Published online Dec 21, 2007. doi: 10.3748/wjg.v13.i47.6365
Revised: September 11, 2007
Accepted: November 29, 2007
Published online: December 21, 2007
AIM: To investigate the expression of coxsackievirus and adenovirus receptor (CAR) and adenovirus-mediated reporter gene transfer in five human colon cancer cell lines.
METHODS: Expression of CAR-specific mRNA and protein was analyzed by reverse transcriptase polymerase chain reaction and Western blotting, respectively. Adenovirus-based gene delivery was evaluated by infection of cells with adenoviral vector carrying the green fluorescent protein (GFP) gene.
RESULTS: All the colon cancer cell lines examined (HT29, LS180, SW480, SW948 and SW1116) expressed CAR full-length mRNA and an alternatively-spliced variant that lacks the transmembrane coding exon. All cell lines were detected as CAR-positive by Western blot analysis. Further, all cells we examined were efficiently infected with adenoviral vector-GFP.
CONCLUSION: The data indicated that the five colon cancer cell lines tested expressed adenovirus primary receptor and could be efficiently infected by adenoviral vectors. Therefore, these cell lines will be useful for adenovirus-based gene transfer and research.
- Citation: Abdolazimi Y, Mojarrad M, Pedram M, Modarressi MH. Analysis of the expression of coxsackievirus and adenovirus receptor in five colon cancer cell lines. World J Gastroenterol 2007; 13(47): 6365-6369
- URL: https://www.wjgnet.com/1007-9327/full/v13/i47/6365.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i47.6365
Colorectal cancer is the third most common cancer and among the three top fatal malignancies. In 2007, there were an estimated 153760 new cases and 52180 deaths from colorectal cancer, which corresponds to approximately 10% of all newly diagnosed cancers in the United States[1,2]. Gene therapy has been considered a potential innovative approach for the treatment of colorectal cancer[3,4]. To date, over 600 gene therapy clinical trials have been initiated in the US, 60% of which are cancer-related[5]. Currently, adenoviral vectors are the most frequently administered vectors[6]. This is because of their advantageous features such as a broad range of cell targets, the ability to be produced in high titers, and their accommodation of relatively large segments of DNA[7,8].
Adenovirus initiates infection as well as does adenovector-mediated gene transfer by attachment of the fiber knob to a cell surface receptor, the coxsackievirus and adenovirus receptor (CAR)[9]. CAR, the primary high-affinity receptor for adenovirus, is a 46-ku transmembrane glycoprotein and belongs to the immunoglobulin superfamily[10,11]. The human CAR gene, CXADR, encodes a 365-amino acid protein, and four pseudogenes have been reported on chromosomes 15, 18 and 21 (two copies)[12]. Alternatively spliced variants of CAR have also been identified[13]. Thoelen et al has described three splice variants (β-, δ- and δ-transcripts) of the CAR gene in addition to the normal α-transcript.
Expression of CAR has been studied in numerous cell lines including those of head and neck carcinoma[14], renal cell carcinoma[15], bladder cancer[16], ovarian cancer[17,18], cervical cancer[19], lung and pancreatic cancer[20], prostate cancer[21] and glioma[22]. In all these studies, high expression of CAR correlated with increased adenoviral infection efficiency; cells lacking or expressing low levels of CAR were resistant to adenovirus infection.
Since CAR expression has not been studied in colon cancer cell lines, we examined the expression of CAR at the mRNA and protein levels, along with its splice variants in five human colon cancer cell lines. Adenovirus-mediated reporter gene transfer was also evaluated.
Five human colon cancer cell lines HT29, SW480, SW948, SW1116 and LS180, as well as the human embryonic kidney (HEK) cell line (CAR-positive) and Chinese hamster ovary (CHO) cells (CAR-negative) (ATCC numbers: HTB-38, CCL-228, CCL-237, CCL-233, CL-187, CRL-1573, CCL-61) were obtained from the National Cell Bank of Iran (NCBI). Cells were cultured in RPMI 1640, Dulbecco’s Modified Eagle’s Medium, Minimum Essential Medium, or L-15 medium. All cells were maintained with 100 U/mL penicillin, 100 μg/mL streptomycin and 100 mL/L fetal bovine serum (FBS). Cell lines were cultured either in 10-cm culture plates or in flasks at 37°C and in 50 mL/L CO2 in a humidified incubator.
Total cellular RNA was extracted from each cell line by Biozol reagent (Bioflux, Japan). Two micrograms of total RNA were converted to cDNA by reverse transcription, using Moloney murine leukemia virus reverse transcriptase (Fermentas, Lithuania) and random primers (Roche, Germany) as described previously[23], with minor modifications. Since pseudogenes have been identified for the CAR gene, the CAR mRNA sequence was aligned with the pseudogenes (multiple alignment at http://bioinfo.genopole-toulouse.prd.fr/multalin/multalin.htm). CAR primers (CARf, 5'-CGTGCTCCTGTGCGGAGTAGT-3'; CARr, 5'-GACCCATCCTTGCTCTGTGCT-3') were designed in exons 1 and 7 with intervening introns of approximately 51 Kb, in a way that would align only with CAR mRNA and mismatched the pseudogenes in at least one nucleotide at the 3'-ends. Phosphoglucomutase-1 (PGM-1), human house keeping gene primers (PGMf, 5'-TCCGACTGAGCGGCACTGGGAGTGC-3'; PGMr, 5'-GCCCGCAGGTCCTCTTTCCCTCACA-3') or murine β-actin house keeping gene primers (actinf, 5'-GAACCCTAAGGCCAACCGTGA-3'; actinr, 5'-AGGAAGAGGATGCGGCAGTGG-3') were used as internal controls.
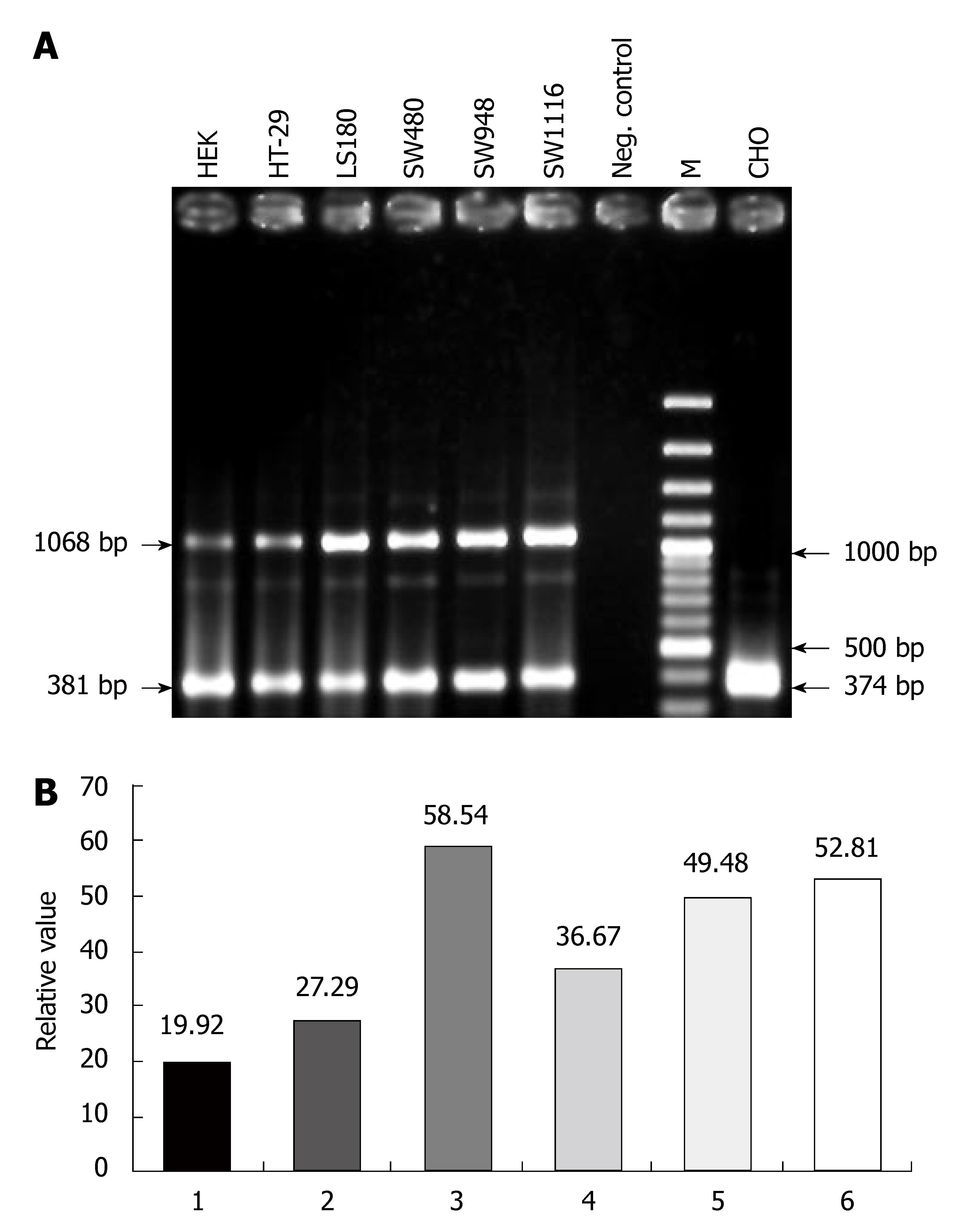
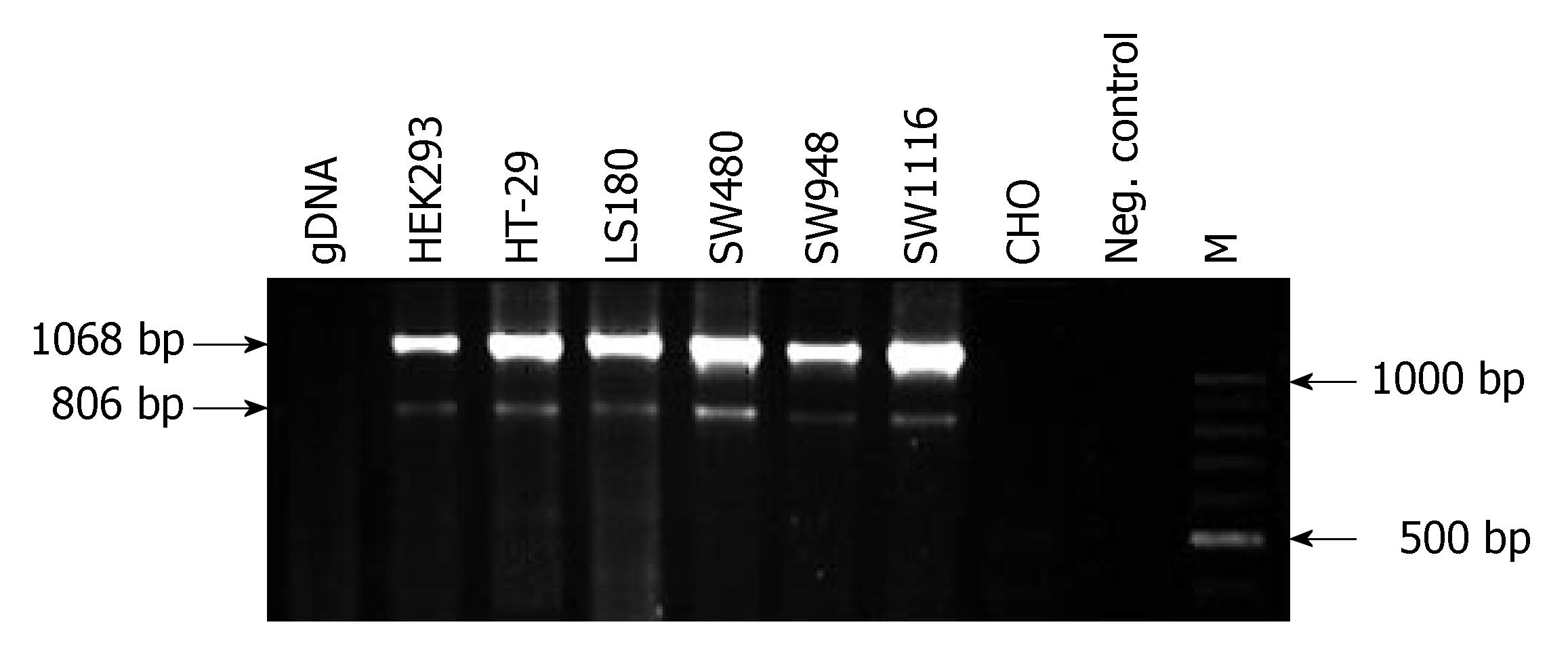
The PCR protocol consisted of an initial denaturation for 5 min at 94°C; followed by 32 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 62°C, and extension for 60 s at 72°C. The final cycle had a prolonged extension time of 7 min at 72°C. PCR products were analyzed by electrophoresis on a 1.5% agarose gel, stained with ethidium bromide and visualized under UV light. The length of the expected product was 1068 (α-transcript) or 806 (β-transcript) bp for the CAR gene and 382 and 374 bp for the PGM1 and β-actin genes, respectively.
Amplification products of the CAR β-transcript were cloned using InsTAclone PCR Cloning Kit (Fermentas) following the manufacturer’s instructions. In brief, PCR products were ligated into a pTZ57R/T vector and transformed by a heat shock method into competent Escherichia coli. Transformants were selected in LB medium containing ampicillin, X-gal and IPTG. The presence of the expected insert was confirmed by PCR and sequencing.
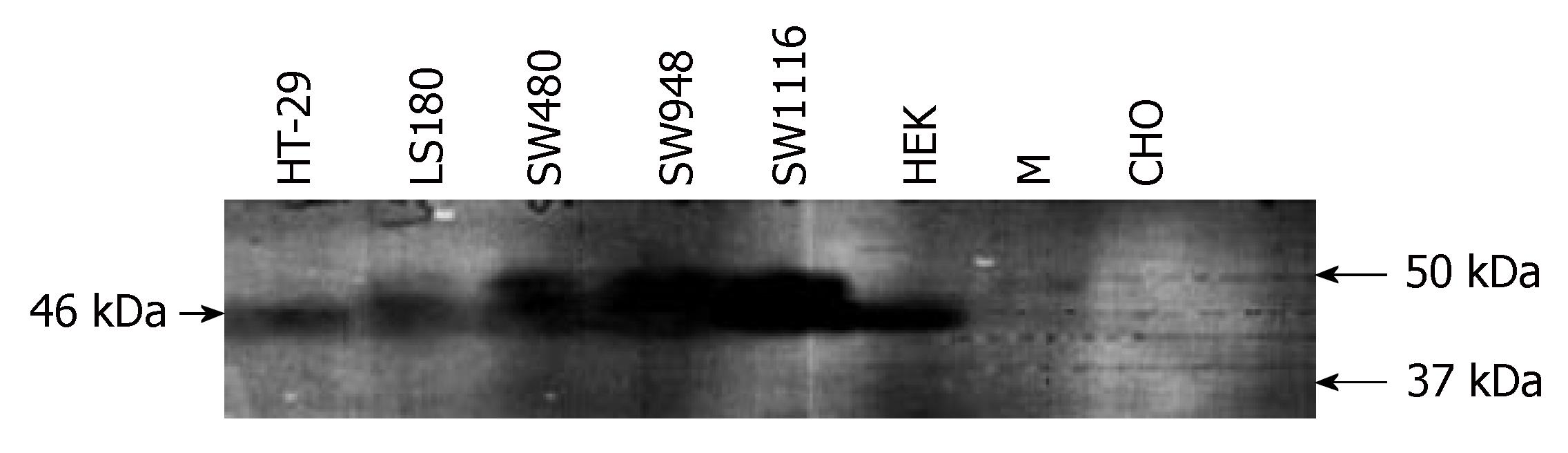
For immunoblotting analysis, cells were lysed for 1 h at 4°C in lysis buffer containing 50 mmol/L Tris (pH 8.0), 150 mmol/L NaCl, 10 mL/L Nonidet-P40 (NP40), 0.1 mL/L SDS and one tablet of complete, mini, EDTA-free protease inhibitor cocktail (Roche, Germany) per 10 mL lysis buffer. After the removal of cell nuclei by centrifugation, protein concentrations were measured by the Bradford protein assay. Equal amounts of protein (15 μg) from each cell lysate were diluted with SDS loading buffer, heated for 5 min at 95°C and electrophoresed on a 10% SDS-polyacrylamide gel. The separated proteins were then electrotransferred to a nitrocellulose membrane, which was blocked with 50 g/L non-fat dried milk in Tris buffered saline-Tween (TBST) buffer to block non-specific binding sites. The blot was then incubated with polyclonal rabbit anti-CAR primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA; 1/1000 dilution in TBST and 0.01 g/mL BSA) with gentle shaking for 1 h at room temperature. After extensive washing, polyclonal anti-rabbit horseradish peroxidase-conjugated secondary antibody (1/1000 dilution in TBST and 0.01 g/mL BSA) was applied for 1 h at room temperature with gentle shaking. Bands were visualized using an ECL kit (Amersham) according to the manufacturer’s instructions.
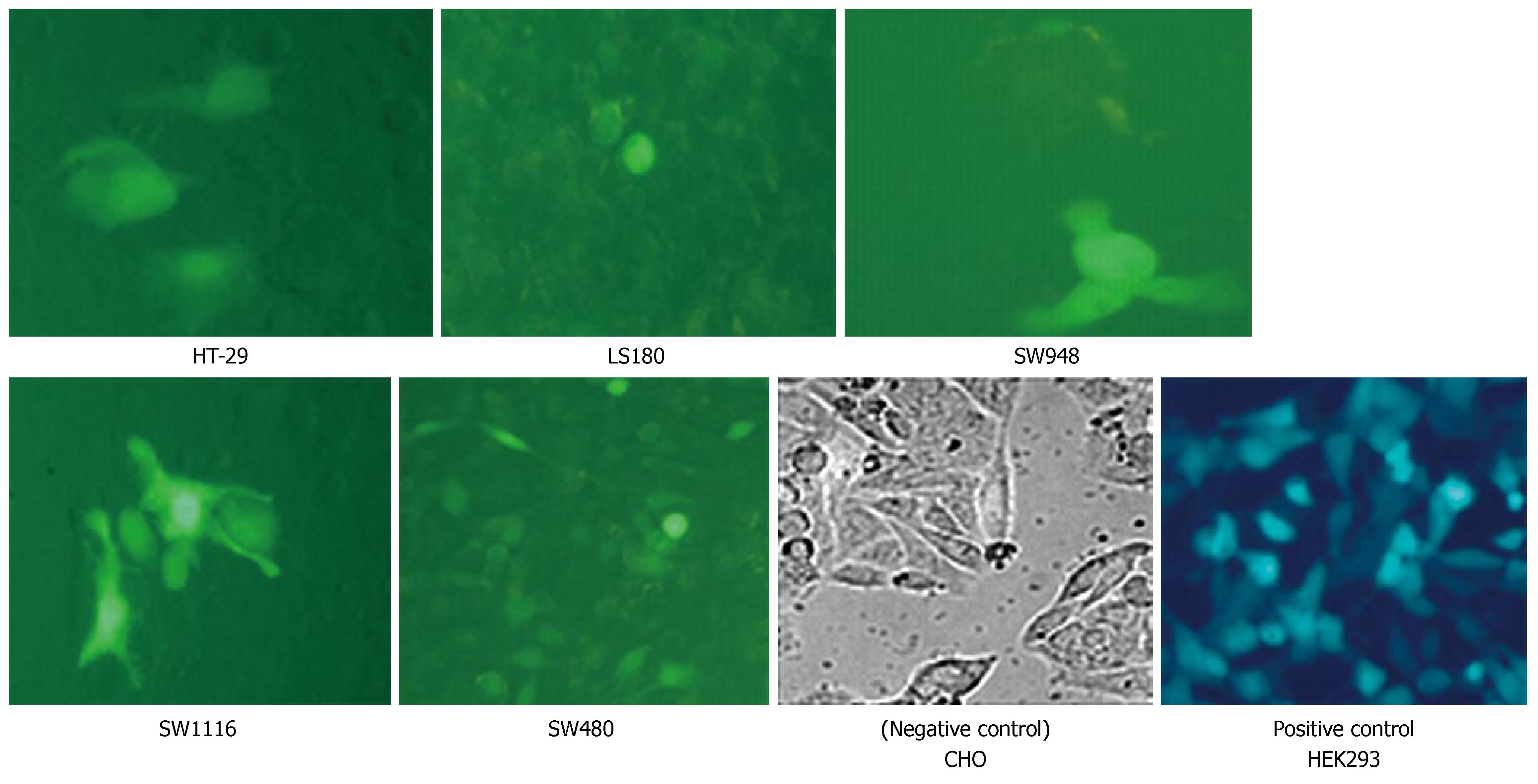
Adenoviral vector expressing the green fluorescent protein (GFP) gene driven by the cytomegalovirus promoter[24] was used to assess the colon cancer cell lines infected with adenoviral vector. Virus titer was determined by optical absorbance at A260 (1 A260 unit = 1012 particles/mL).
To evaluate reporter gene transfer by the adenovirus vector, five colon cancer cell lines were plated in 24-well culture plates at a density of 5 × 104 cells/well and grown at 37°C in a humidified incubator. After overnight incubation, cells were infected with Ad5-GFP at a multiplicity of infection (MOI) of 50 virus particles/cell. Two hours later, cells were washed twice with PBS and 600 μL fresh complete medium was added to every well. After 36 h, cells were evaluated for GFP expression by fluorescence microscopy.
We first examined the expression of CAR by RT-PCR. Integrity was evaluated by running RNA on 1% agarose gel and the quantity of RNA was measured spectrophotometrically. cDNA was then synthesized and CAR specific sequences amplified using CAR-specific primers (CARf, CARr). As shown in Figures 1A and 2, CARf and CARr primers generated a seven-exon-encompassing fragment with the expected length of 1068 bp in all the colon cancer cell lines. These primers could not amplify the pseudogenes (Figure 2, lane 1), and therefore the bands observed in lanes 2-6 were generated by just the amplification of cDNA. Any probable contamination of cDNA with genomic DNA could be disregarded.
We examined the expression of CAR by Western blotting. For immunoblotting, cell lines were cultured, harvested, lysed in appropriate lysis buffer and finally analyzed with anti-CAR antibodies. As shown in Figure 3, a band of about 46 ku was detected in HEK, HT-29, Ls-180, SW480, SW948 and SW1116 cell lines but was not observed in the negative control CHO cells.
After electrophoresis and staining of RT-PCR products, an additional fragment of 806 bp was also observed (Figure 2). This shorter band produced by CAR-specific primers was cloned and sequenced, which revealed that this fragment was the result of an alternative splicing event between exons 4 and 7, lacking exons 5 and 6, of the CAR normal transcript, and was therefore the previously determined β-splice variant. All the colon cell lines studied expressed the β-splice variant although the band intensity of the splice variant was low when compared with the intensity of the normal transcript.
As shown in Figure 4, Ad5-GFP vector entered the cells and expression of the GFP gene indicated the efficient infection of these cells by the adenoviral vector.
Gene therapy is a therapeutic modality for malignant cancers. For this, the recombinant adenovectors Ad2 and Ad5 derived from the human adenovirus subgroup C are attractive gene transfer vehicles for cancer gene therapy[25,26]. As therapeutic efficacy has been demonstrated to correlate with the ability of adenovirus to enter target cells, expression of CAR has been studied extensively.
Several tumor cell lines including those for ovary cancer[17], head and neck carcinoma[14], renal cell carcinoma[15], prostate[21] and bladder[16] cancer have shown relatively low CAR expression; although high expression of CAR mRNA has been reported in osteosarcoma cell lines[27]. In the present study, CAR expression was investigated in five colon cancer cell lines, and it was shown that CAR was expressed in all of them, although some variations in band intensity of RT-PCR products were observed among the different cell lines (Figure 1B). The mismatch between specific primers and CAR pseudogenes made us certain that only cDNA could act as the source of amplification in RT-PCR. The existence of CAR pseudogenes has apparently not been taken into account in previous studies that have investigated CAR expression[18,19,29].
Alternatively spliced products of the CAR gene have been identified; β-variant mRNA has been detected in multiple human tissues such as heart, brain, lung, liver, kidney and pancreas[10], and has also shown to be expressed in osteosarcoma cell lines[27], HeLa cells[28] and musculoskeletal tumors[29]. The β-transcript is predicted to encode a 252-amino-acid protein that can be released from cells to the culture medium because of the absence of a transmembrane domain[28]. CAR cytoplasmic and transmembrane domains are not necessary for virus attachment[30], so the anticipated β-variant protein might be as effective as the full-length variant in binding to adenovirus. In the present study, the β-transcript was demonstrated to be present in colon cell lines, in addition to the normal α-transcript.
To evaluate whether CAR mRNA expression correlated with CAR protein expression, Western blotting was carried out. CAR protein was shown to be present in the colon cell lines investigated; therefore, they could be considered as CAR-positive on the basis of CAR protein expression. In addition, no alternative protein band that could be attributed to the β-transcript-encoded protein was observed. Western blotting was carried out using a commercial polyclonal antibody against amino acids 1 to 300 of the original protein and cell lysates. Thus, the absence of any alternative signal (β-variant protein) could be explained as very low or undetectable intracellular level of this shorter variant and its secretion from the cell surface. However, β-variant protein is not detected in extracts of HeLa cells that express both β- and normal variants[28]. Therefore, the potential translation of CAR alternative transcript and its proposed regulatory role in antivirus defense mechanisms[28] remains to be confirmed.
Experiments with the Ad5 vector carrying the GFP reporter gene showed that the colon cancer cell lines studied were sensitive to adenovirus infection, and after 36 h GFP gene expression was easily detected by fluorescence microscopy.
In conclusion, the results of this study suggested that adenovirus-based gene delivery was efficiently applied to the five colon cancer cell lines. However, to clarify the efficiency of adenoviral-mediated gene transfer in colon tumors, further studies will be required to explore the expression of CAR.
Vectors based on human adenovirus serotypes 2 and 5 show increasing promise as gene delivery vehicles, and currently adenoviral vectors are the most commonly administered vectors for cancer gene therapy. Adenovirus initiates gene transfer by attachment to cell surface receptor, the coxsackievirus and adenovirus receptor (CAR).
Reports show that the efficiency of adenoviral infection is dependent on the expression of CAR on target cells. However, adenovirus-mediated gene transfer is in practice hindered by the relatively low expression of CAR on tumor cells.
This is believed to be the first report to examine the expression of CAR in human colon cancer cell lines at the mRNA and protein level.
These results might be of potential value in adenovirus-based gene transfer and research in colon cancer cell lines.
Gene therapy: to replace a malfunctioning mutated gene with a normal wild-type gene or to express a therapeutic gene in target cells. A prerequisite for efficient gene therapy is to determine a system for delivering the therapeutic gene to target cells. Adenovirus: non-enveloped, icosahedral, particles with double stranded DNA genome that in humans, is divided into six species (A-F) and 51 serotypes. CAR: primary cellular receptor for attachment of both coxsackie B virus and adenovirus subgroup C to target cells.
This manuscript by Yassan Abdolazimi and co-authors is presented in a clear style. Experimental details are provided accordingly. Relevant controls were included in the study. The authors’ findings are discussed with the relevant literature.
S- Editor Zhu LH L- Editor Kerr C E- Editor Liu Y
| 1. | Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6112] [Cited by in RCA: 5988] [Article Influence: 332.7] [Reference Citation Analysis (0)] |
| 2. | Gong Z, Xie D, Deng Z, Bostick RM, Muga SJ, Hurley TG, Hebert JR. The PPAR{gamma} Pro12Ala polymorphism and risk for incident sporadic colorectal adenomas. Carcinogenesis. 2005;26:579-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 3. | Palmer DH, Chen MJ, Kerr DJ. Gene therapy for colorectal cancer. Br Med Bull. 2002;64:201-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 4. | Chen MJ, Chung-Faye GA, Searle PF, Young LS, Kerr DJ. Gene therapy for colorectal cancer: therapeutic potential. BioDrugs. 2001;15:357-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 5. | Palmer DH, Chen MJ, Kerr DJ. Taking Gene Therapy into the Clinic. J Biomed Biotechnol. 2003;2003:71-77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Wilson DR. Viral-mediated gene transfer for cancer treatment. Curr Pharm Biotechnol. 2002;3:151-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Vorburger SA, Hunt KK. Adenoviral gene therapy. Oncologist. 2002;7:46-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 146] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 8. | Mountain A. Gene therapy: the first decade. Trends Biotechnol. 2000;18:119-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 211] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 9. | Qin M, Chen S, Yu T, Escuadro B, Sharma S, Batra RK. Coxsackievirus adenovirus receptor expression predicts the efficiency of adenoviral gene transfer into non-small cell lung cancer xenografts. Clin Cancer Res. 2003;9:4992-4999. [PubMed] |
| 10. | Tomko RP, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352-3356. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 925] [Cited by in RCA: 935] [Article Influence: 33.4] [Reference Citation Analysis (0)] |
| 11. | Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320-1323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2385] [Cited by in RCA: 2318] [Article Influence: 82.8] [Reference Citation Analysis (0)] |
| 12. | Carson SD. Receptor for the group B coxsackieviruses and adenoviruses: CAR. Rev Med Virol. 2001;11:219-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 54] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 13. | Thoelen I, Magnusson C, Tågerud S, Polacek C, Lindberg M, Van Ranst M. Identification of alternative splice products encoded by the human coxsackie-adenovirus receptor gene. Biochem Biophys Res Commun. 2001;287:216-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 14. | Li D, Duan L, Freimuth P, O'Malley BW. Variability of adenovirus receptor density influences gene transfer efficiency and therapeutic response in head and neck cancer. Clin Cancer Res. 1999;5:4175-4181. [PubMed] |
| 15. | Haviv YS, Blackwell JL, Kanerva A, Nagi P, Krasnykh V, Dmitriev I, Wang M, Naito S, Lei X, Hemminki A. Adenoviral gene therapy for renal cancer requires retargeting to alternative cellular receptors. Cancer Res. 2002;62:4273-4281. [PubMed] |
| 16. | Li Y, Pong RC, Bergelson JM, Hall MC, Sagalowsky AI, Tseng CP, Wang Z, Hsieh JT. Loss of adenoviral receptor expression in human bladder cancer cells: a potential impact on the efficacy of gene therapy. Cancer Res. 1999;59:325-330. [PubMed] |
| 17. | Kim M, Zinn KR, Barnett BG, Sumerel LA, Krasnykh V, Curiel DT, Douglas JT. The therapeutic efficacy of adenoviral vectors for cancer gene therapy is limited by a low level of primary adenovirus receptors on tumour cells. Eur J Cancer. 2002;38:1917-1926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 102] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 18. | Kim JS, Lee SH, Cho YS, Choi JJ, Kim YH, Lee JH. Enhancement of the adenoviral sensitivity of human ovarian cancer cells by transient expression of coxsackievirus and adenovirus receptor (CAR). Gynecol Oncol. 2002;85:260-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 32] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 19. | Kim JS, Lee SH, Cho YS, Kim YH, Lee JH. Ectopic expression of the coxsackievirus and adenovirus receptor increases susceptibility to adenoviral infection in the human cervical cancer cell line, SiHa. Biochem Biophys Res Commun. 2001;288:240-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 20. | Pearson AS, Koch PE, Atkinson N, Xiong M, Finberg RW, Roth JA, Fang B. Factors limiting adenovirus-mediated gene transfer into human lung and pancreatic cancer cell lines. Clin Cancer Res. 1999;5:4208-4213. [PubMed] |
| 21. | Okegawa T, Li Y, Pong RC, Bergelson JM, Zhou J, Hsieh JT. The dual impact of coxsackie and adenovirus receptor expression on human prostate cancer gene therapy. Cancer Res. 2000;60:5031-5036. [PubMed] |
| 22. | Fuxe J, Liu L, Malin S, Philipson L, Collins VP, Pettersson RF. Expression of the coxsackie and adenovirus receptor in human astrocytic tumors and xenografts. Int J Cancer. 2003;103:723-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 77] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 23. | Mobasheri MB, Modarressi MH, Shabani M, Asgarian H, Sharifian RA, Vossough P, Shokri F. Expression of the testis-specific gene, TSGA10, in Iranian patients with acute lymphoblastic leukemia (ALL). Leuk Res. 2006;30:883-889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Modarressi MH, Cheng M, Tarnasky HA, Lamarche-Vane N, de Rooij DG, Ruan Y, van der Hoorn FA. A novel testicular RhoGAP-domain protein induces apoptosis. Biol Reprod. 2004;71:1980-1990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 25. | Young LS, Searle PF, Onion D, Mautner V. Viral gene therapy strategies: from basic science to clinical application. J Pathol. 2006;208:299-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 206] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 26. | Glasgow JN, Everts M, Curiel DT. Transductional targeting of adenovirus vectors for gene therapy. Cancer Gene Ther. 2006;13:830-844. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 92] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 27. | Kawashima H, Ogose A, Yoshizawa T, Kuwano R, Hotta Y, Hotta T, Hatano H, Kawashima H, Endo N. Expression of the coxsackievirus and adenovirus receptor in musculoskeletal tumors and mesenchymal tissues: efficacy of adenoviral gene therapy for osteosarcoma. Cancer Sci. 2003;94:70-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 28. | Dörner A, Xiong D, Couch K, Yajima T, Knowlton KU. Alternatively spliced soluble coxsackie-adenovirus receptors inhibit coxsackievirus infection. J Biol Chem. 2004;279:18497-18503. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Gu W, Ogose A, Kawashima H, Ito M, Ito T, Matsuba A, Kitahara H, Hotta T, Tokunaga K, Hatano H. High-level expression of the coxsackievirus and adenovirus receptor messenger RNA in osteosarcoma, Ewing's sarcoma, and benign neurogenic tumors among musculoskeletal tumors. Clin Cancer Res. 2004;10:3831-3838. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.3] [Reference Citation Analysis (0)] |