Published online Feb 21, 2007. doi: 10.3748/wjg.v13.i7.1010
Revised: December 28, 2006
Accepted: January 23, 2006
Published online: February 21, 2007
AIM: To elucidate the role of Rab23 in hepatocellular carcinoma (HCC) by assessing the expression of Rab23 in HCC tissue and in HCC cell lines.
METHODS: Primary tumors (n = 100) were stained with Rab23 antibodies using immunohistochemistry and in situ hybridization in tissue microarrays. Relationships between gene expression and pathology parameters were analysed. The biological significance of Rab23 in Hep-3B cells was examined by knocking down Rab23 gene expression. We designed a pair of double-stranded RNAs against human rab23 and transfected siRNA into Hep-3B cells. Rab23 expression in these cells was examined using RT-PCR and Western blots. We investigated cell growth by MTT assays and fluorescence-activated cell sorting.
RESULTS: High cytoplasmic and nuclear expression of Rab23 was found in 38 of 71 (53.5%) and in 49 of 68 HCC patients (72%) respectively, which correlated with tumor size. HCC cell lines expressed Rab23. In Hep3B cells, siRNA for Rab23 decreased Rab23 mRNA by 4.5-fold and protein expression by 2-fold. Survival rates at 24 and 48 h for Hep-3B cells transfected with siRNA were lower and about 30% Hep-3B cells were apoptotic. Knocking down rab23 suppressed Hep3B cell growth, suggesting that rab23 could play an important role in Hep3B cell growth.
CONCLUSION: Rab23 is overexpressed and/or activated in HCC. Rab23 may be both a HCC predictor and a target for treating HCC.
- Citation: Liu YJ, Wang Q, Li W, Huang XH, Zhen MC, Huang SH, Chen LZ, Xue L, Zhang HW. Rab23 is a potential biological target for treating hepatocellular carcinoma. World J Gastroenterol 2007; 13(7): 1010-1017
- URL: https://www.wjgnet.com/1007-9327/full/v13/i7/1010.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i7.1010
Hepatocellular carcinoma (HCC) is the most frequent primary malignant tumor of the liver. HCC is the third leading global cause of cancer-related death. Over 54% of HCC cases occur in China. Hepatitis B virus is a common causal agent of liver cancer, and other factors have also been found. However, the molecular mechanisms contributing to tumor progression to HCC remain unknown[1].
In recent years, targeted therapies for a variety of HCCs have achieved clinically significant response rates and have given oncologists the chance to develop individual-based strategies for patient therapy. One rationale for such therapeutic approaches is to detect target molecules in the tumor. There is a growing list of such target molecules, the levels of which are now routinely estimated by IHC staining in biopsy specimens or surgically removing tumor material. Such assays not only generate important data for therapeutic decisions, but also provide information relevant to the prognosis of patients[2].
An example is targeting of the hedgehog (Hh) pathway. Dysregulation of this pathway has been implicated in the genesis of cancers from multiple tissue types[3]. Moreover, from embryogenesis to adulthood, skin and gastrointestinal progenitors are regulated by Hh signaling[4-6]. This pathway is activated when sonic hedgehog (SHH) or Indian hedgehog (IHH) ligands bind to their receptor, patched (PTC). When unoccupied by ligand, PTC is a tumor suppressor that binds to and represses smoothened (SMO)[7], preventing the SMO proto-oncoprotein from activating downstream transcription factors, such as GLi1. Conversely, when ligand binds to PTC, SMO is released and GLi1 is activated, resulting in the transcription of target genes including PTC and Gli1[7]. Hh signaling is now known to play a critical role in the gastrointestinal tract in both health and disease. Most recently, two research groups have simultaneously reported abnormal activation of the SHH pathway in HCC[8,9]. Although the investigators paid much attention to the relationships between hedgehog signaling and HCC, little is known about this pathway in carcinogenesis.
One regulator of SHH signaling is Rab23. In 1994, the full-length cDNA encoding Rab23, a novel Ras-related small GTPase, was isolated using the sequence of a previously described GTPases[10]. Northern analysis revealed that Rab23 mRNA is predominantly expressed in the brain, which places the protein together with Rab3a and Rab15, in the group of small GTPases characteristic of the nervous system[11]. Rab23, a new member of the RAB family expressed in retina, is composed of 7 exons spanning a 34 kb domain of genomic DNA. It is located in the pericentromeric region of chromosome 6 between microsatellite markers D6S257 and D6S1659, within the critical region of RP25[12] and localized at the plasma membrane and the endocytic pathway[12,13]. Rab23 is a negative regulator of SHH[13,14]. It acts on upstream of Gli transcription factors in patterning neural cell types in the spinal cord. The primary target of Rab23 is the Gli2 activator. Rab23 and Gli3 repressors have additive effects on patterning. Analysis of Gli3 protein suggests that Rab23 also has a role in promoting the production of Gli3 repressor. Although the patched and smoothened membrane proteins can change subcellular location in response to SHH, analysis demonstrates that Rab23 does not work through either patched or smoothened membrane protein. Instead, Rab23 appears to regulate subcellular localization of essential components of the hedgehog pathway that act both on downstream of smoothened and on upstream of Gli proteins[15].
Until now there have no reports about Rab23 expression in HCC or even in other human tumors. Since Rab23 is a negative regulator of SHH signaling which can induce malignant carcinoma, dysregulation of Rab23 also may result in HCC. If it is true, we may be able to find a new target for diagnosis and treatment of HCC. In the present study, we evaluated the hypothesis that increases in Rab23 signaling induce hepatocarcinogenesis by regulating Hh signaling in HCC tissues.
We compared Rab23 expression in non-neoplastic and malignant human livers by IHC and in situ hybridization using tissue microarrays. Furthermore, since it has been confirmed that there is hedgehog signaling in Hep-3B cells[8,9], we examined the biological significance of the Rab23 gene in Hep-3B cells by knocking down Rab23 gene expression. To identify a possible role of Rab23 in HCC, we designed a pair of double-stranded RNAs (dsRNAs) against human Rab23, and transfected siRNAs into Hep-3B cells. We examined Rab23 expression in these cells using RT-PCR and Western blots. We investigated cell growth by MTT assays and fluorescence-activated cell sorting.
HCC samples from 100 patients were used in this study. All of them were from Sun Yat-Sen University (Guangzhou, China) and selected over a 4-year period (1993-1997). Twenty-five tumor adjacent liver tissue samples were also used. None of the patients received chemotherapy or radiation therapy. Each specimen was fixed in alcoholic formalin for 8-12 h and embedded in paraffin.
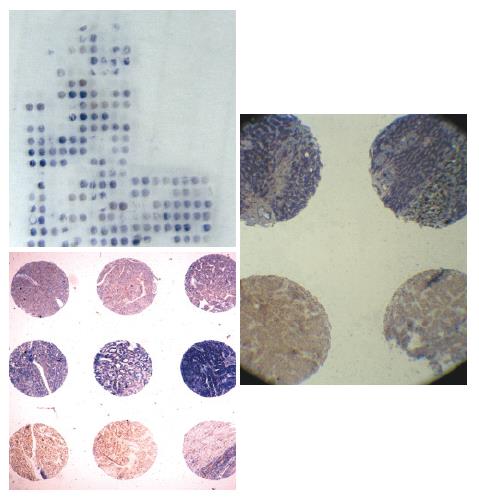
A hundred archived formalin-fixed and paraffin-embedded anonymous, representative specimens of HCC were also assessed. Non-necrotic areas in tumor for coring into a tissue microarray (TMA) were marked using an indelible pen on a hematoxylin and eosin (HE) stained whole section from donor blocks. TMAs were assembled from formalin-fixed and paraffin-embedded tumor tissue samples as previously described[16] by using a precision instrument (Beecher Instruments, Silver Spring, MD). Cores (600 μm in diameter) were randomly arrayed in triplicate across the recipient block with asymmetrical placement for orientation. TMA coordinates and clinicopathological data were stored for reference. TMA slides were sectioned and mounted on Superfrost®/Plus microscope slides as 6-μm sections (Figure 1). Hematoxylin and eosin (H&E) stained TMA sections were used as morphological references.
Paraffin-embedded tissue sections were deparaffinized. Slides from each case were exposed to affinity purified rabbit anti-Rab23 primary antibody (BD Biosciences, San Jose, CA, USA) and diluted in PBS for 2 h at room temperature. Detection was carried out using Elivision TM Plus Polymer HRP (Mouse/Rabbit) immunohistochemistry kits (Maixin.Bio, Fuzhou,China). Location of peroxidase was visualized with diaminobenzidione(DAB). Hematoxylin was used for counterstaining. Sections were thoroughly washed with PBS between steps. Negative controls (absence of primary antibody) were run for each experiment. We defined a positive sample as one in which more than one third of one tissue chip was stained brown.
In situ hybridization was performed according to the manufacturer’s instructions (Roche Molecular Biochemicals, Mannheim, Germany) and the published in situ hybridization protocol[17,18]. Sense and antisense probes were obtained by T3 or T7 in vitro transcription using a DIG RNA labeling kit from Roche (Mannheim, Germany). Rab23 (AY585189) was cloned into BamH1 of pBluescriptSK+. Tissue sections (8-μm thick) were mounted onto poly-L-lysine slides. Following deparaffinization, tissue sections were rehydrated in a series of ethanol dilutions. To enhance the signal and facilitate probe penetration, sections were immersed in 0.3% Triton X-100 solution for 15 min at room temperature and in proteinase K solution (20 μg/mL) for 20 min at 37°C, respectively. Sections were incubated with 4% (w/v) paraformaldehyde/phosphate-buffered saline (PBS) for 5 min at 4°C. After washed with PBS, slides were incubated with prehybridization solution (50% formamide, 50% 4 × SSC) for 2 h at 37°C. Probe was added to each tissue section at a concentration of 1 μg/mL and hybridized overnight at 42°C. After high stringency washing, the sections were incubated with an alkaline phosphatase-conjugated sheep anti-digoxigenin antibody, which catalyzes a color reaction with nitro-blue-tetrazolium (NBT)/5-bromo-4-chloro-3-indolyl phosphate (BCIP) substrate (Roche, Mannheim, Germany). A blue color indicated strong hybridization.
Hep-3B cells kindly donated by Professor Shi-Gang Xiong (Department of Pathology, Keck School of Medicine, University of Southern California, USA.), were cultured in Dulbecco’s minimal essential medium (DMEM; Gibco BRL, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (FBS; Bio-Whittaker, Walkersville, MD, USA), 100 IU/mL penicillin, and 100 μg/mL streptomycin. Cultures were incubated at 37°C in a humidified atmosphere containing 50 mL/L CO2, and the medium was changed twice a week. Cell viability was tested by trypan blue exclusion. All experiments were performed with Hep-3B cells from passages 6-15, which were exposed to serum-free culture medium containing 0.1% bovine serum albumin for 24 h.
Hep-3B cells were harvested by trypsinization and seeded 24 h prior to transfection, on 6-well plates at a density of 300 000 cells/well in 2 mL DMEM with 10% FBS. When the Hep-3B cells were at 90%-95% confluence, the culture medium was replaced with 1 mL of DMEM with or without 10% FBS. Following an overnight incubation, cells received 1 mL of complete medium and were incubated until 72 h post transfection.
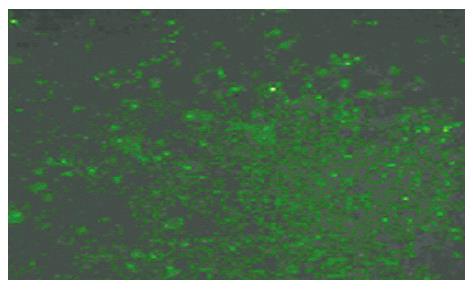
According to the design principle of siRNA[19-21], short interfering RNA for Rab23 and negative control siRNA were designed. The target gene sequence of Rab23 is 5`-3` CCA GAA CTA ACG CAT TCA TCA A. The siRNA sequence (siRNA Duplex) is CCA GAA CUA ACG CAU UCA A dT dA dAdT GGU CUU GAU UGC GUA AGU U. The sequence of negative control siRNA is: sense 5`-UUC UCC GAA CGU GUC ACG UTT-3`, Anti-sense 5`-ACG UGA CAC GUU CGG AGA ATT-3`. Short interfering RNAs (siRNAs) for Rab23 were purchased from Guangzhou Ribobio CO., Ltd (Guangzhou, China) and negative control siRNAs were purchased from Shanghai GenePharma Co., Ltd (Shanghai, China). Double-strand RNA (final concentration of 50 and 100 nmol/L) in OPTI-MEM (80 μL) and oligofectamine (4 μL) in OPTI-MEM (15 μL) were mixed and allowed to stand at room temperature for 20 min according to the manufacturer’s directions (Invitrogen, Carlsbad, USA). The mixture was added directly into a culture of Hep-3B cells (1 × 105/mL) in OPTI-MEM (0.4 mL). After 24 h, the consequences of transfection were watched through fluorescence of the cells (Figure 2). Transfection efficiency was normalized by monitoring luciferase activity from total cell lysates using the firefly luciferase (FLuc) assay kit (Promega, madison, USA). Cells were analyzed after 12 h by MTT assay. After 48 h they were analyzed by fluorescence-activated cell sorting, Western blots and RT-PCR . All experiments were performed in triplicate.
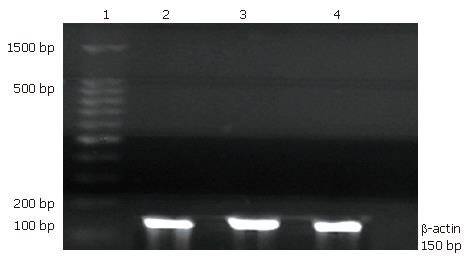
Two-step real-time RT-PCR was performed to compare the expression of Rab23 in normal liver tissue samples and Hep-3B cell lines. Total RNA was extracted using TRIzol reagent (Sigma) according to the manufacturer’s instructions. Reverse transcription with oligo (dT) priming was used to generate cDNAs from total RNA (2 μg) extracts. The synthesized cDNAs for Rab23 and β-actin were amplified using specific sets of primers. Optimal oligonucleotide primers were designed using Beacon Designer 2.0 software (MWG Biotech, High Point, NC). The primers used to amplify human Rab23 were synthesized according to the following sequences: forward primer: 5′ GTA GTA GCC GAA GTG GGA 3′, reverse primer: 5′ CCT TTG TTT GTT GGGTCT C 3′ according to ABO34244 in the gene bank. β-actin primer was designed based on published cDNA sequences: forward primer 5′TCA TCA CCA TTG GCA ATG AG3′, reverse primer 5′ CAC TGT GTT GGC GTA CAG GT 3′. Each PCR mixture contained the appropriate set of forward and reverse primers (0.2 μmol/L), each dNTP at 0.25 mmol/L, 1.25 U Taq polymerase, and 2.5 mmol/L MgCl2 in a PCR buffer. The PCR procedure consisted of 28 cycles of denaturation at 95°C for 1 min, annealing at 58°C for 1 min, and extension at 72°C for 1 min, with initial denaturation of sample cDNA at 95°C for 3 min and an additional extension period of 10 min after the last cycle. The PCR products were subjected to 1.5% agarose gel electrophoresis stained with ethidium bromide and quantitated by densitometry using an Image Master VDS system and associated software (Pharmacia, USA).
Cells were washed with cold PBS and lysed by the addition of a lysis buffer containing 1% Nonidet P-40, 50 mmol/L Tris (pH 7.5), 150 mmol/L NaCl, 0.1% SDS, and protease inhibitor cocktail (Boehringer Mannhein, Lewes, U.K.) for 20 min at 4°C. Insoluble materials were removed by centrifugation at 15 000 r/min for 15 min at 4°C. The supernatant was saved and the protein concentration was determined using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). Cell extracts (50 μg/lane) were separated via 10% gel electrophoresis and electroblotted onto PVDF membranes. Nonspecific binding sites were blocked by incubating nitrocellulose sheets for 1 h in phosphate-buffered saline containing 5% low-fat dry milk. Membranes were probed with primary antibodies overnight at 4°C, followed by a secondary horseradish peroxidase-conjugated second antibody. Blots were developed using an enhanced chemiluminescence detection system (ECL, Amersham Pharmacia Biotech) according to the manufacturer’s instructions.
The cell proliferation and viability were determined by MTT assay as previously described[18]. Briefly, cells (5 × 103 cells per well) were seeded in 96-well microtiter plates (Nunc, Denmark). After different treatments for 12 , 24 , 48 h or 72 h, an aliquot (50 μL) of MTT (Sigma) solution (2 mg/mL in PBS) was added to each well and the plates were incubated for an additional 4 h at 37°C. MTT solution in medium was aspirated off. To achieve solubilization of the formazan crystal formed in viable cells, 150 μL DMSO was added to each well. The absorbance was read at 490 nm on a Dias automatic microwell plate reader with DMSO as the blank.
After treatment for 48 h, cells were washed with cold sterile phosphate-buffered saline (PBS), and harvested in 70% cold alcohol solution for immediate analysis in a Becton Dickinson FACScan flow cytometer (Becton Dickinson, San Jose, CA). Data were acquired and analyzed using CellQuest software (BD Biosciences). The flow cytometer was calibrated daily with CaliBRITE 3 (BD biosciences) for fluorescence sensitivity and spectral overlap.
For immunohistochemistry and in situ hybridization results, Fisher’s exact test or a binomial proportion analysis was used. P < 0.05 was considered statistically significant. Results of RT-PCR, Western blotting and MTT assays were expressed as mean ± SE of at least three separate experiments. Results were analyzed by one-way analysis of variance (ANOVA) followed by the Student-Neumann-Keuls test. Again, differences with P values of < 0.05 were considered statistically significant.
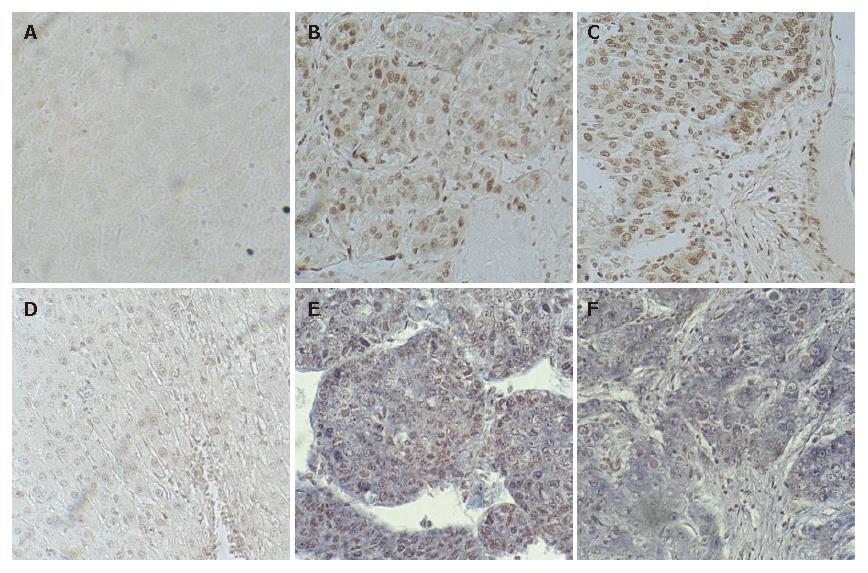
The expression of Rab23 in normal liver tissues has not been reported. Slight or no membranous staining of Rab23 in normal liver tissue was detected in our study. There was also no cytoplasmic or nuclear staining of Rab23 in normal liver tissues (Figure 3A). Conversely, expression of Rab23 was found in HCCs, and the expression rate was 53.5% (38 of 71 samples were positive, Table 1). Most samples expressed Rab23 in nuclei of HCC cells (Figure 3B and C). The results of expression of Rab23 correlated with tumor size (P < 0.01, Table 1), suggesting that Rab23 might be a useful prognostic indicator in HCC .
| In-situ hybridization | Iimmunohistochemistry | |||||
| Pos | Neg | P | Pos | Neg | P | |
| Normal | 0 | 5 | 0 | 5 | ||
| Paracarcinoma | 0 | 18 | 0 | 18 | ||
| HCC | 38 | 33 | 48 | 19 | ||
| Tumor | ||||||
| Differentiation | ||||||
| Well | 4 | 3 | 6 | 1 | ||
| Mod and poor | 34 | 30 | 0.3028 | 42 | 18 | 0.2681 |
| Metastasis | ||||||
| Yes | 9 | 5 | 10 | 4 | ||
| No | 29 | 28 | 0.2663 | 38 | 15 | 0.2591 |
| Tumor size | ||||||
| Small | 3 | 11 | 4 | 13 | ||
| Large | 35 | 22 | 0.0072 | 35 | 15 | 0.001 |
| Age | ||||||
| ≥ 50 yr | 14 | 10 | 15 | 9 | ||
| < 50 yr | 24 | 23 | 0.5613 | 33 | 10 | 0.2149 |
| Sex | ||||||
| Male | 29 | 26 | 44 | 15 | ||
| Female | 9 | 7 | 0.8036 | 4 | 4 | 0.1156 |
| HBsAg | ||||||
| Pos | 34 | 31 | 46 | 17 | ||
| Neg | 4 | 2 | 0.2721 | 2 | 2 | 0.2517 |
| AFP | ||||||
| ≥ 20 μg/L | 26 | 24 | 35 | 12 | ||
| < 20 μg/L | 12 | 9 | 0.6917 | 13 | 7 | 0.4314 |
No expression of Rab23 could be detected by in situ hybridization in five normal liver tissue samples (Figure 3D). To confirm this result, we also checked the transcripts by RT-PCR, and got the same result (data not shown).
To assess the frequency of Rab23 activation in HCC, we examined its expression in tissue samples derived from 100 different HCC cases (Table 1). We found positive staining for Rab23 in 38 of 71 (72%) HCC cases (Figure 3E and F). Further analyses again showed that activation of Rab23 correlated with tumor size (P < 0.01, Table 1).
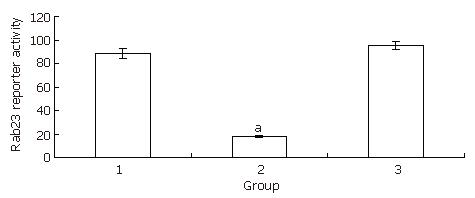
Hep-3B cells were divided into three groups: a transfected group, a blank control group, and a negative control group. After 24 h of treatment with siRNA, Hep-3B cells were harvested and total RNA was extracted. RT-PCR was done as described previously[18]. The expression levels in the transfected group decreased almost 4.5 fold compared with the blank control group. There was no statistical significance between the blank control group and the negative control group (Figure 4,Figure 5, Figure 6).
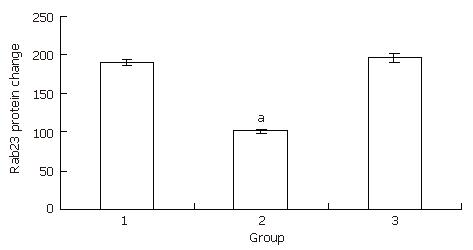
We used the same method for preparing Hep-3B cells for Western blots as for preparing cells for RT-PCR. After 24 h of treatment, Hep-3B cells were harvested and protein was extracted. Western blots were run according to a protocol described previously[22]. The expression levels in the transfected group decreased to almost half the level of the blank control group. There was no statistical significance between the blank control group and the negative control group (Figure 7, Figure 8).
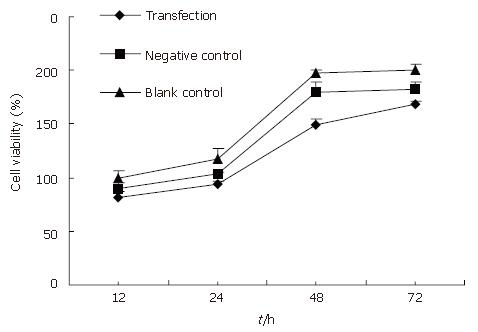
Here we determined whether inhibition of the expression of Rab23 in Hep-3B cells also inhibits the proliferation of these cells by MTT assay. Forty-eight hours and 72 h after adding siRNA, the survival rate of the transfected group decreased compared with controls (P < 0.05, Figure 9). Again, there was no significant difference between the blank and negative control groups.
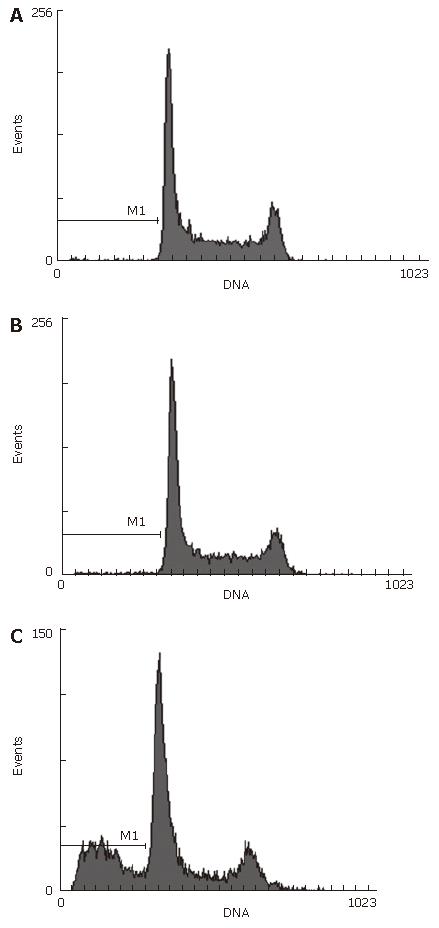
Forty-eight hours after siRNA treatment, Hep-3B cells were tested by fluorescence- activated cell sorting. Apoptosis skewness was seen in the transfected group. The apoptotic rate of Hep-3B was 30% in the blank control group but 0% in the negative control groups (Figure 10).
The worldwide incidence of liver cancer is expected to rise over the next decade. This is serious because the prevalence and mortality rate of HCC are rather high at present[23]. Furthermore, the mechanisms underlying the initiation and progression of HCC remain elusive. One candidate mechanism involves the hedgehog (Hh) pathway. Dysregulation of this pathway has been implicated in the genesis of several kinds of cancer derived from multiple tissue types[3], including HCC[8,9]. One aspect of Hh signaling is negative regulation of this pathway by Rab23[13,15]. Rab23 acts on upstream of Gli transcription factors in patterning neural cell types in the spinal cord. The primary target of Rab23 is the Gli2 activator. Rab23 and Gli3 repressor have additive effects on pattering. Analysis of the Gli3 protein suggests that Rab23 also has a role in promoting the expression of Gli3 repressor. Although the membrane proteins patched and smoothened can change subcellular location in response to SHH, analysis demonstrates that Rab23 does not work through either patched or smoothened. Instead, Rab23 appears to regulate subcellular localization of essential components of the hedgehog pathway that act both on downstream of smoothened and on upstream of Gli proteins[14].
There are no reports about studies of Rab23 in HCC or even in any other human tumors. Since Rab23 is a negative regulator of SHH which can induce malignant carcinoma, Rab23 may also contribute to tumorigenesis in HCC.
Our results identify one potential mechanism underlying hepatocarcinogenesis, namely dysregulation of Rab23 and SHH signaling. In this study, we found that the aberrant expression of Rab23 was a general event during the development of HCC, and that the expression of the Rab23 gene correlated with tumor size. These findings are strongly supported by our studies of Hep3B cells in which Rab23 was silenced by siRNA, which inhibited cell proliferation and increased apoptosis. Further studies are now needed to identify the mechanisms by which Rab23 expression might contribute to dysregulation of SHH signaling and HCC tumorigenesis. Rab23 plays an important role in tumorigenesis of HCC and other human tumors. Also, Rab23 may be a new biological target for prognosis and treatment of HCC.
The authors thank Dr. Ningning Wang, Department of Medical Statistics and Epidemics, School of Public Health, Sun Yat-Sen University, for her valuable advice on statistics.
S- Editor Liu Y L- Editor Wang XL E- Editor Ma WH
| 1. | Tien LT, Ito M, Nakao M, Niino D, Serik M, Nakashima M, Wen CY, Yatsuhashi H, Ishibashi H. Expression of beta-catenin in hepatocellular carcinoma. World J Gastroenterol. 2005;11:2398-2401. [PubMed] |
| 2. | Iimuro Y, Fujimoto J. Strategy of gene therapy for liver cirrohosis and hepatocellular carcinoma. J Hepatobiliary Pancreat Surg. 2003;10:45-47. [PubMed] |
| 3. | Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432:324-331. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 851] [Cited by in RCA: 845] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 4. | Clatworthy JP, Subramanian V. Stem cells and the regulation of proliferation, differentiation and patterning in the intestinal epithelium: emerging insights from gene expression patterns, transgenic and gene ablation studies. Mech Dev. 2001;101:3-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 78] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 5. | Ruiz i Altaba A, Sánchez P, Dahmane N. Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat Rev Cancer. 2002;2:361-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 557] [Cited by in RCA: 563] [Article Influence: 24.5] [Reference Citation Analysis (0)] |
| 6. | van den Brink GR, Bleuming SA, Hardwick JC, Schepman BL, Offerhaus GJ, Keller JJ, Nielsen C, Gaffield W, van Deventer SJ, Roberts DJ. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat Genet. 2004;36:277-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 273] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 7. | Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349-354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1040] [Cited by in RCA: 1035] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 8. | Sicklick JK, Li YX, Jayaraman A, Kannangai R, Qi Y, Vivekanandan P, Ludlow JW, Owzar K, Chen W, Torbenson MS. Dysregulation of the Hedgehog pathway in human hepatocarcinogenesis. Carcinogenesis. 2006;27:748-757. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 202] [Cited by in RCA: 215] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 9. | Huang S, He J, Zhang X, Bian Y, Yang L, Xie G, Zhang K, Tang W, Stelter AA, Wang Q. Activation of the hedgehog pathway in human hepatocellular carcinomas. Carcinogenesis. 2006;27:1334-1340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 184] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 10. | Chavrier P, Simons K, Zerial M. The complexity of the Rab and Rho GTP-binding protein subfamilies revealed by a PCR cloning approach. Gene. 1992;112:261-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 97] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 11. | Olkkonen VM, Peterson JR, Dupree P, Lütcke A, Zerial M, Simons K. Isolation of a mouse cDNA encoding Rab23, a small novel GTPase expressed predominantly in the brain. Gene. 1994;138:207-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 39] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 12. | Marcos I, Borrego S, Antiñolo G. Molecular cloning and characterization of human RAB23, a member of the group of Rab GTPases. Int J Mol Med. 2003;12:983-987. [PubMed] |
| 13. | Evans TM, Ferguson C, Wainwright BJ, Parton RG, Wicking C. Rab23, a negative regulator of hedgehog signaling, localizes to the plasma membrane and the endocytic pathway. Traffic. 2003;4:869-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 103] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 14. | Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194-198. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 294] [Cited by in RCA: 340] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 15. | Eggenschwiler JT, Bulgakov OV, Qin J, Li T, Anderson KV. Mouse Rab23 regulates hedgehog signaling from smoothened to Gli proteins. Dev Biol. 2006;290:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 117] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 16. | Packeisen J, Korsching E, Herbst H, Boecker W, Buerger H. Demystified...tissue microarray technology. Mol Pathol. 2003;56:198-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 90] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 17. | Ma X, Sheng T, Zhang Y, Zhang X, He J, Huang S, Chen K, Sultz J, Adegboyega PA, Zhang H. Hedgehog signaling is activated in subsets of esophageal cancers. Int J Cancer. 2006;118:139-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 119] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 18. | Ma X, Chen K, Huang S, Zhang X, Adegboyega PA, Evers BM, Zhang H, Xie J. Frequent activation of the hedgehog pathway in advanced gastric adenocarcinomas. Carcinogenesis. 2005;26:1698-1705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 145] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 19. | Amarzguioui M, Prydz H. An algorithm for selection of functional siRNA sequences. Biochem Biophys Res Commun. 2004;316:1050-1058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 270] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 20. | Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326-330. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1436] [Cited by in RCA: 1488] [Article Influence: 70.9] [Reference Citation Analysis (0)] |
| 21. | Ui-Tei K, Naito Y, Takahashi F, Haraguchi T, Ohki-Hamazaki H, Juni A, Ueda R, Saigo K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 536] [Cited by in RCA: 563] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 22. | Li W, Zhang J, Huang Q, Zhu H, Zhang X. Long-term administering low anticoagulant activity heparin can lessen rat hepatic fibrosis induced by either CCl(4) or porcine serum injection. Hepatol Res. 2006;36:115-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Marrero JA. Hepatocellular carcinoma. Curr Opin Gastroenterol. 2005;21:308-312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |