, Lai C, Terrell TG. Activin induces cell death in hepatocytes in vivo and in vitro. Hepatology. 1993;18:347-356. |
| 48. | Yasuda H, Mine T, Shibata H, Eto Y, Hasegawa Y, Takeuchi T, Asano S, Kojima I. Activin A: an autocrine inhibitor of initiation of DNA synthesis in rat hepatocytes. J Clin Invest. 1993;92:1491-1496. |
| 49. | Takabe K, Lebrun JJ, Nagashima Y, Ichikawa Y, Mitsuhashi M, Momiyama N, Ishikawa T, Shimada H, Vale WW. Interruption of activin A autocrine regulation by antisense oligodeoxynucleotides accelerates liver tumor cell proliferation. Endocrinology. 1999;140:3125-3132. |
| 50. | Gold EJ, Zhang X, Wheatley AM, Mellor SL, Cranfield M, Risbridger GP, Groome NP, Fleming JS. betaA- and betaC-activin, follistatin, activin receptor mRNA and betaC-activin peptide expression during rat liver regeneration. J Mol Endocrinol. 2005;34:505-515. |
| 51. | Date M, Matsuzaki K, Matsushita M, Tahashi Y, Sakitani K, Inoue K. Differential regulation of activin A for hepatocyte growth and fibronectin synthesis in rat liver injury. J Hepatol. 2000;32:251-260. |
| 52. | Zhang YQ, Shibata H, Schrewe H, Kojima I. Reciprocal expression of mRNA for inhibin betaC and betaA subunits in hepatocytes. Endocr J. 1997;44:759-764. |
| 53. | Wada W, Kuwano H, Hasegawa Y, Kojima I. The dependence of transforming growth factor-beta-induced collagen production on autocrine factor activin A in hepatic stellate cells. Endocrinology. 2004;145:2753-2759. |
| 54. | Endo D, Kogure K, Hasegawa Y, Maku-uchi M, Kojima I. Activin A augments vascular endothelial growth factor activity in promoting branching tubulogenesis in hepatic sinusoidal endothelial cells. J Hepatol. 2004;40:399-404. |
| 55. | De Bleser PJ, Niki T, Xu G, Rogiers V, Geerts A. Localization and cellular sources of activins in normal and fibrotic rat liver. Hepatology. 1997;26:905-912. |
| 56. | Gold EJ, Francis RJ, Zimmermann A, Mellor SL, Cranfield M, Risbridger GP, Groome NP, Wheatley AM, Fleming JS. Changes in activin and activin receptor subunit expression in rat liver during the development of CCl4-induced cirrhosis. Mol Cell Endocrinol. 2003;201:143-153. |
| 57. | Huang X, Li DG, Wang ZR, Wei HS, Cheng JL, Zhan YT, Zhou X, Xu QF, Li X, Lu HM. Expression changes of activin A in the development of hepatic fibrosis. World J Gastroenterol. 2001;7:37-41. |
| 58. | Sugiyama M, Ichida T, Sato T, Ishikawa T, Matsuda Y, Asakura H. Expression of activin A is increased in cirrhotic and fibrotic rat livers. Gastroenterology. 1998;114:550-558. |
| 59. | Patella S, Phillips DJ, de Kretser DM, Evans LW, Groome NP, Sievert W. Characterization of serum activin-A and follistatin and their relation to virological and histological determinants in chronic viral hepatitis. J Hepatol. 2001;34:576-583. |
| 60. | Yuen MF, Norris S, Evans LW, Langley PG, Hughes RD. Transforming growth factor-beta 1, activin and follistatin in patients with hepatocellular carcinoma and patients with alcoholic cirrhosis. Scand J Gastroenterol. 2002;37:233-238. |
| 61. | Pirisi M, Fabris C, Luisi S, Santuz M, Toniutto P, Vitulli D, Federico E, Del Forno M, Mattiuzzo M, Branca B. Evaluation of circulating activin-A as a serum marker of hepatocellular carcinoma. Cancer Detect Prev. 2000;24:150-155. |
| 62. | Grusch M, Drucker C, Peter-Vorosmarty B, Erlach N, Lackner A, Losert A, Macheiner D, Schneider WJ, Hermann M, Groome NP. Deregulation of the activin/follistatin system in hepatocarcinogenesis. J Hepatol. 2006;45:673-680. |
| 63. | Wagner K, Peters M, Scholz A, Benckert C, Ruderisch HS, Wiedenmann B, Rosewicz S. Activin A stimulates vascular endothelial growth factor gene transcription in human hepatocellular carcinoma cells. Gastroenterology. 2004;126:1828-1843. |
| 64. | Thompson TB, Cook RW, Chapman SC, Jardetzky TS, Woodruff TK. Beta A versus beta B: is it merely a matter of expression? Mol Cell Endocrinol. 2004;225:9-17. |
| 65. | Vassalli A, Matzuk MM, Gardner HA, Lee KF, Jaenisch R. Activin/inhibin beta B subunit gene disruption leads to defects in eyelid development and female reproduction. Genes Dev. 1994;8:414-427. |
| 66. | Brown CW, Houston-Hawkins DE, Woodruff TK, Matzuk MM. Insertion of Inhbb into the Inhba locus rescues the Inhba-null phenotype and reveals new activin functions. Nat Genet. 2000;25:453-457. |
| 67. | Kobayashi T, Niimi S, Fukuoka M, Hayakawa T. Regulation of inhibin beta chains and follistatin mRNA levels during rat hepatocyte growth induced by the peroxisome proliferator di-n-butyl phthalate. Biol Pharm Bull. 2002;25:1214-1216. |
| 68. | Niimi S, Horikawa M, Seki T, Ariga T, Kobayashi T, Hayakawa T. Effect of activins AB and B on DNA synthesis stimulated by epidermal growth factor in primary cultured rat hepatocytes. Biol Pharm Bull. 2002;25:437-440. |
| 69. | Hotten G, Neidhardt H, Schneider C, Pohl J. Cloning of a new member of the TGF-beta family: a putative new activin beta C chain. Biochem Biophys Res Commun. 1995;206:608-613. |
| 70. | Mellor SL, Cranfield M, Ries R, Pedersen J, Cancilla B, de Kretser D, Groome NP, Mason AJ, Risbridger GP. Localization of activin beta(A)-, beta(B)-, and beta(C)-subunits in humanprostate and evidence for formation of new activin heterodimers of beta(C)-subunit. J Clin Endocrinol Metab. 2000;85:4851-4858. |
| 71. | Ushiro Y, Hashimoto O, Seki M, Hachiya A, Shoji H, Hasegawa Y. Analysis of the function of activin betaC subunit using recombinant protein. J Reprod Dev. 2006;52:487-495. |
| 72. | Esquela AF, Zimmers TA, Koniaris LG, Sitzmann JV, Lee SJ. Transient down-regulation of inhibin-betaC expression following partial hepatectomy. Biochem Biophys Res Commun. 1997;235:553-556. |
| 73. | Takamura K, Tsuchida K, Miyake H, Tashiro S, Sugino H. Activin and activin receptor expression changes in liver regeneration in rat. J Surg Res. 2005;126:3-11. |
| 74. | Vejda S, Erlach N, Peter B, Drucker C, Rossmanith W, Pohl J, Schulte-Hermann R, Grusch M. Expression of activins C and E induces apoptosis in human and rat hepatoma cells. Carcinogenesis. 2003;24:1801-1809. |
| 75. | Huang X, Li D, Lu H, Wang Z, Wei H, Wang Y, Zhang J, Xu Q. Expression of activins, follistatin mRNA in the development of hepatic fibrosis. Zhonghua Ganzangbing Zazhi. 2002;10:85-88. |
| 76. | Lau AL, Kumar TR, Nishimori K, Bonadio J, Matzuk MM. Activin betaC and betaE genes are not essential for mouse liver growth, differentiation, and regeneration. Mol Cell Biol. 2000;20:6127-6137. |
| 77. | Chabicovsky M, Herkner K, Rossmanith W. Overexpression of activin beta(C) or activin beta(E) in the mouse liver inhibits regenerative deoxyribonucleic acid synthesis of hepatic cells. Endocrinology. 2003;144:3497-3504. |
| 78. | Wada W, Medina J, Hasegawa Y, Kuwano H, Kojima I. Adenovirus-mediated overexpression of the activin betaC subunit accelerates liver regeneration in partially hepatectomized rats. J Hepatol. 2005;43:823-828. |
| 79. | Mellor SL, Ball EM, O’Connor AE, Ethier JF, Cranfield M, Schmitt JF, Phillips DJ, Groome NP, Risbridger GP. Activin betaC-subunit heterodimers provide a new mechanism of regulating activin levels in the prostate. Endocrinology. 2003;144:4410-4419. |
| 80. | Butler CM, Gold EJ, Risbridger GP. Should activin betaC be more than a fading snapshot in the activin/TGFbeta family album? Cytokine Growth Factor Rev. 2005;16:377-385. |
| 81. | O’Bryan MK, Sebire KL, Gerdprasert O, Hedger MP, Hearn MT, de Kretser DM. Cloning and regulation of the rat activin betaE subunit. J Mol Endocrinol. 2000;24:409-418. |
| 82. | Atienzar F, Gerets H, Dufrane S, Tilmant K, Cornet M, Dhalluin S, Ruty B, Rose G, Canning M. Determination of phospholipidosis potential based on gene expression analysis in HepG2 cells. Toxicol Sci. 2007;96:101-114. |
| 83. | Wada W, Medina JJ, Kuwano H, Kojima I. Comparison of the function of the beta(C) and beta(E) subunits of activin in AML12 hepatocytes. Endocr J. 2005;52:169-175. |
| 84. | Hashimoto O, Ushiro Y, Sekiyama K, Yamaguchi O, Yoshioka K, Mutoh K, Hasegawa Y. Impaired growth of pancreatic exocrine cells in transgenic mice expressing human activin betaE subunit. Biochem Biophys Res Commun. 2006;341:416-424. |
| 85. | Chow C, Wong N, To KF, Lo KW. Activin beta E (INHBE), a RASSF1A target gene is downregulated in hepatocellular carcinoma. Proceedings of the. New York: Humana Press 2007; 26. |
| 86. | Chow LS, Lam CW, Chan SY, Tsao SW, To KF, Tong SF, Hung WK, Dammann R, Huang DP, Lo KW. Identification of RASSF1A modulated genes in nasopharyngeal carcinoma. Oncogene. 2006;25:310-316. |
| 87. | Macheiner D, Heller G, Kappel S, Bichler C, Stattner S, Ziegler B, Kandioler D, Wrba F, Schulte-Hermann R, Zochbauer-Muller S. NORE1B, a candidate tumor suppressor, is epigenetically silenced in human hepatocellular carcinoma. J Hepatol. 2006;45:81-89. |
| 88. | Schagdarsurengin U, Wilkens L, Steinemann D, Flemming P, Kreipe HH, Pfeifer GP, Schlegelberger B, Dammann R. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene. 2003;22:1866-1871. |
| 89. | Welt C, Sidis Y, Keutmann H, Schneyer A. Activins, inhibins, and follistatins: from endocrinology to signaling. A paradigm for the new millennium. Exp Biol Med (Maywood). 2002;227:724-752. |
| 90. | Matzuk MM, Finegold MJ, Su JG, Hsueh AJ, Bradley A. Alpha-inhibin is a tumour-suppressor gene with gonadal specificity in mice. Nature. 1992;360:313-319. |
| 91. | Xu J, McKeehan K, Matsuzaki K, McKeehan WL. Inhibin antagonizes inhibition of liver cell growth by activin by a dominant-negative mechanism. J Biol Chem. 1995;270:6308-6313. |
| 92. | Renshaw AA, Granter SR. A comparison of A103 and inhibin reactivity in adrenal cortical tumors: distinction from hepatocellular carcinoma and renal tumors. Mod Pathol. 1998;11:1160-1164. |
| 93. | Pan CC, Chen PC, Tsay SH, Ho DM. Differential immuno-profiles of hepatocellular carcinoma, renal cell carcinoma, and adrenocortical carcinoma: a systemic immunohistochemical survey using tissue array technique. Appl Immunohistochem Mol Morphol. 2005;13:347-352. |
| 94. | Michel U, Rao A, Findlay JK. Rat follistatin: ontogeny of steady-state mRNA levels in different tissues predicts organ-specific functions. Biochem Biophys Res Commun. 1991;180:223-230. |
| 95. | Sugino K, Kurosawa N, Nakamura T, Takio K, Shimasaki S, Ling N, Titani K, Sugino H. Molecular heterogeneity of follistatin, an activin-binding protein. Higher affinity of the carboxyl-terminal truncated forms for heparan sulfate proteoglycans on the ovarian granulosa cell. J Biol Chem. 1993;268:15579-15587. |
| 96. | Schneyer AL, Rzucidlo DA, Sluss PM, Crowley WF Jr. Characterization of unique binding kinetics of follistatin and activin or inhibin in serum. Endocrinology. 1994;135:667-674. |
| 97. | Harrington AE, Morris-Triggs SA, Ruotolo BT, Robinson CV, Ohnuma S, Hyvonen M. Structural basis for the inhibition of activin signalling by follistatin. EMBO J. 2006;25:1035-1045. |
| 98. | de Winter JP, ten Dijke P, de Vries CJ, van Achterberg TA, Sugino H, de Waele P, Huylebroeck D, Verschueren K, van den Eijnden-van Raaij AJ. Follistatins neutralize activin bioactivity by inhibition of activin binding to its type II receptors. Mol Cell Endocrinol. 1996;116:105-114. |
| 99. | Thompson TB, Lerch TF, Cook RW, Woodruff TK, Jardetzky TS. The structure of the follistatin:activin complex reveals antagonism of both type I and type II receptor binding. Dev Cell. 2005;9:535-543. |
| 100. | Shimasaki S, Koga M, Esch F, Cooksey K, Mercado M, Koba A, Ueno N, Ying SY, Ling N, Guillemin R. Primary structure of the human follistatin precursor and its genomic organization. Proc Natl Acad Sci USA. 1988;85:4218-4222. |
| 101. | Keutmann HT, Schneyer AL, Sidis Y. The role of follistatin domains in follistatin biological action. Mol Endocrinol. 2004;18:228-240. |
| 102. | Schneyer A, Schoen A, Quigg A, Sidis Y. Differential binding and neutralization of activins A and B by follistatin and follistatin like-3 (FSTL-3/FSRP/FLRG). Endocrinology. 2003;144:1671-1674. |
| 103. | Iemura S, Yamamoto TS, Takagi C, Uchiyama H, Natsume T, Shimasaki S, Sugino H, Ueno N. Direct binding of follistatin to a complex of bone-morphogenetic protein and its receptor inhibits ventral and epidermal cell fates in early Xenopus embryo. Proc Natl Acad Sci USA. 1998;95:9337-9342. |
| 104. | Amthor H, Nicholas G, McKinnell I, Kemp CF, Sharma M, Kambadur R, Patel K. Follistatin complexes Myostatin and antagonises Myostatin-mediated inhibition of myogenesis. Dev Biol. 2004;270:19-30. |
| 105. | Glister C, Kemp CF, Knight PG. Bone morphogenetic protein (BMP) ligands and receptors in bovine ovarian follicle cells: actions of BMP-4, -6 and -7 on granulosa cells and differential modulation of Smad-1 phosphorylation by follistatin. Reproduction. 2004;127:239-254. |
| 106. | Kogure K, Zhang YQ, Maeshima A, Suzuki K, Kuwano H, Kojima I. The role of activin and transforming growth factor-beta in the regulation of organ mass in the rat liver. Hepatology. 2000;31:916-921. |
| 107. | Takabe K, Wang L, Leal AM, Macconell LA, Wiater E, Tomiya T, Ohno A, Verma IM, Vale W. Adenovirus-mediated overexpression of follistatin enlarges intact liver of adult rats. Hepatology. 2003;38:1107-1115. |
| 108. | Kogure K, Omata W, Kanzaki M, Zhang YQ, Yasuda H, Mine T, Kojima I. A single intraportal administration of follistatin accelerates liver regeneration in partially hepatectomized rats. Gastroenterology. 1995;108:1136-1142. |
| 109. | Kogure K, Zhang YQ, Shibata H, Kojima I. Immediate onset of DNA synthesis in remnant rat liver after 90% hepatectomy by an administration of follistatin. J Hepatol. 1998;29:977-984. |
| 110. | Endo D, Maku-Uchi M, Kojima I. Activin or follistatin: which is more beneficial to support liver regeneration after massive hepatectomy? Endocr J. 2006;53:73-78. |
| 111. | Patella S, Phillips DJ, Tchongue J, de Kretser DM, Sievert W. Follistatin attenuates early liver fibrosis: effects on hepatic stellate cell activation and hepatocyte apoptosis. Am J Physiol Gastrointest Liver Physiol. 2006;290:G137-G144. |
| 112. | Rossmanith W, Chabicovsky M, Grasl-Kraupp B, Peter B, Schausberger E, Schulte-Hermann R. Follistatin overexpression in rodent liver tumors: a possible mechanism to overcome activin growth control. Mol Carcinog. 2002;35:1-5. |
| 113. | Fuwii M, Ishikawa M, Iuchi M, Tashiro S. Effect of follistatin on rat liver regeneration and tumor growth after portal occlusion. Hepatogastroenterology. 2005;52:833-838. |
| 114. | Mashima H, Kanzaki M, Nobusawa R, Zhang YQ, Suzuki M, Mine T, Kojima I. Derangements in the activin-follistatin system in hepatoma cells. Gastroenterology. 1995;108:834-840. |
| 115. | Tsuchida K, Arai KY, Kuramoto Y, Yamakawa N, Hasegawa Y, Sugino H. Identification and characterization of a novel follistatin-like protein as a binding protein for the TGF-beta family. J Biol Chem. 2000;275:40788-40796. |
| 116. | Ullman CG, Perkins SJ. The Factor I and follistatin domain families: the return of a prodigal son. Biochem J. 1997;326:939-941. |
| 117. | Hayette S, Gadoux M, Martel S, Bertrand S, Tigaud I, Magaud JP, Rimokh R. FLRG (follistatin-related gene), a new target of chromosomal rearrangement in malignant blood disorders. Oncogene. 1998;16:2949-2954. |
| 118. | Tortoriello DV, Sidis Y, Holtzman DA, Holmes WE, Schneyer AL. Human follistatin-related protein: a structural homologue of follistatin with nuclear localization. Endocrinology. 2001;142:3426-3434. |
| 119. | Bartholin L, Maguer-Satta V, Hayette S, Martel S, Gadoux M, Corbo L, Magaud JP, Rimokh R. Transcription activation of FLRG and follistatin by activin A, through Smad proteins, participates in a negative feedback loop to modulate activin A function. Oncogene. 2002;21:2227-2235. |
| 120. | Ichikawa T, Zhang YQ, Kogure K, Hasegawa Y, Takagi H, Mori M, Kojima I. Transforming growth factor beta and activin tonically inhibit DNA synthesis in the rat liver. Hepatology. 2001;34:918-925. |
| 121. | Chen W, Woodruff TK, Mayo KE. Activin A-induced HepG2 liver cell apoptosis: involvement of activin receptors and smad proteins. Endocrinology. 2000;141:1263-1272. |
| 122. | Kim BC, van Gelder H, Kim TA, Lee HJ, Baik KG, Chun HH, Lee DA, Choi KS, Kim SJ. Activin receptor-like kinase-7 induces apoptosis through activation of MAPKs in a Smad3-dependent mechanism in hepatoma cells. J Biol Chem. 2004;279:28458-28465. |
| 123. | Jung B, Doctolero RT, Tajima A, Nguyen AK, Keku T, Sandler RS, Carethers JM. Loss of activin receptor type 2 protein expression in microsatellite unstable colon cancers. Gastroenterology. 2004;126:654-659. |
| 124. | Hempen PM, Zhang L, Bansal RK, Iacobuzio-Donahue CA, Murphy KM, Maitra A, Vogelstein B, Whitehead RH, Markowitz SD, Willson JK. Evidence of selection for clones having genetic inactivation of the activin A type II receptor (ACVR2) gene in gastrointestinal cancers. Cancer Res. 2003;63:994-999. |
| 125. | Su GH, Bansal R, Murphy KM, Montgomery E, Yeo CJ, Hruban RH, Kern SE. ACVR1B (ALK4, activin receptor type 1B) gene mutations in pancreatic carcinoma. Proc Natl Acad Sci USA. 2001;98:3254-3257. |
| 126. | Rossi MR, Ionov Y, Bakin AV, Cowell JK. Truncating mutations in the ACVR2 gene attenuates activin signaling in prostate cancer cells. Cancer Genet Cytogenet. 2005;163:123-129. |
| 127. | Bianco C, Strizzi L, Normanno N, Khan N, Salomon DS. Cripto-1: an oncofetal gene with many faces. Curr Top Dev Biol. 2005;67:85-133. |
| 128. | Adkins HB, Bianco C, Schiffer SG, Rayhorn P, Zafari M, Cheung AE, Orozco O, Olson D, De Luca A, Chen LL. Antibody blockade of the Cripto CFC domain suppresses tumor cell growth in vivo. J Clin Invest. 2003;112:575-587. |
| 129. | Gray PC, Harrison CA, Vale W. Cripto forms a complex with activin and type II activin receptors and can block activin signaling. Proc Natl Acad Sci USA. 2003;100:5193-5198. |
| 130. | Gray PC, Shani G, Aung K, Kelber J, Vale W. Cripto binds transforming growth factor beta (TGF-beta) and inhibits TGF-beta signaling. Mol Cell Biol. 2006;26:9268-9278. |
| 131. | Baldassarre G, Tucci M, Lembo G, Pacifico FM, Dono R, Lago CT, Barra A, Bianco C, Viglietto G, Salomon D. A truncated form of teratocarcinoma-derived growth factor-1 (cripto-1) mRNA expressed in human colon carcinoma cell lines and tumors. Tumour Biol. 2001;22:286-293. |
| 132. | Hamada S, Watanabe K, Hirota M, Bianco C, Strizzi L, Mancino M, Gonzales M, Salomon DS. beta-Catenin/TCF/LEF regulate expression of the short form human Cripto-1. Biochem Biophys Res Commun. 2007;355:240-244. |
| 133. | Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massague J, Niehrs C. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480-485. |
| 134. | Seki E, De Minicis S, Osterreicher CH, Kluwe J, Osawa Y, Brenner DA, Schwabe RF. TLR4 enhances TGF-beta signaling and hepatic fibrosis. Nat Med. 2007;13:1324-1332. |
| 135. | Sekiya T, Adachi S, Kohu K, Yamada T, Higuchi O, Furukawa Y, Nakamura Y, Nakamura T, Tashiro K, Kuhara S. Identification of BMP and activin membrane-bound inhibitor (BAMBI), an inhibitor of transforming growth factor-beta signaling, as a target of the beta-catenin pathway in colorectal tumor cells. J Biol Chem. 2004;279:6840-6846. |
| 136. | Shoji H, Tsuchida K, Kishi H, Yamakawa N, Matsuzaki T, Liu Z, Nakamura T, Sugino H. Identification and characterization of a PDZ protein that interacts with activin type II receptors. J Biol Chem. 2000;275:5485-5492. |
| 137. | Matsuzaki T, Hanai S, Kishi H, Liu Z, Bao Y, Kikuchi A, Tsuchida K, Sugino H. Regulation of endocytosis of activin type II receptors by a novel PDZ protein through Ral/Ral-binding protein 1-dependent pathway. J Biol Chem. 2002;277:19008-19018. |
| 138. | Liu ZH, Tsuchida K, Matsuzaki T, Bao YL, Kurisaki A, Sugino H. Characterization of isoforms of activin receptor-interacting protein 2 that augment activin signaling. J Endocrinol. 2006;189:409-421. |
| 139. | Zhang HJ, Tai GX, Zhou J, Ma D, Liu ZH. Regulation of activin receptor-interacting protein 2 expression in mouse hepatoma Hepa1-6 cells and its relationship with collagen type IV. World J Gastroenterol. 2007;13:5501-5505. |
| 140. | Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL. MADR2 maps to 18q21 and encodes a TGFbeta-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543-552. |
| 141. | Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science. 1996;271:350-353. |
| 142. | Yakicier MC, Irmak MB, Romano A, Kew M, Ozturk M. Smad2 and Smad4 gene mutations in hepatocellular carcinoma. Oncogene. 1999;18:4879-4883. |
| 143. | Imamura T, Takase M, Nishihara A, Oeda E, Hanai J, Kawabata M, Miyazono K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature. 1997;389:622-626. |
| 144. | Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631-635. |
| 145. | Kanamaru C, Yasuda H, Takeda M, Ueda N, Suzuki J, Tsuchida T, Mashima H, Ohnishi H, Fujita T. Smad7 is induced by norepinephrine and protects rat hepatocytes from activin A-induced growth inhibition. J Biol Chem. 2001;276:45636-45641. |
| 146. | Ji GZ, Wang XH, Miao L, Liu Z, Zhang P, Zhang FM, Yang JB. Role of transforming growth factor-beta1-smad signal transduction pathway in patients with hepatocellular carcinoma. World J Gastroenterol. 2006;12:644-648. |
| 147. | Park YN, Chae KJ, Oh BK, Choi J, Choi KS, Park C. Expression of Smad7 in hepatocellular carcinoma and dysplastic nodules: resistance mechanism to transforming growth factor-beta. Hepatogastroenterology. 2004;51:396-400. |
| 148. | Kawate S, Ohwada S, Hamada K, Koyama T, Takenoshita S, Morishita Y, Hagiwara K. Mutational analysis of the Smad6 and Smad7 genes in hepatocellular carcinoma. Int J Mol Med. 2001;8:49-52. |
| 149. | Tsuchida K, Nakatani M, Uezumi A, Murakami T, Cui X. Signal transduction pathway through activin receptors as a therapeutic target of musculoskeletal diseases and cancer. Endocr J. 55(1):11-21. |
| 150. | Piek E, Roberts AB. Suppressor and oncogenic roles of transforming growth factor-beta and its signaling pathways in tumorigenesis. Adv Cancer Res. 2001;83:1-54. |
| 151. | Mullauer L, Grasl-Kraupp B, Bursch W, Schulte-Hermann R. Transforming growth factor beta 1-induced cell death in preneoplastic foci of rat liver and sensitization by the antiestrogen tamoxifen. Hepatology. 1996;23:840-847. |
| 152. | Chabicovsky M, Wastl U, Taper H, Grasl-Kraupp B, Schulte-Hermann R, Bursch W. Induction of apoptosis in mouse liver adenoma and carcinoma in vivo by transforming growth factor-beta1. J Cancer Res Clin Oncol. 2003;129:536-542. |
| 153. | Abou-Shady M, Baer HU, Friess H, Berberat P, Zimmermann A, Graber H, Gold LI, Korc M, Buchler MW. Transforming growth factor betas and their signaling receptors in human hepatocellular carcinoma. Am J Surg. 1999;177:209-215. |
| 154. | Jeruss JS, Sturgis CD, Rademaker AW, Woodruff TK. Down-regulation of activin, activin receptors, and Smads in high-grade breast cancer. Cancer Res. 2003;63:3783-3790. |
| 155. | Yoshinaga K, Yamashita K, Mimori K, Tanaka F, Inoue H, Mori M. Activin a causes cancer cell aggressiveness in esophageal squamous cell carcinoma cells. Ann Surg Oncol. 2008;15:96-103. |
| 156. | Razanajaona D, Joguet S, Ay AS, Treilleux I, Goddard-Leon S, Bartholin L, Rimokh R. Silencing of FLRG, an antagonist of activin, inhibits human breast tumor cell growth. Cancer Res. 2007;67:7223-7229. |