Published online Dec 28, 2007. doi: 10.3748/wjg.v13.i48.6581
Revised: November 9, 2007
Accepted: November 26, 2007
Published online: December 28, 2007
AIM: To observe the gene silencing mediated by the specific shRNA targeted against β-catenin and its effect on cell proliferation and cycle distribution in the human colon cancer cell line Colo205.
METHODS: Two shRNA plasmid vectors against β-catenin were constructed and transfected into Colo205 cells with LipofectamineTM2000. The down-regulations of β-catenin, c-myc and cyclinD1 expressions were detected by RT-PCR and western blot analysis. The cell proliferation inhibitions were determined by MTT assay and soft agar colony formation assay. The effect of these two β-catenin shRNAs on cell cycle distribution and apoptosis was examined by flow cytometry.
RESULTS: These two shRNA vectors targeted against β-catenin efficiently suppressed the expression of β-catenin and its down stream genes, c-myc and cyclinD1. The expression inhibition rates were around 40%-50% either at the mRNA or at the protein level. The shRNA-mediated gene silencing of β-catenin resulted in significant inhibition of cell growth both on the culture plates and in the soft agar. Moreover, the cancer cells showed significant G0/G1 arrest and increased apoptosis at 72 h post transfection due to gene silencing.
CONCLUSION: These specific shRNAs targeted against β-catenin could have a gene silencing effect and block the WNT signaling pathway. They could inhibit cell growth, increase apoptosis, and induce cell cycle arrest in Colo205 cells. ShRNA interference against β-catenin is of potential value in gene therapy of colon cancer.
- Citation: Huang WS, Wang JP, Wang T, Fang JY, Lan P, Ma JP. ShRNA-mediated gene silencing of β-catenin inhibits growth of human colon cancer cells. World J Gastroenterol 2007; 13(48): 6581-6587
- URL: https://www.wjgnet.com/1007-9327/full/v13/i48/6581.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i48.6581
Colon cancer is still a serious disease and an abnormal WNT signaling transduction pathway is considered to play an important role during its carcinogenic process. Mutations that activate the WNT pathway have been implicated in colon cancer by turning on genes encoding oncoproteins and cell-cycle regulators. In the absence of the WNT signal, β-catenin is localized in cell-cell junctions, but very low in the cytoplasm and nucleus because excessive β-catenin in cytoplasm is targeted to proteasome-mediated degradation by a complex containing GSK-3β, Axin and APC[1-4]. The binding of WNT proteins to their receptor, frizzled, stabilizes β-catenin by inhibiting the activity of the GSK-3β. Then β-catenin associates with members of the TCF/LEF family and migrates to the nucleus, where this complex activates the transcription of genes, such as c-myc and cyclinD1, which promote proliferation and regulate the cell cycle[5-7]. So, the key step in activating the WNT pathway is the stabilization of β-catenin and its translocation into the nucleus, which may serve as a potential target for colon cancer therapy.
RNA interference (RNAi) is a post-transcriptional gene-silencing mechanism, which was first discovered by Fire and his colleagues in 1998[8]. RNAi is often exploited for specific gene silencing, which is initiated by introduction into cells of double-stranded RNA (dsRNA) that are homologous to the sequence of the target gene. In order to avoid the nonspecific global shutdown of protein synthesis inside the cells and adapt to mammalian cells, the short duplexes of synthetic 21-23 nt RNAs were annealed and then introduced into mammalian cells. This was called small interfering RNA technique (siRNA)[9-11]. In this study, two shRNA plasmid vectors against β-catenin, which could persistently generate siRNA inside cells, were constructed and transfected into the colon cancer cell line Colo205. Its effect on cell proliferation and cycle distribution in this cell line was investigated.
The human colon cancer cell line Colo205, which was kindly provided by my team colleague, Dr Liyong, was cultured in DMEM (Gibco, USA), and supplemented with 10% FBS (Sigma, USA), 100 units/mL peni cillin G, and 100 μg/mL streptomycin (Gibco, USA) at 37°C in a humidified 5% CO2/95% air atmosphere.
Plasmid vector Pgenesil was purchased from Wuhan Genesil Biotechnology Co Ltd. Two different targeted sequences were designed to be homologous to the β-catenin mRNA consensus sequence (GeneBank NM_001904). A negative control sequence was also designed as the same process, which had no homology with human beings or mice. The complementary oligonucleotides encoded a hairpin structure with a 21-mer stem and a 9-bp loop. The stem was derived from the mRNA target site. The loop sequence separated the two complementary domains. All the sequences were transcribed with DNA polymerase III U6 promoter in plasmid Pgenesil. These two β-catenin plasmids were named Pgenesil-CAT1 and Pgenesil-CAT2 respectively. The negative control plasmid was named Pgenesil-Neg. (Table 1) The transformation of these plasmids into competent cells DH5α and extraction of plasmids followed the routine process.
| Name | Sequence of siRNA | Target sites |
| Pgenesil-CAT1 | AACAGTCTTACCTGGACTCTG | 0290-0310 |
| Pgenesil-CAT2 | AAAGGCAATCCTGAGGAAGAG | 0355-0375 |
| Pgenesil-Neg | GACTTCATAAGGCGCATGC | No homology |
Approximately, 1 × 106 cells/well were plated in 6-well plates in medium containing 10% FBS to grow overnight to 80%-90% confluency. Transfection of the shRNA oligonucleotides was performed by using LipofectamineTM2000 (Invitrogen, USA). Colo205 cells were divided into blank control group, negative control group and two test groups (CAT1 and CAT2). Only LipofectamineTM2000 was used for transfection in the blank control group. Plasmid Pgenesil-Neg was used for transfection in the negative control group. Plasmids Pgenesil-CAT1 and Pgenesil-CAT2 were used for transfection in the test groups (CAT1 and CAT2) respectively. In the above three groups, the cells were all transfected with the mixture of plasmid and Lipofectamine TM2000 (1:3) in 2 mL serum-free medium. At 6 h post-transfection, 500 μL FBS/well was added. At 26 h after transfection, the medium was replaced by normal medium containing 10% FBS and antibiotics up to 72 h post-transfection.
At forty-eight hours post-transfection, 1 × 106 cells were collected and total RNA was extracted, using the Trizol reagent (Invitrogen, USA) under its operational instruction. The concentration and purity of the total RNA were detected with UV spectrophotometer analysis at 260 nm and the electrophoresis detection showed good quality of purified RNA. RT-PCR was performed by the two-step method. Synthesis of cDNA was performed by using the cDNA synthesis kit (Fermentas, USA) and following the manufacturer’s instructions. For quantitative analysis of β-catenin, c-myc and cyclinD1 mRNA, the expression of house keeping gene, β-actin mRNA, was used as an internal standard. Primer pairs were designed accordin g to the sequences in the GenBank (Table 2).
| Goal gene | Upstream primer | Downstream primer | PCR frag(bp) |
| β-actin | 5'-TCCTGTGGATCCACGAAACT-3' | 5'-GAAGCATTTGCGGTGGACGAT-3' | 330 |
| β-catenin | 5'-AGATGCAGCAACTAAACAGGA-3' | 5'-GTACTACATTTTAAGCCATCT-3' | 290 |
| C-myc | 5'-TACCCTCTCAACGACAGCAG-3' | 5'-TCTTGACA TTCTCCTCGGTG-3' | 477 |
| CyclinD1 | 5'-GCCAACCTCCTCAACGACCGG-3' | 5'-GTCCATGTTCTGCTGGGCCTG-3' | 744 |
The PCR fragments were separated and visualized in 1.5% agarose gels stained with ethidium bromide. Semiquantitative analysis was performed with the Bio-1D gel analysis software. All experiments were done in triplicate. The ratio of the photodensity of the RT-PCR product of goal gene and β-actin was used to identify the expression intensity of goal gene. The inhibition rate of goal gene expression was calculated with the following formula: inhibition rate of goal gene expression (IR) = (1 - E1/E2) × 100%. E1: the expression intensity of goal gene in the observation group; E2: the expression intensity of goal gene in the blank control group.
The cells were divided and transfected as above for 48 h. After collection from the culture medium, the cells were washed three times with PBS and lysed in 150 μL of ice-cold Tris buffer (50 mmol/L, pH 7.5). The Tris buffer contains 5 mmol/L edetic acid, 150 mmol/L NaCl, 0.1% NP-40, 0.1% SDS, 2.0 g/L aprotinin, 0.02% NaN3, 0.2 mmol/L PMSF, and antiprotease mixture. Then the cells were sonicated, and centrifuged at 12 000 ×g for 15 min. The Coomassie brilliant blue G-250 was used to determine the concentration of protein in each lysate. Loading buffer was added to each lysate, which was subsequently boiled for 3 min and then electrophoresed by SDS-PAGE. The proteins were mixed with 2 × loading buffer with the same volume before electrophoresis. After transferring onto nitrocellulose, proteins were incubated with antibodies (β-catenin, c-myc and cyclinD1, purchased from Santa Cruz, USA) and then with peroxidase-conjugated secondary antibody (Bolster, China). Detection was performed with an enhanced chemiluminescent agent. Signals were detected with the ECL Test Kit (Maixin, China). The β-actin staining served as the internal standard for all membranes. The inhibition rate calculating formula is the same as that described in the RT-PCR section. All experiments were done in triplicate.
The transfected cells were seeded in 96-well plates (1 × 104/mL) and allowed to attach for about 24 h. The MTT (Sigma, USA) was dissolved in PBS at a concentration of 5 mg/mL and filtered. Ten microlitres of stock solution were added to 100 microlitres of medium in each well. Then, the plates were incubated for 4 h at 37°C. After loading of MTT, the medium was replaced with 100 μL DMSO and left for 30 min at room temperature for colour development. The 96-well plates were read by an enzyme-linked immunosorbent assay reader (570 nm, DG-3022A, USA) to determine absorbance values (A). The absorbance values were determined at 36, 48, 60, 72 h after transfection in each group respectively. The experiments were repeated three times. The Inhibition rate was calculated by the following formula. Inhibition rate (IR) = [1 - A1/A2] × 100% A1: absorbance value of the observation group A2: absorbance value of the control group.
At 24 h after transfection as above, the cells of different groups were mixed with the above mentioned culture media containing 0.4% agar. Then 1000 cells were resuspended and seeded in triplicate into 6-well plates coated with 0.5% agar in DMEM containing 10% FBS. After 10 d incubation, the colonies were counted. Colony efficiency (CE) was determined by counting the number of colonies greater than 60 μm in diameter at × 100 magnification in each plate.
Cells of each group seeded in culture plates were transfected as described above. After incubation for 72 h, cells were harvested by trypsinization and centrifugation, washed with cold PBS and fixed overnight with 80% ethanol at -20°C. The fixed cells were collected, washed and resuspended in PBS containing 10 μg/mL propidium iodide (Sigma, USA) and 100 μg/mL RNase A (Takara, Dalian, China), then incubated at 4°C for at least 30 min avoiding light to eliminate the intracellular RNA. Subsequent analyses were performed by flow cytometry (Becton Dickinson, USA). The experiments were repeated three times.
The results were expressed as mean ± SD. The data were treated by one-way analysis of variance and Student’s t-test to determine statistical significance with SPSS 11.5 statistic software. P < 0.05 was considered statistically significant.
The multiclone sites of plasmid Pgenesil were as follows: HindIII-ShRNA-BamHI-U6Promotor-EcoRI-SalI-XbaI-DraIII.
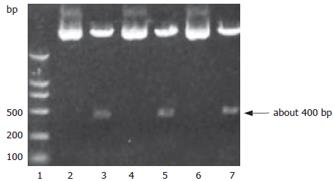
A SalI site for plasmid Pgenesil was designed in the inserted fragments between the sites of BamHI and HindIII. If the insertion was correct, a band about 400 bp should be cut off by SalI. The results of digestion with restriction endonucleases and sequencing showed correct plasmids (Figure 1).
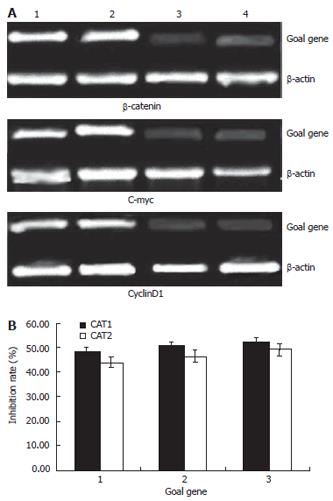
The mRNA expression intensities of goal genes, inhibited by specific shRNAs in the colon cancer cells, were analyzed by semiquantitive RT-PCR. The mRNA levels were normalized by internal control β-actin (Figure 2). At forty-eight hours post-transfection, the expression intensities of β-catenin mRNAs in the blank control, negative control and test groups (CAT1, CAT2) were 0.98 ± 0.02, 0.96 ± 0.03, 0.51 ± 0.03 and 0.55 ± 0.01, respectively. The other genes’ mRNA expression intensities were shown in the table (Table 3). The statistical analysis showed that β-catenin mRNAs of Colo205 cells in the CAT1 and CAT2 groups were down regulated significantly after transfection with either plasmids Pgenesil-CAT1 or Pgenesil-CAT2, compared with that in the blank group (P < 0.05). The inhibition rates were 47.89% and 43.91% in the CAT1 and CAT2 group, respectively (Figure 2). But there is no significant difference between these two groups. The plasmid Pgenesil-Neg had no significant inhibitive effect on the expression of β-catenin mRNA (P > 0.05, vs blank) (Table 3). The statistical results of gene c-myc and cyclinD1 were similar to that of β-catenin. (Table 3).
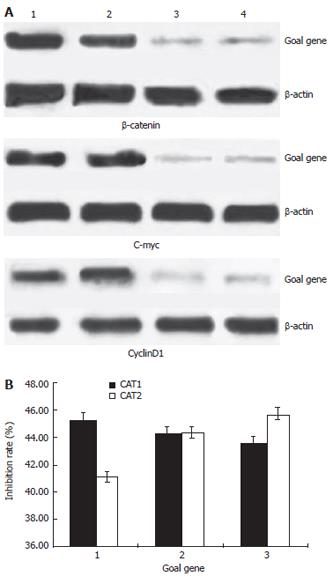
The down-regulated efficiencies of protein expressions inhibited by specific shRNAs in Colo205 cells were analyzed by western blot. The protein levels were normalized by internal control β-actin. (Figure 3) At forty-eight hours post-transfection, all the protein expression ratios (protein vsβ-actin) were calculated and shown in Figure 3 and Table 4. For example, the ratio of β-catenin /β-actin were 0.95 ± 0.02, 0.89 ± 0.04, 0.52 ± 0.02 and 0.56 ± 0.03, in the blank control, negative control and test groups (CAT1,CAT2) respectively. The statistical analysis showed that both the plasmid Pgenesil-CAT1 and Pgenesil-CAT2 could have a significant down-regulation effect on the protein expression of β-catenin in Colo205 cells (P < 0.05, vs blank). But there is no significant difference between these two groups. The similar statistical results could be detected on the protein expression of gene c-myc and cyclinD1 (P < 0.05, vs blank). The inhibition rates were around 40%-50%. Reasonably, the plasmid Pgenesil-Neg had no inhibitive effect on any gene of β-catenin, c-myc and cyclinD1 (P > 0.05, vs blank).
The proliferation of Colo205 cells inhibited by specific shRNAs was analyzed by the MTT assay. The results were shown in Table 5. The statistical analysis showed that both the plasmids Pgenesil-CAT1 and Pgenesil-CAT2 could have significant inhibitive effects on the proliferation of Colo205 cells, compared with the blank group at 72 h after transfection (P < 0.05, vs Blank). But the plasmids Pgenesil-Neg had no significant inhibitive effect on cell proliferation (P > 0.05, vs Blank).
The anchorage-independent proliferation of Colo205 cells inhibited by specific shRNAs was analyzed by soft agar colony formation assays. The average numbers of colonies in the blank, negative, CAT1, CAT2 group were 46, 43, 9 and 11 respectively. The Colo205 cells in the CAT1 and CAT2 groups formed significantly less colonies (4-5 fold decrease) in soft agar than the cells in the blank and negative control groups did (P < 0.05, vs Blank). At the same time we observed that most of the colonies in the CAT1 and CAT2 groups were much smaller than those in the blank and negative control groups. All these results suggested that reductions in β-catenin protein level induced by the β-catenin gene silencing decreased the abilities of Colo205 cells to form colonies in soft agar.
In comparison to the blank control group, the CAT1 and CAT2 groups both showed cell accumulation in the G0/G1 phase and reduction in the phases of S and G2/M. Moreover, the cells with sub- G1 DNA content also increased and apoptosis rates were around 5% in these two groups. No statistically significant difference was found between the negative control group and the blank control group (Table 6 ).
RNA interference is a ubiquitous mechanism of eukaryotic gene regulation and an excellent strategy for specific gene silencing. The specificity of RNAi is determined by 21-23 nt RNA duplexes, referred to as micro-RNA (miRNA) or small interfering RNAs (siRNA)[12]. ShRNA is formed by hairpin structures and stretches of double-stranded RNA, which will be cleaved by the ribonuclease dicer to produce mature miRNA inside the targeted cells. After unwinding, one of the strands becomes incorporated into the RNA-induced silencing complex (RISC) and guides the destruction or repression of complementary mRNA[13]. Recently the vector-based approach of shRNA interference has been developed in order to achieve stable, long-term, and highly specific suppression of gene expression in mammalian cells[14]. These shRNA expression vectors have many advantages: they can be stably introduced into cells and persistently effective, either as selectable plasmids or as retroviruses. They are relatively cheap to generate[15,16]. These vectors are often under the control of an RNA polymerase III promoter such as U6 or H1[17]. They can transcribe and generate siRNA continuously and the gene silencing effect can last persistently inside the cells. These findings have opened a broad new avenue for the analysis of gene function and gene therapy[18].
The coordination of cell proliferation and apoptosis in cells is one of the hot spots in cancer research. The balance between cell proliferation and apoptosis, and the distribution of cell cycles play very important roles in the carcinogenesis process of colon cancer. The WNT/β-catenin signaling transduction pathway is a highly conservative pathway in the development of colon cancer, which regulates intestinal epithelial proliferation and patterning[19,20]. Mutations that activate the WNT signaling pathway cause the hyper-proliferation of intestinal crypt progenitors and a series of consequential changes finally lead to the occurrence of colon cancer[21,22]. β-catenin is the key component in this pathway. The increased levels of β-catenin frequently found in both premalignant and malignant cells are associated with increased rates of cellular proliferation[23]. Moreover, modest over-expression of β-catenin in epithelial cells can lead to increased proliferation and result in transformation[24-26]. It has been reported that the excessive expression of stabilized β-catenin leads to tumorigenesis in the colorectum, central nervous system, skin and other tissues[27-30]. So, β-catenin has attracted much attention as gene therapy target for carcinomas, including colon cancer. β-catenin has been successfully down-regulated by RNAi in some previous study. For example, Udit N Verma et al synthesized siRNA and interfere with the β-catenin gene in the cell line SW480 and HCT116[31]. However, the down-regulation of β-catenin expression by RNAi in the colon cancer Colo205 cell line has not been reported before. Furthermore their research used synthesized small interference RNA as a gene silencing tool, which just transiently suppressed the gene expression and was often limited to cells that are easily transfected[17]. In our study, we did differently by using the vector-based RNA interference technique. According to the design of the shRNA, we selected two shRNA sequences from the β-catenin (CTNNB1) gene and used the plasmid Pgenesil as the vector. The plasmid Pgenesil vector contains the EGFP gene that makes it convenient to observe the result of transfection. When successfully transfected into the colon cancer cells, the vector Pgenesil could transcribe and generate the interfering RNAs continually under the control of the U6 promotor. So the concentration of intracellular interfering RNA oligonucleotides can remain stable long after transfection. That is what we thought to be the mechanism for the persistently inhibitive effect of the vector-based interference technique. This is the major advantage against the common siRNA technique.
In this study, these two β-catenin specific shRNAs showed their evident effect on silencing the β-catenin gene. Our results demonstrated that after transfection with the β-catenin specific shRNA vectors, β-catenin mRNA and protein expression were significantly suppressed, detected by RT-PCR and western blot. At the same time, we could observe that the genes c-myc and cyclinD were also significantly depressed both at the mRNA and protein levels due to the β-catenin gene silencing. This result confirmed that the c-myc and cyclinD1 genes are the downstream genes regulated by β-catenin. Gene silencing of β-catenin can sequentially lead to the silence of c-myc and cyclinD. Furthermore, no significant differences of β-catenin mRNA or protein expression were observed between these two vectors (CAT1 and CAT2). On the other hand, the negative control plasmid had no inhibitory effect on β-catenin mRNA and protein expression. Neither did it in the genes c-myc and cyclinD1. These results together confirmed that the inhibitions of β-catenin and its downstream genes were specifically induced by these shRNAs aimed at β-catenin. The MTT assays demonstrated that the proliferation curves of the cells were similar in the blank and negative control groups. Viability of cells transfected with the negative control vectors did not significantly decline compared with that of the blank control group. Thus the vector plasmid Pgenesil could be used as a safe and non-toxic carrier for shRNA interference. However, cell proliferation in test groups, which were transfected with β-catenin specific shRNAs, showed increasingly slow down in relation to the time after transfection. At 72 h post-transfection, the inhibitive rates of cell proliferation were 54% and 47% in the CAT1 and CAT2 group respectively, which have significant differences compared with that of the blank control group. As for the anchorage-independent proliferation, we could also detect the significant reduction of colonies in either amounts or sizes in soft agar in the test groups. The MTT and soft agar assay together revealed that these two β-catenin specific shRNAs possessed the anti-proliferation function. Our flow cytometry results indicated that the cell growth inhibition by β-catenin shRNA was due to the cell cycle G0/G1 arrest and induction of apoptosis. This was somewhat controversial to Udit N Verma’s report, which implied that the decreased cell proliferation did not depend on the mechanism of increased apoptosis[31]. We think that the controversy may be due to different cell lines. In our opinion, we think that the imbalance between cell proliferation and apoptosis induced by the β-catenin shRNA could be possibly explained by mechanisms of β-catenin gene silencing and its consequential downstream genes silencing, such as c-myc and cyclinD1, which regulate the cell cycle transition checkpoints and cellular proliferations. As we know, c-myc is a positive regulator of G1-specific cyclin-dependent kinases (CDKs). Even in quiescent cells, c-myc activation is sufficient to induce cell cycle entry in the absence of growth factors[32,33]. CyclinD1 is a regulatory kinase that is a critical modulator of progression through the G1 to S phase transition of the cell cycle. When quiescent cells re-enter the cell cycle and divide, cyclinD1 is the first cyclin to be activated[34,35]. Overexpression of these two genes may stimulate the cells to overcome the cell cycle checkpoints and enhance cell proliferation[36]. Inhibitions of these two genes may result in cell cycle arrest and increased apoptosis, thus inhibiting cell growth. But whether this mechanism is of universality in other colon cancer cell lines or even malignant tumors, further detailed investigations need to be carried out.
In conclusion, our experiments showed that the shRNA targeted against β-catenin could specifically mediate the β-catenin gene silencing and consequentially suppress the expression of its downstream genes, c-myc and cyclinD1. These genes’ silencing effect could efficiently inhibit the growth of colon cancer Colo205 cells. The shRNA interference targeted against β-catenin may have potential therapeutic utility in human colon cancer.
We would like to thank my team colleague, Dr Liyong, for the generous gift of Colo205 cells. We are grateful for Dr Jia-Wei Liao for his technical support.
Abnormal WNT signaling transduction pathway is considered to play an important role during the carcinogenesis process. The key step in activating WNT pathway is the stabilization of β-catenin and its translocation into nucleus, which may serve as a potential target for colon cancer therapy. RNAi is often exploited for specific gene silencing, which provides a strategy for cancer gene therapy. Therefore, we would do some basic research for colon cancer therapy by using this technique.
The coordination of cell proliferation and apoptosis in cells is one of the hotspots in cancer research. The balance between cell proliferation and apoptosis, and the distribution of cell cycles play very important roles in the carcinogenesis process of colon cancer. β-catenin is the key component in this process. Thus β-catenin has attracted much attention as gene therapy target for colon cancer.
In this study, two β-catenin specific shRNAs showed their evident effect on silencing the β-catenin gene. The results also indicated that the cell growth inhibition by β-catenin shRNA was due to the cell cycle G0/G1 arrest and induction of apoptosis, although this was somewhat different from other’s report.
The shRNA interference targeted against β-catenin may have potential therapeutic utility in human colon cancer.
ShRNA: ShRNA is the short form of “short hairpin RNA”. It is formed by hairpin structures and stretches of double-stranded RNA, which will be cleaved by the ribonuclease dicer to produce mature miRNA inside the targeted cells.
This paper investigated the gene silencing mediated by the specific shRNA targeted against β-catenin and its effect on cell proliferation and cycle distribution in human colon cancer cell line Colo205. The authors conclude that The specific shRNAs targeted against β-catenin could have gene silencing effect and block the WNT signaling pathway. They could inhibit cell growth, increase apoptosis, and induce cell cycle arrest in Colo205 cells. ShRNA interference against β-catenin is of potential value in gene therapy of colon cancer.
S- Editor Liu Y L- Editor Alpini GD E- Editor Li HY
| 1. | Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis--a look outside the nucleus. Science. 2000;287:1606-1609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 995] [Cited by in RCA: 989] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 2. | Pennisi E. How a growth control path takes a wrong turn to cancer. Science. 1998;281:1438-1439, 1441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 3. | Polakis P. The many ways of Wnt in cancer. Curr Opin Genet Dev. 2007;17:45-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 664] [Cited by in RCA: 694] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 4. | Polakis P. The oncogenic activation of beta-catenin. Curr Opin Genet Dev. 1999;9:15-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 517] [Cited by in RCA: 520] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 5. | Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023-1026. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1090] [Cited by in RCA: 1097] [Article Influence: 37.8] [Reference Citation Analysis (0)] |
| 6. | Nakamura T, Hamada F, Ishidate T, Anai K, Kawahara K, Toyoshima K, Akiyama T. Axin, an inhibitor of the Wnt signalling pathway, interacts with beta-catenin, GSK-3beta and APC and reduces the beta-catenin level. Genes Cells. 1998;3:395-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 224] [Cited by in RCA: 228] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 7. | Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787-1790. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2986] [Cited by in RCA: 3101] [Article Influence: 110.8] [Reference Citation Analysis (0)] |
| 8. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10522] [Cited by in RCA: 10146] [Article Influence: 375.8] [Reference Citation Analysis (1)] |
| 9. | Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877-6888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1037] [Cited by in RCA: 1033] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 10. | Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA. 2002;99:6047-6052. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 770] [Cited by in RCA: 776] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 11. | Guan HT, Xue XH, Dai ZJ, Wang XJ, Li A, Qin ZY. Down-regulation of survivin expression by small interfering RNA induces pancreatic cancer cell apoptosis and enhances its radiosensitivity. World J Gastroenterol. 2006;12:2901-2907. [PubMed] |
| 12. | Liu F, He CW, Zhang YF, Zhou KY. RNA interference by expression of short hairpin RNAs suppresses bcl-xL gene expression in nasopharyngeal carcinoma cells. Acta Pharmacol Sin. 2005;26:228-234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 13. | Kisielow M, Kleiner S, Nagasawa M, Faisal A, Nagamine Y. Isoform-specific knockdown and expression of adaptor protein ShcA using small interfering RNA. Biochem J. 2002;363:1-5. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 44] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 14. | Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA. 2002;99:5515-5520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 859] [Cited by in RCA: 883] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 15. | McManus MT, Sharp PA. Gene silencing in mammals by small interfering RNAs. Nat Rev Genet. 2002;3:737-747. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1044] [Cited by in RCA: 991] [Article Influence: 43.1] [Reference Citation Analysis (0)] |
| 16. | Shi Y. Mammalian RNAi for the masses. Trends Genet. 2003;19:9-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 204] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 17. | Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3' overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20:497-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 533] [Cited by in RCA: 526] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 18. | Czauderna F, Fechtner M, Aygün H, Arnold W, Klippel A, Giese K, Kaufmann J. Functional studies of the PI(3)-kinase signalling pathway employing synthetic and expressed siRNA. Nucleic Acids Res. 2003;31:670-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 74] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 19. | Cong F, Schweizer L, Chamorro M, Varmus H. Requirement for a nuclear function of beta-catenin in Wnt signaling. Mol Cell Biol. 2003;23:8462-8470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 57] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 20. | Coluccia AM, Benati D, Dekhil H, De Filippo A, Lan C, Gambacorti-Passerini C. SKI-606 decreases growth and motility of colorectal cancer cells by preventing pp60(c-Src)-dependent tyrosine phosphorylation of beta-catenin and its nuclear signaling. Cancer Res. 2006;66:2279-2286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 83] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 21. | Moser AR, Dove WF, Roth KA, Gordon JI. The Min (multiple intestinal neoplasia) mutation: its effect on gut epithelial cell differentiation and interaction with a modifier system. J Cell Biol. 1992;116:1517-1526. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 215] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 22. | Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359:235-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1308] [Cited by in RCA: 1301] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 24. | Morin PJ, Vogelstein B, Kinzler KW. Apoptosis and APC in colorectal tumorigenesis. Proc Natl Acad Sci USA. 1996;93:7950-7954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 358] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 25. | Orford K, Orford CC, Byers SW. Exogenous expression of beta-catenin regulates contact inhibition, anchorage-independent growth, anoikis, and radiation-induced cell cycle arrest. J Cell Biol. 1999;146:855-868. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 211] [Cited by in RCA: 215] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 26. | Zhu AJ, Watt FM. beta-catenin signalling modulates proliferative potential of human epidermal keratinocytes independently of intercellular adhesion. Development. 1999;126:2285-2298. [PubMed] |
| 27. | Zurawel RH, Chiappa SA, Allen C, Raffel C. Sporadic medulloblastomas contain oncogenic beta-catenin mutations. Cancer Res. 1998;58:896-899. [PubMed] |
| 28. | Gat U, DasGupta R, Degenstein L, Fuchs E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95:605-614. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 887] [Cited by in RCA: 855] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 29. | Chan EF, Gat U, McNiff JM, Fuchs E. A common human skin tumour is caused by activating mutations in beta-catenin. Nat Genet. 1999;21:410-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 468] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 30. | van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, van der Horn K, Batlle E, Coudreuse D, Haramis AP. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell. 2002;111:241-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1605] [Cited by in RCA: 1615] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 31. | Verma UN, Surabhi RM, Schmaltieg A, Becerra C, Gaynor RB. Small interfering RNAs directed against beta-catenin inhibit the in vitro and in vivo growth of colon cancer cells. Clin Cancer Res. 2003;9:1291-1300. [PubMed] |
| 32. | Niu ZS, Li BK, Wang M. Expression of p53 and C-myc genes and its clinical relevance in the hepatocellular carcinomatous and pericarcinomatous tissues. World J Gastroenterol. 2002;8:822-826. [PubMed] |
| 33. | Rochlitz CF, Herrmann R, de Kant E. Overexpression and amplification of c-myc during progression of human colorectal cancer. Oncology. 1996;53:448-454. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 34. | Takahashi Y, Kawate S, Watanabe M, Fukushima J, Mori S, Fukusato T. Amplification of c-myc and cyclin D1 genes in primary and metastatic carcinomas of the liver. Pathol Int. 2007;57:437-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 35. | Sato Y, Itoh F, Hareyama M, Satoh M, Hinoda Y, Seto M, Ueda R, Imai K. Association of cyclin D1 expression with factors correlated with tumor progression in human hepatocellular carcinoma. J Gastroenterol. 1999;34:486-493. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 36. | Shiina H, Igawa M, Shigeno K, Terashima M, Deguchi M, Yamanaka M, Ribeiro-Filho L, Kane CJ, Dahiya R. Beta-catenin mutations correlate with over expression of C-myc and cyclin D1 Genes in bladder cancer. J Urol. 2002;168:2220-2226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 40] [Article Influence: 1.7] [Reference Citation Analysis (0)] |