Published online Nov 21, 2007. doi: 10.3748/wjg.v13.i43.5718
Revised: July 31, 2007
Accepted: September 14, 2007
Published online: November 21, 2007
AIM: To investigate the role of SMYD3 in hepatocellular carcinoma (HCC) development and progression and to verify whether its regulation activity was through RIZ1 inactivation.
METHODS: Expression of SMYD3 in HCC cell lines and tissues were measured; silencing of SMYD3 by RNA interference (RNAi) was effectuated, hepatoma cell proliferation, migration and apoptosis were tested, with RIZ1 CpG promoter methylation, and corresponding mRNA expression were investigated.
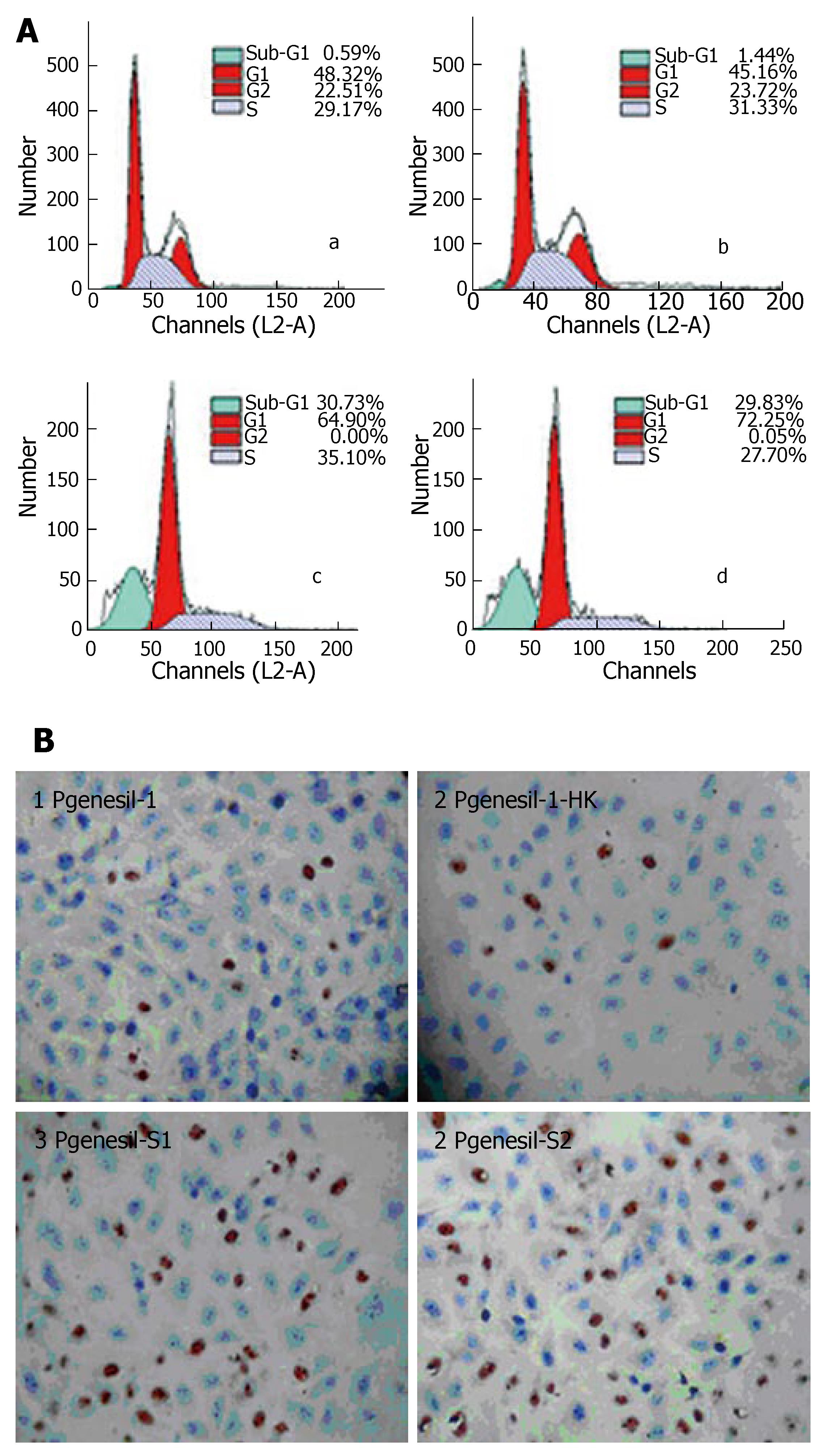
RESULTS: SMYD3 over-expression in HCC was associated with RIZ1 hypermethylation and mRNA down-expression. Suppression of SMYD3 expression de-methylated RIZ1 CpG promoter (P < 0.01) and increased RIZ1 mRNA expression (P < 0.01). Consequently, SMYD3 down-expression with RIZ1 de-methylation strongly inhibited hepatoma cell growth (MTT inhibitory rates: Pgenesil-1-s1 60.95% ± 7.97%, Pgenesil-1-s2 72.14% ± 9.68% vs Pgenesil-1-hk 6.89% ± 4.12%, P < 0.01) and migration (Pgenesil-1-s1 4.24% ± 1.58%, Pgenesil-1-s1 4.87% ± 0.73% vs Pgenesil-1 19.03% ± 4.63%, Pgenesil-1-hk 19.95% ± 5.21%, P < 0.01) and induced apoptosis (FCM subG1 phase Pgenesil-1-s1 19.07% ± 1.78%, Pgenesil-1-s2 17.68% ± 2.36% vs Pgenesil-1 0.47% ± 0.12%, Pgenesil-1-hk 1.46% ± 0.28%, P < 0.01. TUNEL-positive cells: Pgenesil-1-s1 40.24% ± 5.18%, Pgenesil-1-s2 38.48% ± 4.65% vs Pgenesil-1 2.18% ± 1.34%, Pgenesil-1-hk 2.84% ± 1.22%, P < 0.01) in HepG2 cells.
CONCLUSION: These results demonstrate that SMYD3 plays a critical role in the carcinogenesis and progression of HCC. The proliferation, migration induction and apoptosis inhibition activities of SMYD3 may be mediated through RIZ1 CpG promoter hypermethylation.
- Citation: Chen LB, Xu JY, Yang Z, Wang GB. Silencing SMYD3 in hepatoma demethylates RIZI promoter induces apoptosis and inhibits cell proliferation and migration. World J Gastroenterol 2007; 13(43): 5718-5724
- URL: https://www.wjgnet.com/1007-9327/full/v13/i43/5718.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i43.5718
Hepatocellular carcinoma (HCC) is one of the most common malignancies worldwide, generally long-term survival is disappointing. Epigenetics, defined as heritable changes in gene expression that are not coded in the DNA sequence itself, is increasingly linked with tumorigenesis[1,2]. It is well established that DNA methylation, nucleosomal histone post-translational modifications such as methylation, acetylation/deacetylation, phosphorylation ADP-ribosylation, and ubiquitination, and other epigenetic patterns are central to proper gene expression. Increasing evidence shows that disrupting epigenetic patterns can induce carcinogenesis or affect the outcome of cancer[3,4]. Among these, histone methylation is considered to be critical for transcriptional regulation and seems to play an important role in tumor epigenetic modification. Recently, SMYD3 was identified and characterized to specifically methylate histone H3 at lysine 4 (K4), and this activity is indeed dependent on an intact SET domain, an evolutionarily conserved protein module shown to facilitate histone methyltransferase activity[5]. Expression of SMYD3 was frequently enhanced in colorectal carcinoma (CRC), breast cancer tissue, as well as HCC, and elevated SMYD3 expression was involved in the growth of CRC, breast cancer and HCC cells[5-7]. Although transcriptional activation of downstream genes including Nkx2.8 and WNT10B gene has been reported[5,6], the effect of SMYD3 on HCC development and the underlining mechanism remains unclear.
Inactivation of tumor suppressive genes (TSGs) plays an important role in carcinogenesis. RIZ1, a typical TSG with H3-K9 methyltransferase activity, was shown to lose its expression and tumor-suppressing activity in many types of human tumors including HCC[8], CRC[9], breast cancer[10], prostate cancer[11] and gastric cancer[12]. Adenovirus-mediated RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis in breast cancer, liver cancer, and microsatellite instability-positive colon cancer cells. Adenovirus RIZ1 can also inhibit growth of colon cancer xenografts[13]. Previous data suggest that the RIZ locus is a target of frequent deletion in HCC, but a more common way of RIZ inactivation in HCC may not involve mutations that alter peptide sequences, but by CpG promoter hypermethylation[14].
Therefore, we sought to explore whether targeting of SMYD3 expression can inhibit the development of HCC, and determine whether the biological function of SMYD3 in HCC development and progression is mediated through RIZ1 inactivation. We found SMYD3 was significantly overexpressed in HCC cell lines, but not in normal hepatocellular cells and peri-cancerous tissues. Silencing of SMYD3 by RNAi remarkably inhibited HCC cell proliferation and migration, and induced apoptosis in vitro. Furthermore, RIZ1 was under-expressed in HCC cells with its promoter hypermethylated, while inhibition of SMYD3 demethylated RIZ1 and recovered its expression in HCC cells. Our results indicated that SMYD3 plays pro-oncological function in HCC development and progression; thus the activity of SMYD3 in HCC may be partly through its activity on RIZ1 promoter hypermethylation and RIZ1 inactivity.
HCC cell lines HepG2 and Hep3B were purchased from the American Type Culture Collection (Rockville, MD). HCC cell line SMMC-7721 and liver cell line L-02 were generous gifts from Dr. Danhui Weng (HUST, China). The cells were cultured in Dulbecco's modified Eagle's medium (Gibco-BRL, Carlsbad, CA) supplemented with 10% fetal bovine serum (Gibco-BRL).
Total RNA was extracted using Trizol reagent (Takara, Tokyo, Japan). RT-PCR was performed with the Advantage RT-PCR kit (Takara), according to the manufacturer’s protocol. PCR setting: Initial denaturation at 94°C for 3 min before 18 cycles (for GAPDH) or 30 cycles (for SMYD3 and RIZ1) at 94°C for 30 s, 50°C for 45 s and 72°C for 60 s. The sets of primers are listed in Supplementary Information Table S1.
The Pgenesil-1 vector containing the U6 promoter region was purchased from Genesil Biotechnology Corporation (Wuhan, China). Plasmids expressing siRNAs were prepared by cloning double-stranded oligonucleotides into the Pgenesil-1 vector. The sets of primers are listed in Supplementary Information Table S1. Positive clones were identified by restriction digestion and confirmed by sequencing. The resulting plasmids were designated as Pgenesil-1-s1, Pgenesil-1-s2 and negative control Pgenesil-1-hk. Transfection was performed by Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) in accordance with the manufacturer's protocol.
Proteins were prepared by homogenization of cells in lysis buffer (10 mmol/L Tris-HCl, pH 8.0; 140 mmol/L NaCl; 5 mmol/L EDTA; 0.25 g/L NaN3; 10 g/L Triton X-100; 10 g/L deoxycholate; 1 g/L SDS; 0.5 mmol/L PMSF; 1 g/L leupeptin; 1 g/L aprotinin). Protein concentration was determined by the Lowry method (Bio-Rad, Hercules, CA). Proteins were resolved (30 μg per lane) in 120 g/L sodium dodecyl sulfate-polyacrylamide gel electrophoresis and was transblotted onto a PVDF membrane (Amersham, Arlington Heights, IL). Membranes were blocked in 50 mL/L non-fat milk overnight and subsequently incubated with 1:2000 rabbit polyclonal anti-SMYD3 antibody (a generous gift from Dr. Ryuji Hamamoto of the University of Tokyo, Tokyo, Japan), or anti-β-actin (1:5000 dilution, Sigma, St. Louis, MO) for 1 h and then with secondary anti-rabbit IgG-horseradish-peroxidase antibody for 1 h at room temperature. Immunoreactive proteins were visualized by means of enhanced chemiluminescence reagent (Pierce Biotechnology, Rockford, IL) and exposure to autoradiographic film.
MTT experiments were carried out using the cell proliferation kit (Roche Diagnostics, Indianapolis, IN), according to the manufacturer's manual. Briefly, stable transfected and parental HepG2 cells were seeded at a density of 5 × 103 cells/well in 96-well plates and allowed to grow for 48 h and then 20 μL 5 g/L MTT was added. After incubation for 4 h, DMSO 200 g/L was added to each well to dissolve crystals. After 5 min incubation at 37°C, absorbance was measured at 570 nm. Assays were performed in triplicate.
This assay for the invasiveness of cells was based on the principle of Boyden chamber (BD Biosciences, San Diego, CA) and performed according to the manufacturer’s protocol. Briefly, the top compartment was prepared by coating the filter with diluted Matrigel and incubated for 30 min. A suspension of 1 × 104 cells in serum-free medium was inoculated in the upper chamber, and conditioned media obtained from NIH3T3 cells was placed in the lower compartment of the chamber, as a chemoattractant. After 24 h incubation, noninvasive cells were removed with a cotton swab. The cells which migrated through the filter and adhered to the lower surface of the filter were fixed with methanol, stained with hematoxylin, and counted manually in 5 randomly selected microscopic fields. Assays were performed in triplicate. The data were reported as the percentage of cells successfully passing through the Matrigel and filter relative to those migrating through the control filter.
HepG2 cells, 1 × 106, were seeded in 60-mm dishes and transfected with SMYD3-RNAi-plasmids at 60%-80% confluence. Then, 48 h after transfection, cells were deprived of serum for 36 h. Afterwards, cells were harvested, fixed in 700 mL/L ethanol and washed with 1 × PBS and suspended in 50 mg/L propidium iodide. DNA content was determined by flow cytometry using a Becton-Dickinson FACSCalibur (Becton Dickinson, Bedford, MA). Results were analyzed by ModFit LT2.0 and Cellquest software. Assays were performed in triplicate.
Apoptotic cells were identified with the In Situ Cell Death Detection kit (Roche Applied Science, Shanghai, China) using the protocol recommended by the manufacturer. In brief, HepG2 cells were grown on coverslips. The next day, cells were transfected with SMYD3-RNAi-plasmids. At 48 h after transfection, cells were deprived of serum for 36 h. Coverslips with adherent cells were fixed in 40 g/L paraformaldehyde for 1 h at room temperature and permeabilized with 1 g/L Triton X-100 for 2 min on ice. DNA fragments were labeled with the TdT-mediated dUTP nick end labeling (TUNEL) reaction mixture for 60 min at 37°C in a humidified atmosphere in the dark. Diaminobenzidine was used to mark the apoptotic cells (brown staining). Cells were counted manually in six randomly selected microscopic fields for each sample. The apoptosis index was defined by the percentage of apoptotic cells among the total cells of each sample.
Methylation Specific PCR (MSP) assay was performed according to the procedure described by Fang et al[15]. In brief, 1 μg of the genomic DNA was modified by sodium bisulfide using the CpGenome DNA Modification Kit (Intergen, Purchase, NY) in accordance with the manufacturer's instructions. Modified DNA was amplified by two different primer pairs specific to the unmethylated (U) and methylated (M) RIZ1 sequences, respectively. The sets of primers are listed in Supplementary Information Table S1. The PCR amplification was performed for a total of 45 cycles with an annealing temperature of 68°C and 60°C for M-sequences and U-sequences, respectively. The PCR products were then analyzed using a 35 g/L agarose gel.
SPSS 13.0 was used to analyze the data. Statistical significance was assessed by comparing mean ± SD with Student’s t test for independent groups. P < 0.05 was considered statistically significant.
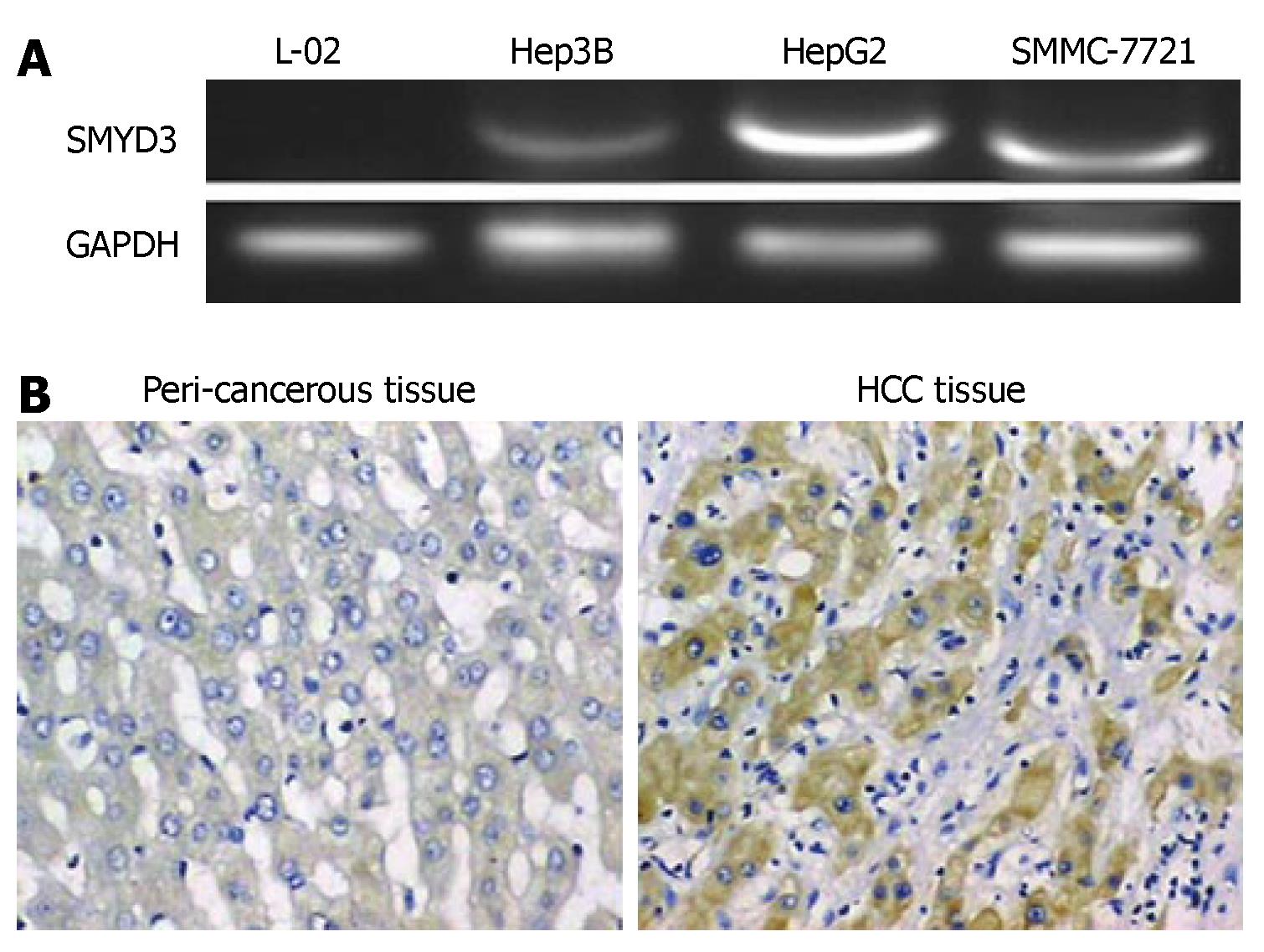
To determine whether SMYD3 gene is overexpressed in HCC cell lines, we compared the level of SMYD3 gene expression in normal human hepatocellular cell line L-02 to that of three HCC cell lines. RT-PCR analysis revealed that SMYD3 was overexpressed in all HCC cells, but absent in L-02 cells (Figure 1). These data indicate that enhanced SMYD3 expression is involved in a majority of HCC.
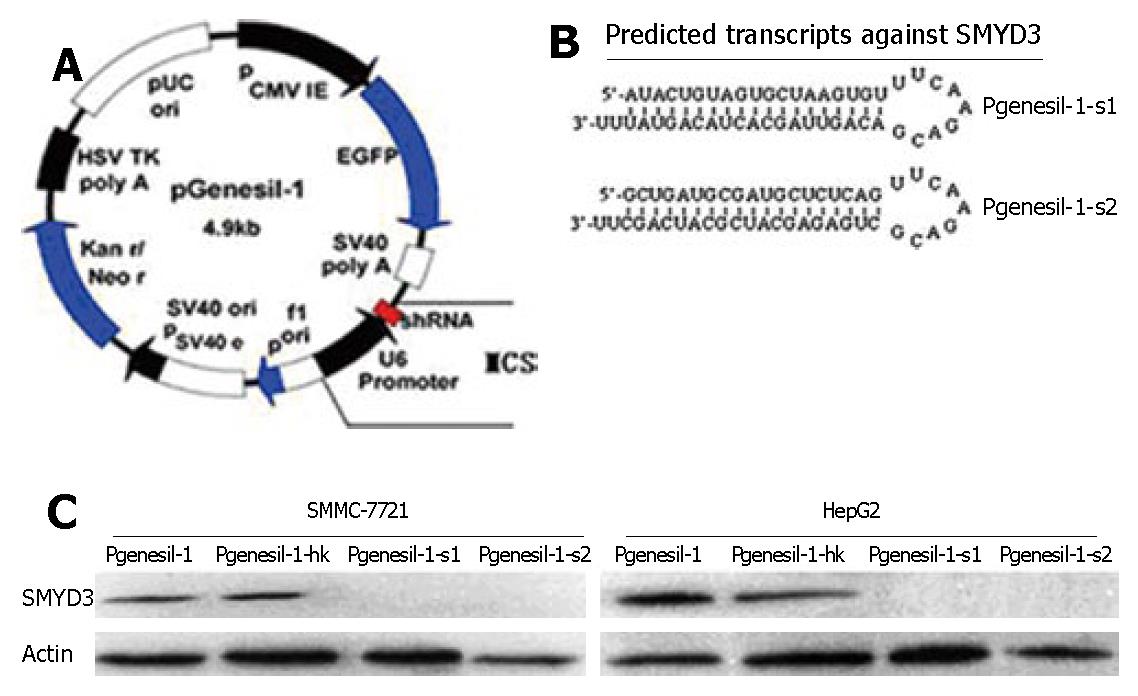
To inhibit SMYD3 expression in HCC cells, we used a DNA-based siRNA method (Figure 2A). Two siRNAs targeting different 21 sequences of human SMYD3 were cloned into Pgensil-1 vector to express RNA, which is expected to fold back to form a hairpin loop structure after being transcribed. The hairpin dsRNA can then be further cleaved by Dicer to generate a 21-nucleotide siRNA, the active form for the RNAi effect (Figure 2B). We used HepG2 and SMMC-7721 cells, which overexpress SMYD3 abundantly. Forty eight hours after transfection, Pgenesil-1-s1 and Pgenesil-1-s2 markedly knocked down SMYD3 expression in these cells, as determined by Western blot analysis. The specificity of RNAi targeting SMYD3 was shown by transfection with Pgenesil-1-hk which had no effect on SMYD3 expression (Figure 2C).
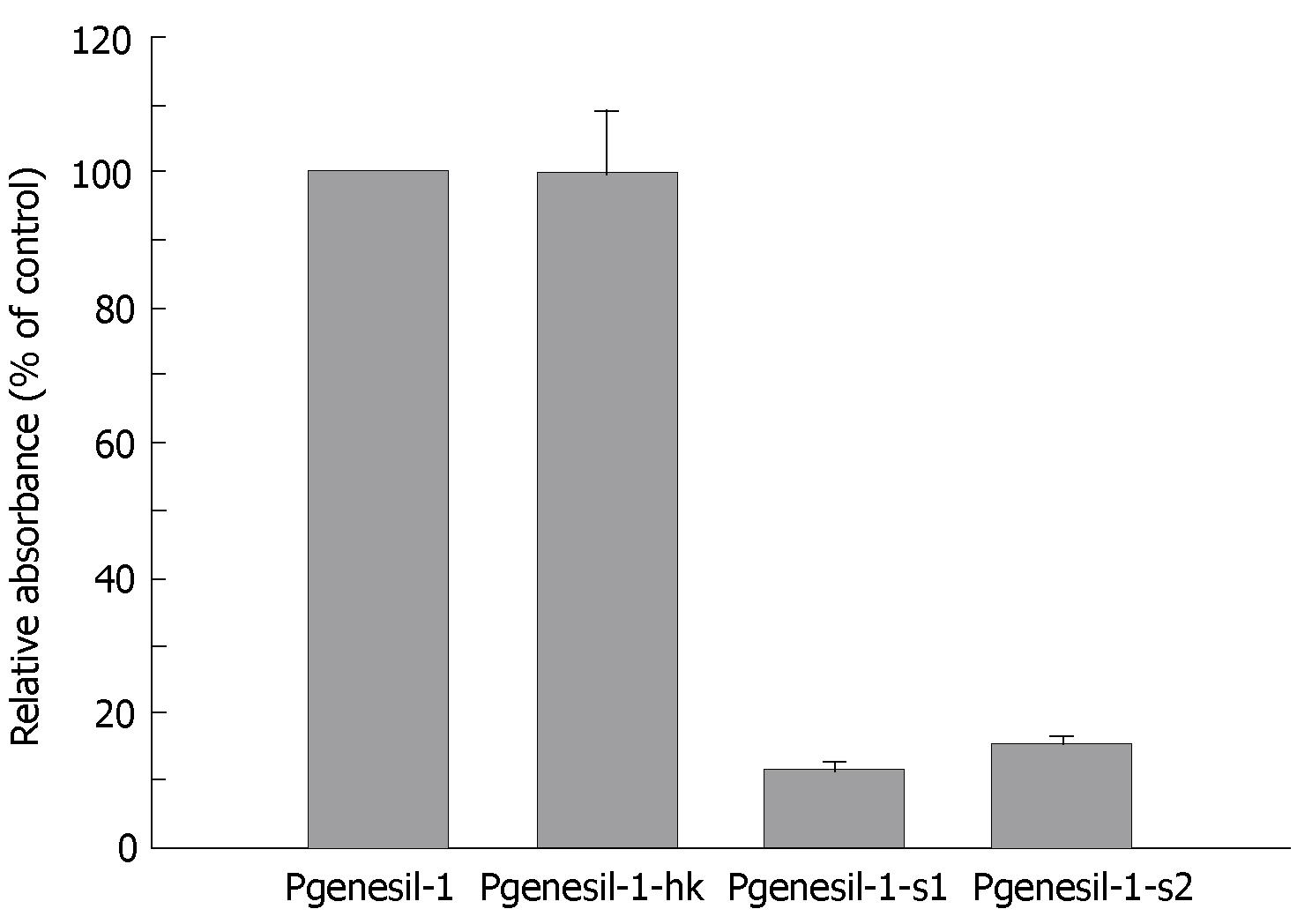
MTT assay was used to examine the inhibitory effect of RNAi against SMYD3 on cell growth in HepG2 cells. Suppression of SMYD3 expression significantly inhibited cell growth compared to the cells transfected with control plasmid Pgenesil-1. Pgenesil-1-hk, which did not suppress expression of SMYD3, was shown to have little inhibitory effect on growth of HepG2 cells, compared to Pgenesil-1 (Figure 3). The growth inhibitory effect of the plasmids was consistent with their gene silencing effect. Therefore, SMYD3 RNAi significantly suppressed the growth of HeG2 cells in vitro, indicating that SMYD3 may be involved in the regulation of cell proliferation.
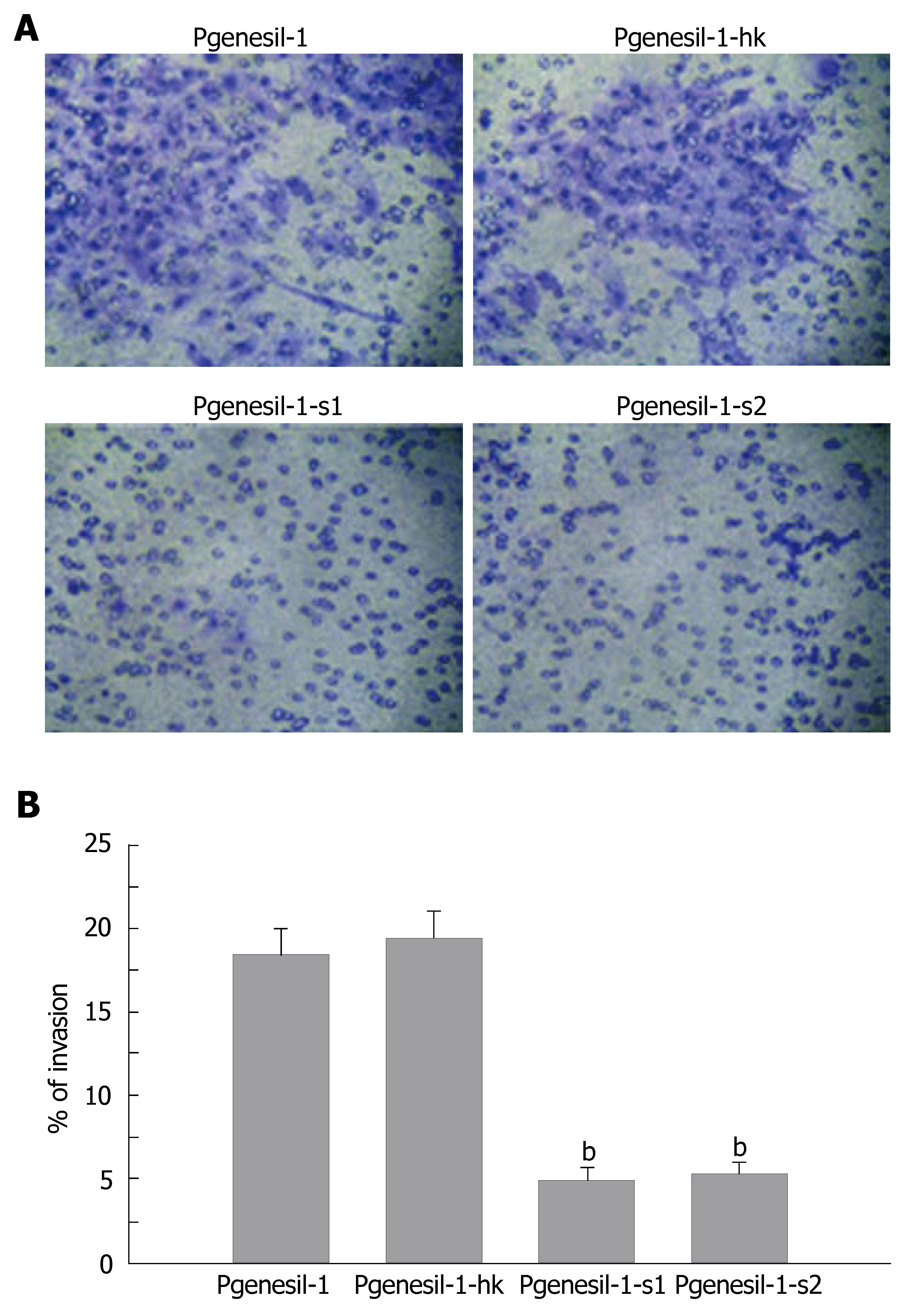
The HepG2 cell line has been characterized as a highly invasive hepatocellular cancer cell line. To determine whether SMYD3 gene knockdown by RNAi could reduce its invasive potential, an in vitro invasion assay was performed. As shown in Figure 4, HepG2 cells were greatly deprived of invasiveness by depletion of SMYD3 gene expression. The percentage of invasive cells was decreased by 3-4 fold in Pgenesil-1-s1 and Pgenesil-1-s2 treated groups, compared to Pgenesil-1 treated groups. No significant decrease of invasive cells was observed in Pgenesil-1-hk treated cells. These results demonstrate that SMYD3 not only plays a pivotal role in the process of development, but it also is involved in tumor cell migration.
Flow cytometry analysis showed that after 48 h of transfection and 36 h serum deprivation, the number of cells in the sub G1 phase was 19.07 ± 1.78% and 17.68 ± 2.36% in HepG2 cells transfected with Pgenesil-1-s1 and Pgenesil-1-s2, respectively, while only 0.47% ± 0.12% and 1.46% ± 0.28% cells were observed in HepG2 cells transfected with Pgenesil-1 and Pgenesil-1-hk, respectively (Figure 5A). These results possibly suggest the induction of apoptosis. To confirm whether silencing of SMYD3 can induce apoptosis in HCC cells following serum deprivation, TUNEL assay was performed. The number of TUNEL-positive cells was 40.24% ± 5.18% and 38.48% ± 4.65% in HepG2 cells transfected with Pgenesil-1-s1 and Pgenesil-1-s2, respectively. Only 2.18% ± 1.34% and 2.84% ± 1.22% were TUNEL positive in control cells transfected with Pgenesil-1 and Pgenesil-1-hk, P < 0.01, respectively (Figure 5B). These results indicate that inhibition of SMYD3 by RNAi significantly promoted apoptosis in HepG2 cells following serum deprivation.
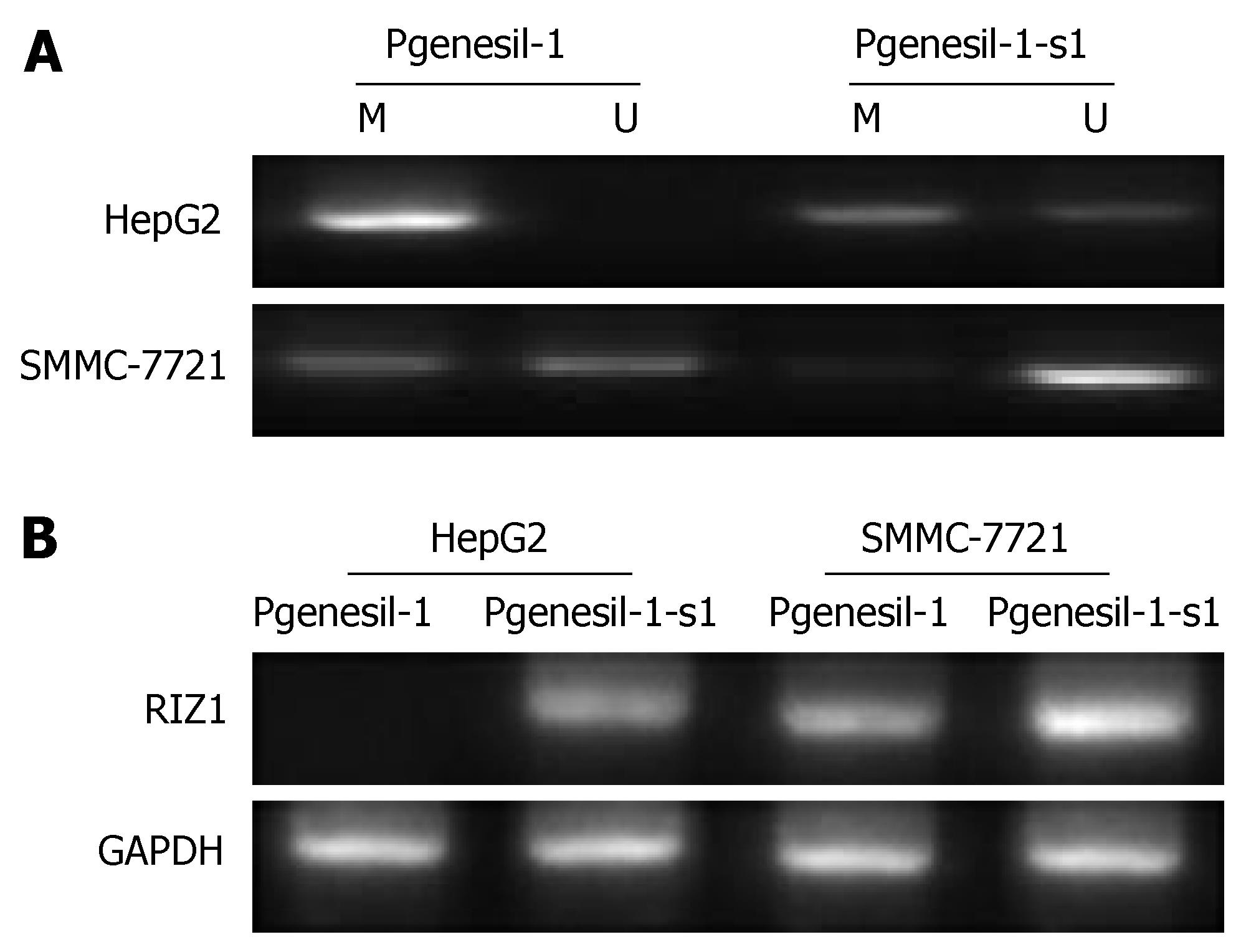
Through MSP and RT-PCR, we observed that the RIZ1 promoter was totally methylated with the lack expression of RIZ1 mRNA in HepG2 cells. After inhibition of SMYD3 by Pgenesil-1-s1, the RIZ1 promoter was found partial methylated in HepG2 cells, consistent with the change of RIZ1 mRNA (Figure 6). These results suggest that the activity of SMYD3 may be regulated through RIZ1 CpG promoter hypermethylation and RIZ1 inactivation.
Among epigenetic regulatory ways, histone methylation, perhaps more than any other form of modification, has demonstrated the power of modifications over DNA-based functions, regulating fundamental processes such as gene transcription and DNA repair[16]. SMYD3, a gene located in 1q44 chromosome, encodes a protein with HMTase activity specifically on histone H3 at K4, and is involved in carcinogenesis and tumor progression[5]. In the present study, we confirmed that SMYD3 is overexpressed in HCC cell lines. To further investigate the function of SMYD3 in HCC carcinogenesis and tumor progression, RNAi technology was applied to specifically knock down SMYD3 expression in HCC cell lines. We demonstrated that depletion of SMYD3 reduced hepatoma cell growth, migration and induced cell apoptosis upon serum deprivation. Our findings suggest that SMYD3 plays pro-oncologic role in HCC development and progression. Interestingly, our results show that the expression level of SMYD3 in Hep3B cells differs significantly from that in HepG2 and SMMC-7721 cells. We hypothesized that the difference is possibly due to the HBV infectious state of the cells that we used; HBV positive in Hep3B, but negative in HepG2 and SMMC-7721 cells. However, this hypothesis needs to be further clarified.
SMYD3 is considered to function through its H3-K4 histone metylation, regulating expression of Nkx2.8[17,18], Wnt10B[19,20] and other genes involved in hepatoma cell-cycle regulation, cell proliferation and apoptosis. SMYD3 also upregulates genes linked to cell adhesion and invasion, such as ITGA5[21], COLQ[22], SELL[23], NEURL[24], and PECAM1[25], which may be responsible for cell migration regulatory role of SMYD3 in our experiment. To investigate whether SMYD3 might act on HCC oncological activity through TSG inactivation, we examined if RIZ1 activity could be regulated by SMYD3 in different hepatoma cells. RIZ1 was found to be downregulated in HepG2 cells with its promoter CpG hypermethylated, which was in line with enhanced SMYD3 expression, while knockdown of SMYD3 demethylated RIZ1 promoter and upregulated RIZ1 expression in these cells. In addition, different SMYD3 expression levels in hepatoma cells was also consistent with RIZ1 promoter methylation and mRNA expression in hepatoma cells in our results (data not shown) and in other results. HepG2 cells have been shown to lack RIZ1 expression due to promoter hypermethylation; in contrast, Hep3B cells do not show RIZ1 promoter hypermethylation and express RIZ1 mRNA[26,27]. These results strongly suggest that SMYD3 plays a role in HCC development and progression, partly through RIZ1 promoter hypermethylation and RIZ1 inactivation.
As a histone/protein methyltransferase, SMYD3 mainly acts on histones or proteins; therefore, the mechanism by which SMYD3 regulate RIZ1 promoter CpG islands needs to be further clarified. SMYD3 includes a putative 428-amino acid protein containing a SET domain (codons 148-239) which is HMT, and a zf-MYND domain (codons 49-87), a typical zinc finger domain. The presence of a MYND-type zinc-finger domain in SMYD3 suggests that SMYD3 can recognize and bind particular sequences present in the promoter region of downstream genes through its MYND zinc finger. The specific SMYD3 binding elements (SBE) in target DNA are 5′-CCCTCC-3′ or 5′-GGAGGG-3′, which are present in the promoter regions of SMYD3 downstream genes, such as Nkx2.8[5]. It is interesting to note that one SBE, 5′-GGAGGG-3′, is present in the promoter region of RIZ1[27]. A typical zinc finger domain in SMYD3 and SBE sequence in RIZ1 promoter strongly suggests that SMYD3 may act on RIZ1 promoter through its zf-MYND domain recognizing and binding to the SBE within the RIZ1 promoter. However, SMYD3 may also recognize SBE in other genes, such as DNA methyltransferases (DNMT), which are crucial for DNA methylation[28], and sequentially regulates RIZ1 promoter hypermethylation. There is other evidence suggesting that histone modification may interact with DNA methylation or may regulate DNA methylation[29]. In mammals, H3K9 methylation and CpG methylation shows a complex interplay in which each mark can influence the activity of the other.
Our results imply that, besides their gene-transacting role, SMYD3 and other H3-K4 methyltransferases might influence carcinogenesis and tumor progression by silencing TSGs through DNA methylation. In mammals, DNA methylation must be catalyzed by DNA methyltransferases, whether SMYD3 directly regulates DNMT expression, or SMYD3 changes the local conformation of RIZ1 promoter to facilitate DNMT congregation needs to be further investigated. Moreover, SMYD3 can di- and tri-methylate H3-K4, if its DNA methylation activity is dependent on H3-K4 transactivation, the H3-K4 hypermethylation patterns also needs to be established.
SMYD3, a H3K4 methyltransferase, was shown to be enhanced expressed in HCC, and involved in the growth of hepatocellular carcinoma (HCC) cells. Although transcriptional activation of downstream genes including Nkx2.8 and WNT10B gene was reported, the effect of SMYD3 on HCC development and the underlining mechanism remain unclear. RIZ1, one typical TSG with H3-K9 methyltransferase activity, was shown to lose its expression and tumor-suppressing activity in HCC, RIZ inactivation in HCC was mainly through its CpG promoter hypermethylation. In this article, whether SMYD3 regulates HCC proliferation, apoptosis and migration through RIZ1 promoter hypermethylation was initially investigated.
In previous studies, histone metylation were thought to be irreversible. However, in recent studies, histone demethylase was found, which means that histone can be hypermethylated or demethylated in regulating gene expression. Since histone modification play important epigenetic regulating roles in gene transcription, this may provide new target for carcinoma therapy through hypermethylating or demethylating histone.
This article suggests H3-T4 histone methyltransferase regulates HCC biology, not only through oncogene transcription, but through interacting with H3-K9 HMT which mainly plays inhibitory role in oncogene expression, and through inhibiting TSG expression by promoter hypermethylation. These findings deepen our understanding of the interaction of different histone modification ways in gene transcription modification.
Clarifying the regulating models of H3-K4 methyltransferase, such as SMYD3, represents a potentiality for understanding oncogene expression and tumor suppressive gene under-expression in carcinogenesis and progress. By deepened understanding the key role of SMYD3 in HCC development may also provide new target for carcinoma treatment.
SMYD3 SET and MYND domain-containing protein 3; RIZ1 retinoblastoma protein-interacting zinc finger gene1; TSG tumor suppressive gene; HMT Histone methyltransferase; DNMT DNA methyltransferases.
The manuscript describes studies showing that several HCC cell lines and one HCC tissue sample express SMYD3 and is well described.
S- Editor Ma N L- Editor Rippe RA E- Editor Yin DH
| 1. | Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2210] [Cited by in RCA: 2202] [Article Influence: 104.9] [Reference Citation Analysis (0)] |
| 2. | Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315-2335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 335] [Cited by in RCA: 340] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 3. | Yu J, Zhang H, Gu J, Lin S, Li J, Lu W, Wang Y, Zhu J. Methylation profiles of thirty four promoter-CpG islands and concordant methylation behaviours of sixteen genes that may contribute to carcinogenesis of astrocytoma. BMC Cancer. 2004;4:65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 95] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 4. | Alaminos M, Dávalos V, Ropero S, Setién F, Paz MF, Herranz M, Fraga MF, Mora J, Cheung NK, Gerald WL. EMP3, a myelin-related gene located in the critical 19q13.3 region, is epigenetically silenced and exhibits features of a candidate tumor suppressor in glioma and neuroblastoma. Cancer Res. 2005;65:2565-2571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 111] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 5. | Hamamoto R, Furukawa Y, Morita M, Iimura Y, Silva FP, Li M, Yagyu R, Nakamura Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat Cell Biol. 2004;6:731-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 549] [Cited by in RCA: 569] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 6. | Hamamoto R, Silva FP, Tsuge M, Nishidate T, Katagiri T, Nakamura Y, Furukawa Y. Enhanced SMYD3 expression is essential for the growth of breast cancer cells. Cancer Sci. 2006;97:113-118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 220] [Cited by in RCA: 219] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 7. | Frank B, Hemminki K, Wappenschmidt B, Klaes R, Meindl A, Schmutzler RK, Bugert P, Untch M, Bartram CR, Burwinkel B. Variable number of tandem repeats polymorphism in the SMYD3 promoter region and the risk of familial breast cancer. Int J Cancer. 2006;118:2917-2918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 8. | Jiang Gl, Liu L, Buyse IM, Simon D, Huang S. Decreased RIZ1 expression but not RIZ2 in hepatoma and suppression of hepatoma tumorigenicity by RIZ1. Int J Cancer. 1999;83:541-546. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 9. | Chadwick RB, Jiang GL, Bennington GA, Yuan B, Johnson CK, Stevens MW, Niemann TH, Peltomaki P, Huang S, de la Chapelle A. Candidate tumor suppressor RIZ is frequently involved in colorectal carcinogenesis. Proc Natl Acad Sci USA. 2000;97:2662-2667. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 121] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 10. | He L, Yu JX, Liu L, Buyse IM, Wang MS, Yang QC, Nakagawara A, Brodeur GM, Shi YE, Huang S. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58:4238-4244. [PubMed] |
| 11. | Suzuki M, Shigematsu H, Shivapurkar N, Reddy J, Miyajima K, Takahashi T, Gazdar AF, Frenkel EP. Methylation of apoptosis related genes in the pathogenesis and prognosis of prostate cancer. Cancer Lett. 2006;242:222-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 43] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 12. | Piao Z, Fang W, Malkhosyan S, Kim H, Horii A, Perucho M, Huang S. Frequent frameshift mutations of RIZ in sporadic gastrointestinal and endometrial carcinomas with microsatellite instability. Cancer Res. 2000;60:4701-4704. [PubMed] |
| 13. | Jiang GL, Huang S. Adenovirus expressing RIZ1 in tumor suppressor gene therapy of microsatellite-unstable colorectal cancers. Cancer Res. 2001;61:1796-1798. [PubMed] |
| 14. | Nishimura T, Nishida N, Itoh T, Komeda T, Fukuda Y, Ikai I, Yamaoka Y, Nakao K. Discrete breakpoint mapping and shortest region of overlap of chromosome arm 1q gain and 1p loss in human hepatocellular carcinoma detected by semiquantitative microsatellite analysis. Genes Chromosomes Cancer. 2005;42:34-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 24] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 15. | Fang W, Piao Z, Buyse IM, Simon D, Sheu JC, Perucho M, Huang S. Preferential loss of a polymorphic RIZ allele in human hepatocellular carcinoma. Br J Cancer. 2001;84:743-747. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 16. | Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006;5:37-50. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 952] [Cited by in RCA: 950] [Article Influence: 50.0] [Reference Citation Analysis (0)] |
| 17. | Apergis GA, Crawford N, Ghosh D, Steppan CM, Vorachek WR, Wen P, Locker J. A novel nk-2-related transcription factor associated with human fetal liver and hepatocellular carcinoma. J Biol Chem. 1998;273:2917-2925. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 43] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 18. | Kajiyama Y, Tian J, Locker J. Regulation of alpha-fetoprotein expression by Nkx2.8. Mol Cell Biol. 2002;22:6122-6130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 28] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Benhaj K, Akcali KC, Ozturk M. Redundant expression of canonical Wnt ligands in human breast cancer cell lines. Oncol Rep. 2006;15:701-707. [PubMed] |
| 20. | Kirikoshi H, Katoh M. Expression of WNT7A in human normal tissues and cancer, and regulation of WNT7A and WNT7B in human cancer. Int J Oncol. 2002;21:895-900. [PubMed] |
| 21. | Koike T, Kimura N, Miyazaki K, Yabuta T, Kumamoto K, Takenoshita S, Chen J, Kobayashi M, Hosokawa M, Taniguchi A. Hypoxia induces adhesion molecules on cancer cells: A missing link between Warburg effect and induction of selectin-ligand carbohydrates. Proc Natl Acad Sci USA. 2004;101:8132-8137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 187] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 22. | Deprez P, Inestrosa NC, Krejci E. Two different heparin-binding domains in the triple-helical domain of ColQ, the collagen tail subunit of synaptic acetylcholinesterase. J Biol Chem. 2003;278:23233-23242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 23. | Johansson FK, Göransson H, Westermark B. Expression analysis of genes involved in brain tumor progression driven by retroviral insertional mutagenesis in mice. Oncogene. 2005;24:3896-3905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 24. | Kondoh N, Wakatsuki T, Ryo A, Hada A, Aihara T, Horiuchi S, Goseki N, Matsubara O, Takenaka K, Shichita M. Identification and characterization of genes associated with human hepatocellular carcinogenesis. Cancer Res. 1999;59:4990-4996. [PubMed] |
| 25. | Frachon S, Gouysse G, Dumortier J, Couvelard A, Nejjari M, Mion F, Berger F, Paliard P, Boillot O, Scoazec JY. Endothelial cell marker expression in dysplastic lesions of the liver: an immunohistochemical study. J Hepatol. 2001;34:850-857. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 57] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 26. | Ricketts SL, Carter JC, Coleman WB. Identification of three 11p11.2 candidate liver tumor suppressors through analysis of known human genes. Mol Carcinog. 2003;36:90-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 27. | Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 471] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 28. | Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 927] [Cited by in RCA: 883] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 29. | Johnson L, Cao X, Jacobsen S. Interplay between two epigenetic marks. DNA methylation and histone H3 lysine 9 methylation. Curr Biol. 2002;12:1360-1367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 342] [Article Influence: 14.9] [Reference Citation Analysis (0)] |