Published online Aug 28, 2007. doi: 10.3748/wjg.v13.i32.4350
Revised: April 23, 2007
Accepted: April 26, 2007
Published online: August 28, 2007
AIM: To explore the genetic diversities of UL144 open reading frame (ORF) of cytomegalovirus DNA detected in colon tissue from infants with Hirschsprung’s disease (HD) by sequencing UL144 DNA in 23 aganglionic colon tissue and 4 urine samples from 25 HD infants.
METHODS: Nest PCR was performed for amplification of the UL144 gene. The UL144 gene was analyzed with softwares, such as DNAclub, BioEdit, PROSITE database, and DNAstar.
RESULTS: The strains from HD patients were distributed among three genotypes of UL144: group 1A (64%), group 2 (24%), and group 3 (12%). The UL144 genotypes between strains from HD and control group were compared by chi square test (χ2 = 1.870, P = 0.393). Strains from the colon were sporadically distributed in UL144 genotypes.
CONCLUSION: There are genetic diversities of UL144 ORF in colon tissue of infants with HD. However, cytomegalovirus UL144 genotypes are not associated with clinical manifestations of HD.
- Citation: Mao ZQ, Huang Y, Sun M, Ruan Q, Qi Y, He R, Huang YJ, Ma YP, Ji YH, Sun ZR, Gao H. Genetic polymorphism of UL144 open reading frame of human cytomegalovirus DNA detected in colon samples from infants with Hirschsprung’s disease. World J Gastroenterol 2007; 13(32): 4350-4354
- URL: https://www.wjgnet.com/1007-9327/full/v13/i32/4350.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i32.4350
Human cytomegalovirus (HCMV) is composed of a large and complicated genome. HCMV infection can cause various clinical manifestations, such as jaundice, hepatomegaly, splenomegaly, purpura, pneumonia, central nervous system impairment. HCMV infection is mild or asymptomatic in immunocompetent hosts. However, serious outcomes may occur in immunocompromised hosts and congenitally-infected infants. It has been shown that glycoprotein B (gB) is associated with the severity and/or tissue-tropism of human cytomegalovirus-infected hosts[1-6], other studies have shown contradictory results[7-9]. HCMV UL144 gene encodes a transmembrane glycoprotein and is a homologue of the herpes simplex virus entry mediator (HveA or HveM), a member of tumor necrosis factor receptor (TNFR) superfamily[10]. Therefore, the locus may play an important role in determining the biological behavior of wild-type HCMV strains. Lurain et al[11] have shown significant strain-specific sequence variability of UL144 by sequencing 45 low passaged clinical isolates from solid organism graft and AIDS adults. The nucleotide sequences of cytomegalovirus UL144 gene can be divided into group 1, group 2 and group 3. Group 1 can be divided into three distinct subgroups (A, B and C). Bale et al[12] have confirmed these results by using isolates from 28 healthy children and 16 congenitally-infected infants subsequently, suggesting that the UL144 genotype is associated with neither the outcomes of intrauterine HCMV infection nor the development and severity of HCMV disease at birth. However, Arav-Bogger et al[13] reported different results with those of Bale and Lurain.
Hirschsprung’s disease (HD) is one of the common malformations in children. Its etiology is obscured. It was reported that congenital HCMV infection is correlated with development of HD[14-17]. To further understand the relationship between genetic diversities of HCMV UL144 ORF in colon samples and clinical outcomes of HD, we sequenced UL144 DNA in 23 aganglionic colon tissue and 4 urine samples from 25 HD infants.
Fifty-five infants with Hirschsprung’s disease were selected from July 2001 to May 2004. The youngest infant was 19- d old and the eldest one was 1.5- year old. Their average age was 8.71 ± 6.8 mo on operation. All patients were diagnosed as HD based on clinical analysis, barium enema examination, and biopsy specimens. HE staining showed that nerve plexus proliferated and no ganglion cells were found on colon mucous membrane under light microscope. Veins became large and congested, and leukomonocyte infiltration was observed on colon mucous membrane of the narrowed segment in some HD infants.
A total of 53 aganglionic colon tissue and 4 urine samples were obtained from 55 infants with HD. Of which, 3 were total colonic type, and 4 long segment HD type, 46 sigmoid rectal type. The specimens were stored at -70°C until use.
Sixteen urine samples from 11 asymptomatic patients, 3 from thrombocytopenia patients, 1 from congenital heart disease patient, and 1 from renal disease infant, were investigated after cytomegalovirus DNA was detected by quantitative PCR[18]. All infants were admitted to Second Affiliated Hospital of China Medical University from March 2001 to September 2004. Cytomegalovirus was detected in all urine samples and 16 of 53 narrow segment colon specimens from HD patients by quantitative PCR. The study was approved by the local ethical committee.
Virus DNA in urine was isolated from the precipitated cells by boiling them in lysis buffer for 15 min. Tissue DNA was extracted with xylene, proteinase K, and phenol chloroform protocol, followed by ethanol precipitation. All extracted DNA preparations were diluted in water as templates used in PCR amplification.
The PCR reaction mixture containing 1 × buffer, 1.5 mmol/L MgCl2, 0.2 mmol/L dNTPs, 150 ng up and down primer respectively, 0.5 U of Taq polymerase (Promega, Madison city, USA), 3.5 μL sample, and ddH2O was added to a final volume of 50 μL. Nest PCR was performed to amplify UL144 when the outside primers yielded either negative or weak results. The sequences of outside primer set designated by Lurain et al[11] as UL144B are as follows: forward (UL144Ca) 5’-CGTATTACAAACCGCGGAGAGGAT-3’, reverse (UL144Cb) 5’-ACTCAGACACGGTTCCGTAA-3’. The inner primers were designed based on the Toledo sequence (GenBank accession No. AY446871) using Primer premier 5.0: forward (UL144Ca2) 5’-AGACACCGTTCGGCCCTAT-3’, reverse (UL144Cb2) 5’-TTTAGTGCAGGAATTGGAA-3’. A 681 bp fragment containing UL144 coding sequence was amplified. The conditions for amplification with all primer sets were at 95°C for 5 min, followed by 30 cycles at 95°C for 45 s, at 54°C for 1 min, and at 72°C for 1 min and 30 s, and a single extension cycle at 72°C for 10 min. PCR products were detected on a 1.5% agarose gel stained with ethidium bromide under UV illumination.
PCR products including the whole length of UL144 open reading frame (ORF) were gel-purified using PCR fragment purification kit (TaKaRa, Dalian city, China) according to the manufacturer’s instructions, and then sequenced directly with the BigDye terminator cycle sequencing kit (Perkin Elmer, Foster city, USA). Sequencing was usually performed on both DNA strands, using the UL144Ca2 and UL144Cb2 primers. Sequencing reactions were performed with a Perkin-Elmer Gene Amp PCR system 2400 (Perkin Elmer, Foster city, USA) at 96°C for 10 s, at 50°C for 5 s, and at 60 °C for 4 min for a total of 30 cycles. The sequencing products were analyzed on an ABI 3700 automated sequencer.
To get accurate sequence data for clinical strain M20, in which the sequence represents lapped spike, UL144 PCR products of clinical strain M20 were cloned into PGEM-T vector (Promega, Madison city, USA) and the UL144 was sequenced using standard M13 + primer.
Nucleotide and amino acid sequences were compared using Program of BioEdit 5.0. Multiple-alignment algorithm in the Megalign program package was used in phylogenetic analysis (Lasergene; DNAstar). Functional motifs were identified from the PROSITE database.
Twenty-seven strains from HD infants and 16 strains from urine sample were sequenced. UL144 ORF DNA sequences from these strains were submitted to GenBank by using program Sequin. The accession number of strains from HD patients is AY999272-AY999296, AY818285, AY818293, respectively. The accession number of strains from control group is AY818269-270, AY818272, AY818276, AY818280, AY818283, AY818286, AY818292, AY818295, AY818302-303, AY818305-306, AF447377, AF447388-89, respectively.
Descriptive statistics were carried out by chi square test or Fisher's exact test with SPSS 10.0 software package. P≤ 0.05 was considered statistically significant.
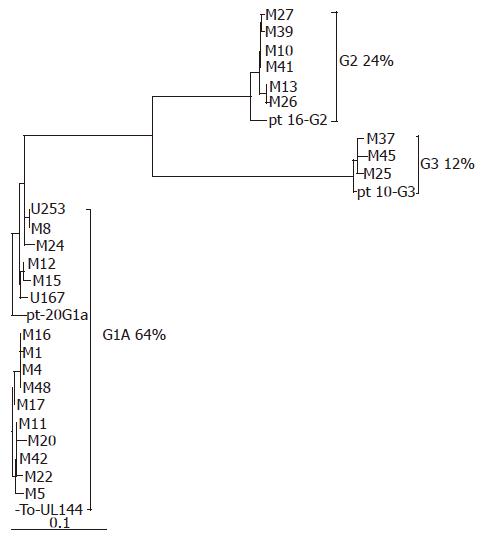
Twenty-three operational and 4 urine samples from HD infants and 16 urine samples from control group yielded positive results of the predicted size when amplified with the UL144Ca/Cb and UL144Ca2/Cb2 primer sets. All the amplified products were sequenced. UL144 ORF was detected using DNAclub. Then, UL144 ORFs of all strains were compared using Clustal W. Alignment comparison revealed that the variations dispersed over the whole ORF but concentrated in the 5’ half. UL144 sequence of various clinical strains was found in 80.4%-99.4% of nucleotides and 79%-100% of amino acids sequence identities compared with that of Toledo. Strains from HD patients presented cytomegalovirus UL144 hypervariability. The cytomegalovirus UL144 sequences from the same patients, but different samples (M25 and U296, M27 and U298) were completely identical. Based on the phylogenetic analysis, the UL144 sequences of strains from HD patients were categorized into three major groups according to the schema classified by Lurain et al[11], referred to as group 1A, group 2, and group 3 (Figure 1). Our results showed that most of the strains belonged to group 1A, amounting to 64% (16 of 25). The minority of infants (3 of 25, 12%) showed strains that conformed to group 3. Twenty-four percent (6 of 25) of the strains were distributed in group 2. No strains were found in group 1B and group 1C. The variability within each genotype ranged 0%-0.6%. Compared with the corresponding groups (groups 1A, 2, and 3) remarked by Lurain[11], the homology was 97.5%-98.9%, 97.9%-98.1%, and 99.1%-99.2%, respectively.
The distribution of UL144 genotypes in urine samples from infants with other diseases except for Hirschsprung’s disease (control group) was found in 6 (37.5%), 1 (6.3%), 5 (31.2%), 4 (25.0%) of genotype group 1A, group 1B, group 2, and group 3, respectively. The UL144 genotypes between strains from infants with Hirschsprung’s disease and those from control group were compared by chi square test (χ2 = 1.870 and P = 0.393, Table 1).
| Source | n | 1A (%) | 1B (%) | 2 (%) | 3 (%) | χ2 | P |
| HD | 25 | 16 (64.0) | 0 (-) | 6 (24.0) | 3 (12.0) | 1.870 | 0.393 |
| C-group | 16 | 6 (37.5) | 1 (6.3) | 5 (31.2) | 4 (25.0) |
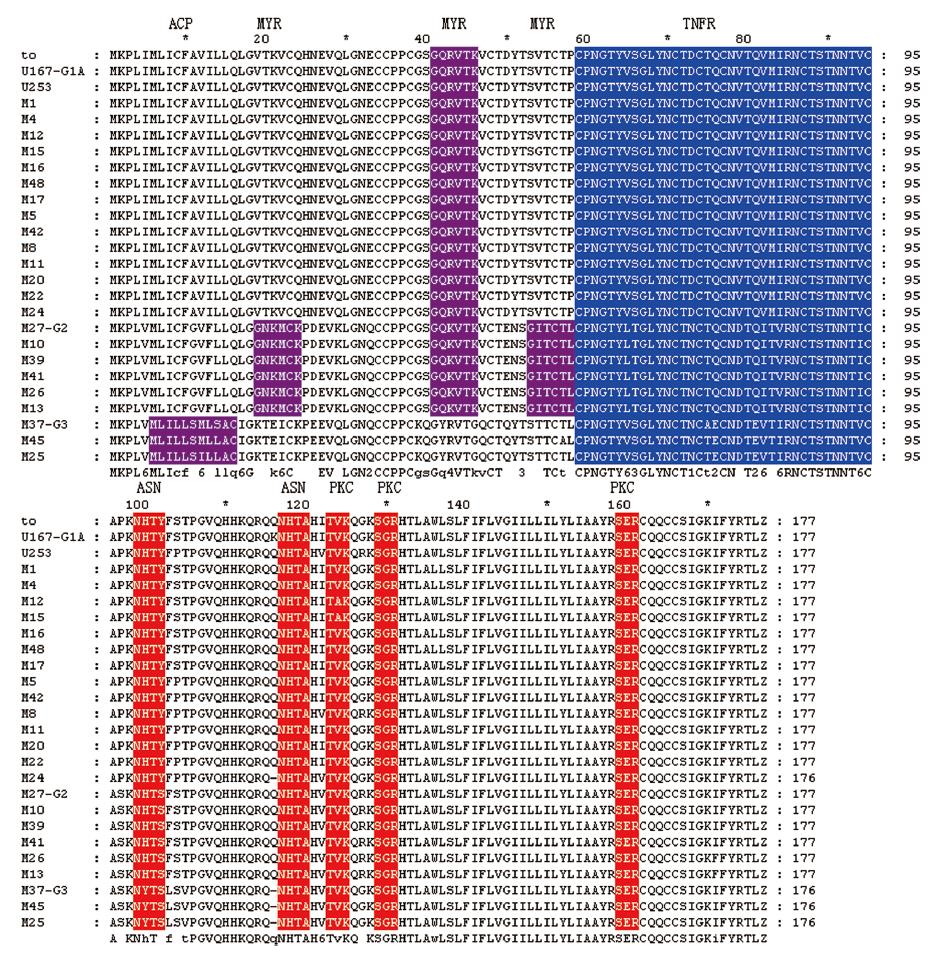
Analysis of posttranslational modification sites of predicted UL144 protein showed that most of the important motifs, such as tumor necrosis factor-like receptor (TNFR), N-myristoylation (MYR), protein kinase C phosphorylaction site (PKC), and N-glycosylation (ASN) were shared by strains from all groups. However, because valine (V) and serine (S) substituted by glycine (G) at 19 and 53 amino acid residues in strains belong to group 2, another two MYR functional sites were added, respectively, in 19-24 and 53-59 amino acid residues. Compared with those in the other two groups, only strains in group 3 had a specific acyl carrier protein site (ACP) but MYR was absent motif in their UL144 putative protein (Figure 2).
Three UL144 genotypes (group 1A, group 2, and group 3) were found among strains in colon samples from infants with Hirschsprung’s disease. With respect to clinical features, strains from Hirschsprung’s disease patients were scattered in the UL144 genotypes. Because the number of samples of the long segment and total colon type was limited, no relationship was found between their clinical phenotypes and UL144 genotypes.
Hirschsprung’s disease has been proved to be a disease of neural crest. It is a developmental disorder of the enteric nervous system (ENS) characterized by the absence of ganglion cells in the myenteric and submucosal plexuses along a variable portion of the distal intestine. It was reported that different genes such as RET, GDNT, EDNRB and EDN3 are involved in the pathogenesis of Hirschsprung’s disease[19]. However, microenvironment factors can also play a role in the pathogenesis of aganglionosis. It was reported that one of the etiologies of Hirschsprung’s disease is cytomegalovirus infection determined by immuno-histochemical and in situ hybridization test, and cytomegalovirus DNA has been detected either in ganglion cells or within myenteric and submucosal plexus along small and large intestine[17,20,21]. It has been widely accepted that Hirschsprung’s Disease is associated with a narrowed bowel segment. In this study, we explored the genetic diversities of cytomegalovirus UL144 ORF in narrowed colon tissue samples from infants with Hirschsprung’s disease.
Based on the nucleotide sequences of UL144 gene, cytomegalovirus strains have been divided into five genotypes by Lurain et al[11]. In their study, the predominant genotype of UL144 was group 3. In the current study, samples were obtained directly from narrow segment colon tissue and urine in patients with Hirschsprung’s disease, without any passage in cell culture. Hence, the strains in bowel tissue specimens from patients could authentically reflect the natural genetic composition of cytomegalovirus infection in end organ. The UL144 gene sequences of cytomegalovirus strains from Hirschsprung’s disease patients could be categorized into three major groups according to the schema of Lurain et al[11], referred to as group 1A in 16 of 25(64%), group 2 in 6 of 25 (24%), and group 3 in 3 of 25 (12%), no strains were found in group 1B and group 1C in patients with Hirschsprung’s disease. Compared with the results of Lurain[11], Bale[12], and Murayama et al[22], the predominant genotype of UL144 in our studied strains was group 1A (64%). The discrepancy may be due to the difference in the studied population.
A number of studies have attempted to correlate cytomegalovirus genetic variants with specific manifestations of cytomegalovirus disease or sites of infection[7,9,23]. However, no definitive association has been established yet. Whether the outcomes of congenital cytomegalovirus infection and tissue tropism are related to UL144 genotype is still controversial. No association has been found between the gene variation and particular diseases[11,12,24]. However, Ara-Boger et al[13] showed that the relatively rare UL144 genotypes A and C, but not the most common genotype B, may be associated with the most serious outcome of cytomegalovirus congenital infection. Genotypes A-C correspond to group 1A, group 3 and group 2 in Lurains’ study, respectively.
It was reported that polymorphism of gB genotypes is related to geographic and demographic composition[25]. However, the results of our previous study[26] are consistent with the reported data. Nonetheless, the proportion of UL144 group 1A is higher in strains from infants with congenital cytomegalovirus infection[13,27], suggesting that cytomegalovirus strains in group 1A have a much stronger pathogenicity in congenital cytomegalovirus infection. In the current study, the predominant UL144 genotype in intestinal tissue samples from infants with Hirschsprung’s disease was group 1A (64%), but the comparison of distribution of gene composition of strains between infants with Hirschsprung’s disease and control group did not reach statistical significance (P = 0.393), suggesting that UL144 gene diversities are not related to the particular disease.
With respect to clinical features, strains from 18 infants with Hirschsprung’s disease of sigmoid rectal type were scattered in all UL144 genotypes. Since the number of samples was small, the association has not been ascertained between UL144 genotype and clinical phenotype of HD. Moreover, the identical sequences of cytomegalovirus UL144 gene from the same patients (M25 and U296, M27 and U298) suggested that DNA sequences in urine samples can reflect cytomegalovirus UL144 genotype of infected organs.
In the current study, analysis of posttranslational modification sites of predicted UL144 protein showed that most of the important motifs, including TNFR, MYR, PKC, and ASN were conserved in all strains, suggesting that it is necessary for these motifs to maintain the HCMV biological functions. However, only the strains in group 2 had two MYR motifs in 19-24 and 53-59 amino acid residues, meanwhile, only the strains in group 3 had a specific ACP but MYR was absent motif in their UL144 putative protein in comparison with those in the other two groups. Further study is required on the relationship between these changes and cytomegalovirus infection, replication and clinical sequelae.
The authors thank Lian-Ying Wang and Ya-Luo Dong for their help in obtaining clinical samples and statistic analysis.
S- Editor Liu Y L- Editor Wang XL E- Editor Yin DH
| 1. | Torok-Storb B, Boeckh M, Hoy C, Leisenring W, Myerson D, Gooley T. Association of specific cytomegalovirus genotypes with death from myelosuppression after marrow transplantation. Blood. 1997;90:2097-2102. [PubMed] |
| 2. | Rosen HR, Corless CL, Rabkin J, Chou S. Association of cytomegalovirus genotype with graft rejection after liver transplantation. Transplantation. 1998;66:1627-1631. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 3. | Shepp DH, Match ME, Ashraf AB, Lipson SM, Millan C, Pergolizzi R. Cytomegalovirus glycoprotein B groups associated with retinitis in AIDS. J Infect Dis. 1996;174:184-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 83] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 4. | Klein M, Schoppel K, Amvrossiadis N, Mach M. Strain-specific neutralization of human cytomegalovirus isolates by human sera. J Virol. 1999;73:878-886. [PubMed] |
| 5. | Rasmussen L, Hong C, Zipeto D, Morris S, Sherman D, Chou S, Miner R, Drew WL, Wolitz R, Dowling A. Cytomegalovirus gB genotype distribution differs in human immunodeficiency virus-infected patients and immunocompromised allograft recipients. J Infect Dis. 1997;175:179-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 55] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 6. | Bongarts A, Von Laer D, Vogelberg C, Ebert K, Van Lunzen J, Garweg J, Vaith P, Hufert FT, Haller O, Meyer-König U. Glycoprotein B genotype of human cytomegalovirus: distribution in HIV-infected patients. Scand J Infect Dis. 1996;28:447-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 7. | Fries BC, Chou S, Boeckh M, Torok-Storb B. Frequency distribution of cytomegalovirus envelope glycoprotein genotypes in bone marrow transplant recipients. J Infect Dis. 1994;169:769-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 115] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 8. | Peek R, Verbraak F, Bruinenberg M, Van der Lelij A, Van den Horn G, Kijlstra A. Cytomegalovirus glycoprotein B genotyping in ocular fluids and blood of AIDS patients with cytomegalovirus retinitis. Invest Ophthalmol Vis Sci. 1998;39:1183-1187. [PubMed] |
| 9. | Chern KC, Chandler DB, Martin DF, Kuppermann BD, Wolitz RA, Margolis TP. Glycoprotein B subtyping of cytomegalovirus (CMV) in the vitreous of patients with AIDS and CMV retinitis. J Infect Dis. 1998;178:1149-1153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 30] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Benedict CA, Butrovich KD, Lurain NS, Corbeil J, Rooney I, Schneider P, Tschopp J, Ware CF. Cutting edge: a novel viral TNF receptor superfamily member in virulent strains of human cytomegalovirus. J Immunol. 1999;162:6967-6970. [PubMed] |
| 11. | Lurain NS, Kapell KS, Huang DD, Short JA, Paintsil J, Winkfield E, Benedict CA, Ware CF, Bremer JW. Human cytomegalovirus UL144 open reading frame: sequence hypervariability in low-passage clinical isolates. J Virol. 1999;73:10040-10050. [PubMed] |
| 12. | Bale JF, Petheram SJ, Robertson M, Murph JR, Demmler G. Human cytomegalovirus a sequence and UL144 variability in strains from infected children. J Med Virol. 2001;65:90-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 13. | Arav-Boger R, Willoughby RE, Pass RF, Zong JC, Jang WJ, Alcendor D, Hayward GS. Polymorphisms of the cytomegalovirus (CMV)-encoded tumor necrosis factor-alpha and beta-chemokine receptors in congenital CMV disease. J Infect Dis. 2002;186:1057-1064. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 81] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 14. | Tam PK, Quint WG, van Velzen D. Hirschsprung's disease: a viral etiology? Pediatr Pathol. 1992;12:807-810. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 14] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 15. | Déchelotte PJ, Mulliez NM, Bouvier RJ, Vanlieféringhen PC, Lémery DJ. Pseudo-meconium ileus due to cytomegalovirus infection: a report of three cases. Pediatr Pathol. 1992;12:73-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 16. | Dimmick JE, Bove KE. Cytomegalovirus infection of the bowel in infancy: pathogenetic and diagnostic significance. Pediatr Pathol. 1984;2:95-102. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 20] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 17. | Asabe K, Nagasaki A, Sato K, Nakayama M. Intestinal obstruction caused by congenital cytomegalovirus infection: report of a case. Surg Today. 2003;33:764-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 18. | He R, Liu LQ, Lu SM, Ruan Q. Quantitative detection of HUMAN CYTOMEGALOVIRUS-DNA from urine in infants by FQ-PCR. Zhonghua Xiaoer neike Zazhi. 2001;12:739-742. In China. |
| 19. | Martucciello G, Ceccherini I, Lerone M, Jasonni V. Pathogenesis of Hirschsprung's disease. J Pediatr Surg. 2000;35:1017-1025. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 78] [Cited by in RCA: 60] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 20. | Wang Lianying, Li Zheng, Ruan Qiang. Congenital megacolon and cytomegalovirus infection in infants. Zhonghua Xiaoer Waike Zazhi. 1996;17:3-5. |
| 21. | Chen Leiling, Hu Tingzhe, Liu Jihong. Human cytomegalovirus infection and Hirschsprung's disease. Zhonghua Xiaoer Waike Zazhi. 2002;23:225-227. |
| 22. | Murayama T, Takegoshi M, Tanuma J, Eizuru Y. Analysis of human cytomegalovirus UL144 variability in low-passage clinical isolates in Japan. Intervirology. 2005;48:201-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 23. | Pignatelli S, Dal Monte P, Rossini G, Landini MP. Genetic polymorphisms among human cytomegalovirus (HCMV) wild-type strains. Rev Med Virol. 2003;14:383-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 113] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 24. | Picone O, Costa JM, Chaix ML, Ville Y, Rouzioux C, Leruez-Ville M. Human cytomegalovirus UL144 gene polymorphisms in congenital infections. J Clin Microbiol. 2005;43:25-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 41] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 25. | Zipeto D, Hong C, Gerna G, Zavattoni M, Katzenstein D, Merigan TC, Rasmussen L. Geographic and demographic differences in the frequency of human cytomegalovirus gB genotypes 1-4 in immunocompromised patients. AIDS Res Hum Retroviruses. 1998;14:533-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 26. | He R, Ruan Q, Xia C, Liu LQ, Lü SM, Lu Y, Qi Y, Ma YP, Liu Q, Ji YH. Sequence variability of human cytomegalovirus UL144 open reading frame in low-passage clinical isolates. Chin Med Sci J. 2004;19:293-297. [PubMed] |
| 27. | Tanaka K, Numazaki K, Tsutsumi H. Human cytomegalovirus genetic variability in strains isolated from Japanese children during 1983-2003. J Med Virol. 2005;76:356-360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |