Published online Sep 28, 2007. doi: 10.3748/wjg.v13.i36.4818
Revised: July 2, 2007
Accepted: July 9, 2007
Published online: September 28, 2007
Hepatitis C virus (HCV) needs to tightly manipulate host defences in order to establish infection. The innate immune response slows down viral replication by activating cytokines such as the type I interferons (IFN-α/β), which trigger the synthesis of antiviral proteins and modulate the adaptive immune system. HCV has therefore developed a number of countermeasures to stay ahead of the IFN system. Here, I will attempt to summarize the current state of research regarding IFN responses against HCV and the viral escape strategies. Particular emphasis will be put on the newly discovered mechanisms HCV employs to avoid the induction of IFN in infected cells.
- Citation: Weber F. Interaction of hepatitis C virus with the type I interferon system. World J Gastroenterol 2007; 13(36): 4818-4823
- URL: https://www.wjgnet.com/1007-9327/full/v13/i36/4818.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i36.4818
The type I interferon system which mainly involves IFN-α and -β is a powerful and universal intracellular defence system against viruses. Knockout mice which are unresponsive to IFN-α/β due to targeted deletions in the type I IFN receptor quickly succumb to viral infections although they have a regular adaptive immune system[1,2]. Likewise, humans with genetic defects in STAT-1, which is involved in the signaling cascade of the IFN system, die of viral disease at an early age[3].
All nucleated cells of the mammalian body are able to synthesize and secrete type I IFNs in response to virus infection. Secreted IFNs are then recognized by neighboring cells and cause them to express potent antiviral proteins[4,5]. As a result, virus multiplication is slowed down or even stopped, and the organism buys time for the establishment of an adaptive immune response.
Type I IFNs are classified according to their amino acid sequence and comprise a large number (at least 13) of IFN-α subtypes and a single IFN-β[6], as well as some additional family members[7,8]. Expression patterns, i.e. which IFNs will be synthesized at which time point, mostly depend on the particular cell type.
Fibroblasts secrete mainly IFN-β as an initial response to infection but switch to IFN-α during the subsequent amplification phase of the IFN response[9]. By contrast, dendritic cells, which play an important role in immunosurveillance, directly secrete high levels of IFN-α subtypes[10,11].
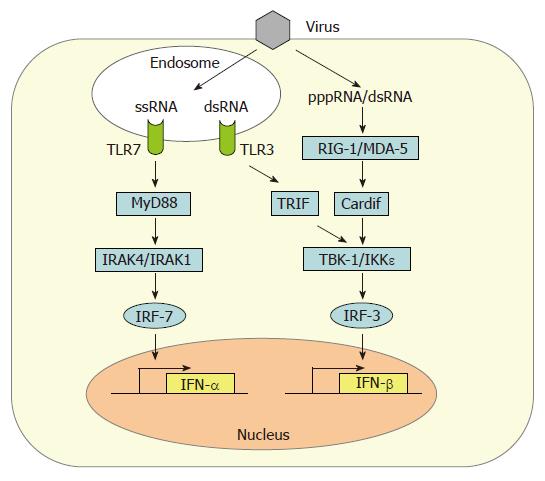
Induction of IFN-β gene expression in fibroblasts occurs by the intracellular, so-called “classic pathway” (Figure 1). In infected cells, a signaling chain is activated by viral RNA molecules which are generated during genome transcription and replication[12]. Two intracellular RNA helicases, RIG-I[13] and MDA5[14], act as sentinels for viral RNA[15-17]. Then, a recently discovered adaptor protein binds to RIG-I and MDA5 and mediates the signal to downstream factors. It is called either Cardif for “CARD adaptor inducing IFN-β”[18], IPS-1 for “interferon-β-promoter stimulator 1”[19], MAVS for “mitochondrial antiviral signaling” molecule[20], or VISA for “virus-induced signaling adaptor”[21]. Cardif/IPS-1/MAVS/VISA activates two IκB kinase (IKK)-related kinases, IKKε and TANK-binding kinase-1 (TBK-1), which phosphorylate the transcription factor IRF-3[22,23]. IRF-3 is a member of the IFN regulatory factor (IRF) family and plays a central role in the activation of the IFN-β promoter[24]. Phosphorylated IRF-3 homo-dimerizes and moves into the nucleus where it recruits the transcriptional coactivators p300 and CREB-binding protein (CBP) to initiate IFN-β mRNA synthesis[24,25]. This first-wave IFN triggers expression of a related factor, IRF-7, which in fibroblasts is only present in low amounts[26]. IRF-7 can be activated the same way as IRF-3[27-29], leading to a positive-feedback loop that initiates the synthesis of several IFN-α subtypes as the second-wave IFNs[9,30]. In addition, NF-κB and AP-1 are recruited in a dsRNA-dependent way[31,32]. Together these transcription factors strongly upregulate IFN-β gene expression.
Until very recently, it was assumed that the main trigger of intracellular cytokine induction by all viruses is double-stranded RNA (dsRNA) which supposedly forms as a by-product of genome replication. However, we have recently found that some viruses do not produce substantial amounts of dsRNA[33]. Instead, ssRNA containing a 5’ triphosphate group is much more potent than dsRNA in activating RIG-I-dependent IFN induction[34-36].
Among the cells of the lymphatic system, myeloid dendritic cells (mDCs)[11] and, most prominently, plasmacytoid dendritic cells (pDCs)[10] are the main IFN producers. In addition to the classical, intracellular pathway of IFN induction described above, pDCs sense the presence of viruses by the extracytoplasmic toll-like receptors (TLRs)[37-39]. It is thought that TLRs serve as sensors for viral infection of phagocytosed cells[40]. Human pDCs mostly express TLR7 and TLR9 which recognize viral single-stranded(ss) RNA and dsDNA, respectively[41], whereas mDCs express TLR3 which responds to dsRNA[42]. Upon activation, TLRs signal through different intracellular adaptor molecules such as MyD88 (TLR7 and 9) or TRIF (TLR3) to induce IFN transcription[41]. Interestingly, DCs already contain high levels of IRF-7[43,44], thus explaining their ability to rapidly produce high amounts of alpha-IFNs. Furthermore, TLR7 and TLR9 are retained in the endosomes of pDCs to allow prolonged IFN induction signaling[45].
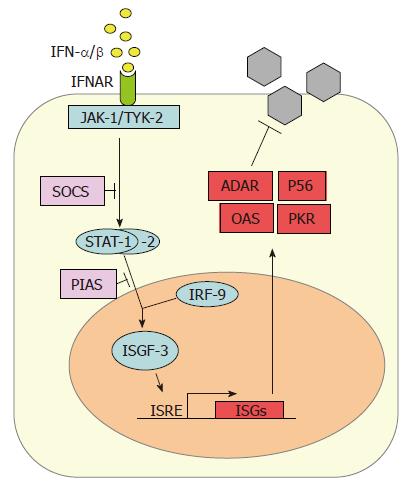
IFN-α/β subtypes all bind to and activate a common type I IFN receptor. It consists of two subunits (IFNAR-1 and IFNAR-2) and is present on virtually all host cells[5,6]. Binding of IFN-α/β leads to heterodimerization of the IFNAR subunits and to conformational changes in the intracellular parts of the receptor which activate the so-called JAK-STAT signaling pathway (Figure 2). The signal transducer and activator of transcription (STAT) proteins are latent cytoplasmic transcription factors which become phosphorylated by the Janus kinase (JAK) family members JAK-1 and TYK-2[46]. Phosphorylated STAT-1 and STAT-2 recruit a third factor, IRF-9 (also called p48), to form a complex known as IFN stimulated gene factor 3 (ISGF-3). The ISGF-3 heterotrimer translocates to the nucleus and binds to IFN-stimulated response elements (ISRE) in the promoter regions of IFN-stimulated genes (ISGs), thereby inducing their transcription.
Several specialized proteins serve as negative regulators and inhibitors of the JAK-STAT pathway. For example, the suppressor of cytokine signaling (SOCS) proteins specifically prevent STAT activation by binding to activated cytokine receptors, inhibiting the activity of JAKs, and targeting bound signaling proteins for proteasomal degradation[47]. Also, the protein inhibitor of activated STAT (PIAS) family members function as small ubiquitin-like modifier (SUMO) E3 ligases and inhibit the transcriptional activity of STATs[48].
IFN-α combined with ribavirin is the standard treatment for HCV infection, and its effect can be potentiated by co-adminitration of IFN-γ[49,50]. IFN-α/β activates the expression of more than 300 IFN-stimulated genes (ISGs) which have antiviral, antiproliferative, and immunomodulatory functions[51,52]. IFN-induced proteins include enzymes, transcription factors, cell surface glycoproteins, cytokines, chemokines and a large number of factors that need to be further characterized. Up to now, only a few antiviral proteins have been characterized in detail. Type I IFNs are known to be effective against HCV replicon systems[53,54], and several IFN-induced proteins have documented anti-HCV activity, namely protein kinase R (PKR)[55], the RNA-specific adenosine deaminase 1 (ADAR 1)[56], the 2’-5’ oligoadenylate synthetases (2-5 OAS) / RNaseL system[57], and P56[58].
PKR, ADAR1, and 2-5 OAS are constitutively expressed in normal cells in a latent, inactive form. Basal mRNA levels are upregulated by IFN-α/β and these enzymes need to be activated by viral dsRNA. PKR is a serine-threonine kinase that phosphorylates the alpha subunit of the eukaryotic translation initiation factor eIF2[59]. As a consequence, translation of cellular and viral mRNAs is blocked. ADAR 1 catalyzes the deamination of adenosine on target dsRNAs to yield inosine. As a result the secondary structure is destabilized due to a change from an AU base pair to the less stable IU base pair and mutations accumulate within the viral genome[5]. The 2-5 OAS catalyzes the synthesis of short 2’-5’ oligoadenylates that activate the latent endoribonuclease RNaseL[60]. RNaseL, in turn, then degrades both viral and cellular RNAs, leading to viral inhibition[61]. P56 binds the eukaryotic initiation factor 3e (eIF3e) subunit of the eukaryotic translation initiation factor eIF3. It functions as an inhibitor of translation initiation at the level of eIF3 ternary complex formation and is likely to suppress viral RNA translation[62,63].
Several recent studies have clarified that the RNA of HCV is a potent trigger of IFN induction, leading to the establishment of an antiviral state. Therefore, in order to establish infection and to persist in the human host, HCV has been forced to evolve efficient counterstrategies. Intracellular IFN induction by HCV appears to be mostly mediated by RIG-I binding to viral RNA[64]. Extracellularly, no specific TLR has been identified yet, but by deduction from data on related flaviviruses, TLR3 and TLR7 would be the most obvious candidates. The dsRNA-binding TLR3 was shown to be activated by West Nile virus[65], and the ssRNA-binding TLR7 is activated by Dengue virus[66]. Moreover, TLR7 can elicit HCV immunity, and a synthetic TLR7 agonist reduced HCV mRNA and protein levels in HuH-7 hepatocytes[67]. It is important to note that TRL7 is expressed in hepatocytes of normal as well as HCV-infected people[67]. Thus, TRL7 may indeed play a role during natural infection.
On the other hand, HCV is capable of disturbing the IFN response at multiple levels[68,69]. With respect to IFN induction, it was recently discovered that the NS3/4A protease specifically cleaves Cardif[18] as well as TRIF[70,71]. Since both these adaptor proteins are important for IFN induction via the classical intracellular pathway (Cardif) and the TLR3-driven endosomal pathway (TRIF), NS3/4A is the key factor of HCV to disturb IRF-3 activation[72] which would otherwise result in IFN gene transcription. In addition, NS3 directly interacts with TBK1 to inhibit its association with IRF-3 and its activation[73].
With respect to the IFN response, it was shown that expression of the full-length virus genome or the core protein suppresses IFN signal transduction[74,75]. Most likely, this is due to an up-regulation of protein phosphatase 2A by ER stress[76], resulting in association of STAT1 with its inhibitor PIAS1[77]. Moreover, for the core protein it was shown that it interferes with the JAK/STAT pathway[78] and is able to activate the JAK-STAT signaling inhibitor SOCS-3[79], further contributing to the HCV-induced block of IFN signaling.
HCV also directly counteracts the antiviral IFN response. The NS5A protein, which confers a multitude of functions in virus replication[80], also plays a key role in escape from the antiviral action of IFN. A stretch of 40 amino acids on NS5A, termed the IFN sensitivity region (ISDR), was correlated with responsiveness to IFN therapy[81-83]. Moreover, NS5A was shown to directly bind to and repress PKR, and this interaction involved the ISDR[84]. However, other groups did not find a connection between viral IFN susceptibility and a particular ISDR sequence[85-87], and PKR activity was not affected by expression of the HCV genome[88] or NS5A[89], although NS5A clearly reduced the antiviral effects of IFN[89]. A possible solution for this discrepancy could be that ISDR sequence variations affect the efficiency of HCV replication[90,91]. Thus, the correlation between particular ISDR sequences and IFN sensitivity could be caused by differences in HCV replication strength. In addition, NS5A induces IL-8 (also termed CXCL-8), a chemokine which inhibits the antiviral actions of IFN[92]. Elevated IL-8 levels were indeed detected in the sera of IFN non-responders[93]. Moreover, in cell culture CXCL-8 protein levels are positively associated with chronic HCV replication and CXCL-8 removal inhibits HCV replication[94]. Interestingly, CXCL-8 cannot only be induced by NS5A, but also by the HCV RNA-sensitive RIG-I pathway[95].
NS5A also interferes with the 2-5 OAS/RNaseL pathway by binding to 2-5 OAS[96]. Furthermore, the HCV genome sequences of IFN-resistant strains have fewer RNase L recognition sites than those of more IFN-sensitive ones[97], thus allowing escape from nucleolytic cleavage[97]. PKR activity is also modified by the internal ribosome entry site (IRES) of HCV[98] and the E2 protein[99].
The multiple countermeasures of HCV to avoid a fully-fledged IFN response appear to be quite efficient, since 85% of the HCV-infected patients develop a chronic infection, and up to 60% of those patients do not respond to IFN therapy or experience a relapse when therapy is stopped[100]. Our rapidly increasing knowledge about HCV immune escape will certainly lead to a significant improvement in both prevention and therapy for hepatitis C.
S- Editor Ma N L- Editor Roberts SE E- Editor Ma WH
| 1. | Müller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, Aguet M. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918-1921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1978] [Cited by in RCA: 2006] [Article Influence: 64.7] [Reference Citation Analysis (0)] |
| 2. | Weber F, Kochs G, Haller O. Inverse interference: how viruses fight the interferon system. Viral Immunol. 2004;17:498-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 160] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 3. | Dupuis S, Jouanguy E, Al-Hajjar S, Fieschi C, Al-Mohsen IZ, Al-Jumaah S, Yang K, Chapgier A, Eidenschenk C, Eid P. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33:388-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 628] [Cited by in RCA: 621] [Article Influence: 28.2] [Reference Citation Analysis (0)] |
| 4. | Haller O, Kochs G, Weber F. The interferon response circuit: induction and suppression by pathogenic viruses. Virology. 2006;344:119-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 510] [Cited by in RCA: 527] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 5. | Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778-809, table of contents. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2029] [Cited by in RCA: 2003] [Article Influence: 83.5] [Reference Citation Analysis (0)] |
| 6. | Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3048] [Cited by in RCA: 3080] [Article Influence: 114.1] [Reference Citation Analysis (0)] |
| 7. | Roberts RM, Ezashi T, Rosenfeld CS, Ealy AD, Kubisch HM. Evolution of the interferon tau genes and their promoters, and maternal-trophoblast interactions in control of their expression. Reprod Suppl. 2003;61:239-251. [PubMed] |
| 8. | van Pesch V, Lanaya H, Renauld JC, Michiels T. Characterization of the murine alpha interferon gene family. J Virol. 2004;78:8219-8228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 166] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 9. | Marié I, Durbin JE, Levy DE. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660-6669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 833] [Cited by in RCA: 840] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 10. | Colonna M, Krug A, Cella M. Interferon-producing cells: on the front line in immune responses against pathogens. Curr Opin Immunol. 2002;14:373-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 181] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 11. | Diebold SS, Montoya M, Unger H, Alexopoulou L, Roy P, Haswell LE, Al-Shamkhani A, Flavell R, Borrow P, Reis e Sousa C. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature. 2003;424:324-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 443] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 12. | Bowie AG, Fitzgerald KA. RIG-I: tri-ing to discriminate between self and non-self RNA. Trends Immunol. 2007;28:147-150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 46] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730-737. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2934] [Cited by in RCA: 3122] [Article Influence: 148.7] [Reference Citation Analysis (0)] |
| 14. | Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264-17269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 788] [Cited by in RCA: 806] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 15. | Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O. Cell type-specific involvement of RIG-I in antiviral response. Immunity. 2005;23:19-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1024] [Cited by in RCA: 1059] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 16. | Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M, Akira S. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851-2858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1264] [Cited by in RCA: 1281] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 17. | Yoneyama M, Fujita T. Function of RIG-I-like receptors in antiviral innate immunity. J Biol Chem. 2007;282:15315-15318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 232] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 18. | Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167-1172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1976] [Cited by in RCA: 1921] [Article Influence: 96.1] [Reference Citation Analysis (0)] |
| 19. | Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981-988. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2018] [Cited by in RCA: 2056] [Article Influence: 102.8] [Reference Citation Analysis (0)] |
| 20. | Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2373] [Cited by in RCA: 2700] [Article Influence: 135.0] [Reference Citation Analysis (0)] |
| 21. | Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727-740. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1529] [Cited by in RCA: 1554] [Article Influence: 77.7] [Reference Citation Analysis (0)] |
| 22. | Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491-496. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2013] [Cited by in RCA: 2185] [Article Influence: 99.3] [Reference Citation Analysis (0)] |
| 23. | Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148-1151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1317] [Cited by in RCA: 1347] [Article Influence: 61.2] [Reference Citation Analysis (0)] |
| 24. | Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282:15325-15329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 374] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 25. | Suhara W, Yoneyama M, Kitabayashi I, Fujita T. Direct involvement of CREB-binding protein/p300 in sequence-specific DNA binding of virus-activated interferon regulatory factor-3 holocomplex. J Biol Chem. 2002;277:22304-22313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 93] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1074] [Cited by in RCA: 1115] [Article Influence: 44.6] [Reference Citation Analysis (0)] |
| 27. | Iwamura T, Yoneyama M, Yamaguchi K, Suhara W, Mori W, Shiota K, Okabe Y, Namiki H, Fujita T. Induction of IRF-3/-7 kinase and NF-kappaB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells. 2001;6:375-388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 235] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 28. | tenOever BR, Sharma S, Zou W, Sun Q, Grandvaux N, Julkunen I, Hemmi H, Yamamoto M, Akira S, Yeh WC. Activation of TBK1 and IKKvarepsilon kinases by vesicular stomatitis virus infection and the role of viral ribonucleoprotein in the development of interferon antiviral immunity. J Virol. 2004;78:10636-10649. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 158] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 29. | Smith EJ, Marié I, Prakash A, García-Sastre A, Levy DE. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or Ikappa B kinase but is blocked by Vaccinia virus E3L protein. J Biol Chem. 2001;276:8951-8957. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 190] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 30. | Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 455] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 31. | Chu WM, Ostertag D, Li ZW, Chang L, Chen Y, Hu Y, Williams B, Perrault J, Karin M. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity. 1999;11:721-731. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 306] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 32. | Yang YL, Reis LF, Pavlovic J, Aguzzi A, Schäfer R, Kumar A, Williams BR, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6095-6106. [PubMed] |
| 33. | Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. J Virol. 2006;80:5059-5064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 698] [Cited by in RCA: 763] [Article Influence: 40.2] [Reference Citation Analysis (0)] |
| 34. | Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M. 5'-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994-997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1879] [Cited by in RCA: 1898] [Article Influence: 99.9] [Reference Citation Analysis (0)] |
| 35. | Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science. 2006;314:997-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1735] [Cited by in RCA: 1735] [Article Influence: 91.3] [Reference Citation Analysis (0)] |
| 36. | Plumet S, Herschke F, Bourhis JM, Valentin H, Longhi S, Gerlier D. Cytosolic 5'-triphosphate ended viral leader transcript of measles virus as activator of the RIG I-mediated interferon response. PLoS One. 2007;2:e279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 149] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 37. | Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1111] [Cited by in RCA: 1124] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 38. | Bowie AG, Haga IR. The role of Toll-like receptors in the host response to viruses. Mol Immunol. 2005;42:859-867. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 178] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 39. | Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319-15323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 339] [Cited by in RCA: 377] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 40. | Schulz O, Diebold SS, Chen M, Näslund TI, Nolte MA, Alexopoulou L, Azuma YT, Flavell RA, Liljeström P, Reis e Sousa C. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature. 2005;433:887-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 686] [Cited by in RCA: 699] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 41. | Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987-995. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2991] [Cited by in RCA: 3052] [Article Influence: 145.3] [Reference Citation Analysis (0)] |
| 42. | Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4510] [Cited by in RCA: 4639] [Article Influence: 193.3] [Reference Citation Analysis (0)] |
| 43. | Kerkmann M, Rothenfusser S, Hornung V, Towarowski A, Wagner M, Sarris A, Giese T, Endres S, Hartmann G. Activation with CpG-A and CpG-B oligonucleotides reveals two distinct regulatory pathways of type I IFN synthesis in human plasmacytoid dendritic cells. J Immunol. 2003;170:4465-4474. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 268] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 44. | Prakash A, Smith E, Lee CK, Levy DE. Tissue-specific positive feedback requirements for production of type I interferon following virus infection. J Biol Chem. 2005;280:18651-18657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 88] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 45. | Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, Takaoka A, Taya C, Taniguchi T. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434:1035-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 693] [Cited by in RCA: 737] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 46. | Levy DE, Darnell JE. Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651-662. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2306] [Cited by in RCA: 2456] [Article Influence: 106.8] [Reference Citation Analysis (0)] |
| 47. | Kubo M, Hanada T, Yoshimura A. Suppressors of cytokine signaling and immunity. Nat Immunol. 2003;4:1169-1176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 487] [Cited by in RCA: 502] [Article Influence: 23.9] [Reference Citation Analysis (0)] |
| 48. | Shuai K, Liu B. Regulation of gene-activation pathways by PIAS proteins in the immune system. Nat Rev Immunol. 2005;5:593-605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 321] [Cited by in RCA: 334] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 49. | Katayama K, Kasahara A, Sasaki Y, Kashiwagi T, Naito M, Masuzawa M, Katoh M, Yoshihara H, Kamada T, Mukuda T. Immunological response to interferon-gamma priming prior to interferon-alpha treatment in refractory chronic hepatitis C in relation to viral clearance. J Viral Hepat. 2001;8:180-185. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 50. | Okuse C, Rinaudo JA, Farrar K, Wells F, Korba BE. Enhancement of antiviral activity against hepatitis C virus in vitro by interferon combination therapy. Antiviral Res. 2005;65:23-34. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 51. | de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH, Williams BR. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912-920. [PubMed] |
| 52. | Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623-15628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1479] [Cited by in RCA: 1477] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 53. | Guo JT, Bichko VV, Seeger C. Effect of alpha interferon on the hepatitis C virus replicon. J Virol. 2001;75:8516-8523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 383] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 54. | Frese M, Pietschmann T, Moradpour D, Haller O, Bartenschlager R. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J Gen Virol. 2001;82:723-733. [PubMed] |
| 55. | Pflugheber J, Fredericksen B, Sumpter R, Wang C, Ware F, Sodora DL, Gale M. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication. Proc Natl Acad Sci USA. 2002;99:4650-4655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 143] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 56. | Taylor DR, Puig M, Darnell ME, Mihalik K, Feinstone SM. New antiviral pathway that mediates hepatitis C virus replicon interferon sensitivity through ADAR1. J Virol. 2005;79:6291-6298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 135] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 57. | Guo JT, Sohn JA, Zhu Q, Seeger C. Mechanism of the interferon alpha response against hepatitis C virus replicons. Virology. 2004;325:71-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 58. | Wang C, Pflugheber J, Sumpter R, Sodora DL, Hui D, Sen GC, Gale M. Alpha interferon induces distinct translational control programs to suppress hepatitis C virus RNA replication. J Virol. 2003;77:3898-3912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 181] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 59. | Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112-6120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 627] [Cited by in RCA: 641] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 60. | Silverman RH. Fascination with 2-5A-dependent RNase: a unique enzyme that functions in interferon action. J Interferon Res. 1994;14:101-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 71] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 61. | Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C. Interferon action and apoptosis are defective in mice devoid of 2',5'-oligoadenylate-dependent RNase L. EMBO J. 1997;16:6355-6363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 445] [Cited by in RCA: 440] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 62. | Hui DJ, Bhasker CR, Merrick WC, Sen GC. Viral stress-inducible protein p56 inhibits translation by blocking the interaction of eIF3 with the ternary complex eIF2.GTP.Met-tRNAi. J Biol Chem. 2003;278:39477-39482. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 123] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 63. | Terenzi F, Pal S, Sen GC. Induction and mode of action of the viral stress-inducible murine proteins, P56 and P54. Virology. 2005;340:116-124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 56] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 64. | Sumpter R, Loo YM, Foy E, Li K, Yoneyama M, Fujita T, Lemon SM, Gale M. Regulating intracellular antiviral defense and permissiveness to hepatitis C virus RNA replication through a cellular RNA helicase, RIG-I. J Virol. 2005;79:2689-2699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 736] [Cited by in RCA: 724] [Article Influence: 36.2] [Reference Citation Analysis (0)] |
| 65. | Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10:1366-1373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 819] [Cited by in RCA: 846] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 66. | Wang JP, Liu P, Latz E, Golenbock DT, Finberg RW, Libraty DH. Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. J Immunol. 2006;177:7114-7121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 152] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 67. | Lee J, Wu CC, Lee KJ, Chuang TH, Katakura K, Liu YT, Chan M, Tawatao R, Chung M, Shen C. Activation of anti-hepatitis C virus responses via Toll-like receptor 7. Proc Natl Acad Sci USA. 2006;103:1828-1833. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 175] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 68. | Gale M, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 469] [Cited by in RCA: 455] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 69. | Thimme R, Lohmann V, Weber F. A target on the move: innate and adaptive immune escape strategies of hepatitis C virus. Antiviral Res. 2006;69:129-141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 86] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 70. | Breiman A, Grandvaux N, Lin R, Ottone C, Akira S, Yoneyama M, Fujita T, Hiscott J, Meurs EF. Inhibition of RIG-I-dependent signaling to the interferon pathway during hepatitis C virus expression and restoration of signaling by IKKepsilon. J Virol. 2005;79:3969-3978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 135] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 71. | Li K, Foy E, Ferreon JC, Nakamura M, Ferreon AC, Ikeda M, Ray SC, Gale M, Lemon SM. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA. 2005;102:2992-2997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 847] [Cited by in RCA: 817] [Article Influence: 40.9] [Reference Citation Analysis (0)] |
| 72. | Foy E, Li K, Wang C, Sumpter R, Ikeda M, Lemon SM, Gale M. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science. 2003;300:1145-1148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 624] [Cited by in RCA: 597] [Article Influence: 27.1] [Reference Citation Analysis (0)] |
| 73. | Otsuka M, Kato N, Moriyama M, Taniguchi H, Wang Y, Dharel N, Kawabe T, Omata M. Interaction between the HCV NS3 protein and the host TBK1 protein leads to inhibition of cellular antiviral responses. Hepatology. 2005;41:1004-1012. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 95] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 74. | Heim MH, Moradpour D, Blum HE. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J Virol. 1999;73:8469-8475. [PubMed] |
| 75. | Melén K, Fagerlund R, Nyqvist M, Keskinen P, Julkunen I. Expression of hepatitis C virus core protein inhibits interferon-induced nuclear import of STATs. J Med Virol. 2004;73:536-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 78] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 76. | Christen V, Treves S, Duong FH, Heim MH. Activation of endoplasmic reticulum stress response by hepatitis viruses up-regulates protein phosphatase 2A. Hepatology. 2007;46:558-565. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 103] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 77. | Duong FH, Filipowicz M, Tripodi M, La Monica N, Heim MH. Hepatitis C virus inhibits interferon signaling through up-regulation of protein phosphatase 2A. Gastroenterology. 2004;126:263-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 161] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 78. | de Lucas S, Bartolome J, Carreno V. Hepatitis C virus core protein down-regulates transcription of interferon-induced antiviral genes. J Infect Dis. 2005;191:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 79. | Bode JG, Ludwig S, Ehrhardt C, Albrecht U, Erhardt A, Schaper F, Heinrich PC, Häussinger D. IFN-alpha antagonistic activity of HCV core protein involves induction of suppressor of cytokine signaling-3. FASEB J. 2003;17:488-490. [PubMed] |
| 80. | Macdonald A, Harris M. Hepatitis C virus NS5A: tales of a promiscuous protein. J Gen Virol. 2004;85:2485-2502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 288] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 81. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995;96:224-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 452] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 82. | Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77-81. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 752] [Cited by in RCA: 734] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 83. | Witherell GW, Beineke P. Statistical analysis of combined substitutions in nonstructural 5A region of hepatitis C virus and interferon response. J Med Virol. 2001;63:8-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 84. | Gale M, Blakely CM, Kwieciszewski B, Tan SL, Dossett M, Tang NM, Korth MJ, Polyak SJ, Gretch DR, Katze MG. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208-5218. [PubMed] |
| 85. | Aizaki H, Saito S, Ogino T, Miyajima N, Harada T, Matsuura Y, Miyamura T, Kohase M. Suppression of interferon-induced antiviral activity in cells expressing hepatitis C virus proteins. J Interferon Cytokine Res. 2000;20:1111-1120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 86. | Aus dem Siepen M, Lohmann V, Wiese M, Ross S, Roggendorf M, Viazov S. Nonstructural protein 5A does not contribute to the resistance of hepatitis C virus replication to interferon alpha in cell culture. Virology. 2005;336:131-136. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 87. | Paterson M, Laxton CD, Thomas HC, Ackrill AM, Foster GR. Hepatitis C virus NS5A protein inhibits interferon antiviral activity, but the effects do not correlate with clinical response. Gastroenterology. 1999;117:1187-1197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 88. | François C, Duverlie G, Rebouillat D, Khorsi H, Castelain S, Blum HE, Gatignol A, Wychowski C, Moradpour D, Meurs EF. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J Virol. 2000;74:5587-5596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 105] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 89. | Podevin P, Sabile A, Gajardo R, Delhem N, Abadie A, Lozach PY, Beretta L, Bréchot C. Expression of hepatitis C virus NS5A natural mutants in a hepatocytic cell line inhibits the antiviral effect of interferon in a PKR-independent manner. Hepatology. 2001;33:1503-1511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 56] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 90. | Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972-1974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1154] [Cited by in RCA: 1150] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 91. | Appel N, Pietschmann T, Bartenschlager R. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J Virol. 2005;79:3187-3194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 191] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 92. | Polyak SJ, Khabar KS, Paschal DM, Ezelle HJ, Duverlie G, Barber GN, Levy DE, Mukaida N, Gretch DR. Hepatitis C virus nonstructural 5A protein induces interleukin-8, leading to partial inhibition of the interferon-induced antiviral response. J Virol. 2001;75:6095-6106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 239] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 93. | Polyak SJ, Khabar KS, Rezeiq M, Gretch DR. Elevated levels of interleukin-8 in serum are associated with hepatitis C virus infection and resistance to interferon therapy. J Virol. 2001;75:6209-6211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 174] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 94. | Koo BC, McPoland P, Wagoner JP, Kane OJ, Lohmann V, Polyak SJ. Relationships between hepatitis C virus replication and CXCL-8 production in vitro. J Virol. 2006;80:7885-7893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 95. | Wagoner J, Austin M, Green J, Imaizumi T, Casola A, Brasier A, Khabar KS, Wakita T, Gale M, Polyak SJ. Regulation of CXCL-8 (interleukin-8) induction by double-stranded RNA signaling pathways during hepatitis C virus infection. J Virol. 2007;81:309-318. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 58] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 96. | Taguchi T, Nagano-Fujii M, Akutsu M, Kadoya H, Ohgimoto S, Ishido S, Hotta H. Hepatitis C virus NS5A protein interacts with 2',5'-oligoadenylate synthetase and inhibits antiviral activity of IFN in an IFN sensitivity-determining region-independent manner. J Gen Virol. 2004;85:959-969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 92] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 97. | Han JQ, Barton DJ. Activation and evasion of the antiviral 2'-5' oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA. 2002;8:512-525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 85] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 98. | Vyas J, Elia A, Clemens MJ. Inhibition of the protein kinase PKR by the internal ribosome entry site of hepatitis C virus genomic RNA. RNA. 2003;9:858-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 99. | Taylor DR, Shi ST, Romano PR, Barber GN, Lai MM. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 548] [Cited by in RCA: 530] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 100. | Pawlotsky JM. The nature of interferon-alpha resistance in hepatitis C virus infection. Curr Opin Infect Dis. 2003;16:587-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |