Published online Aug 7, 2007. doi: 10.3748/wjg.v13.i29.3985
Revised: April 3, 2007
Accepted: April 7, 2007
Published online: August 7, 2007
AIM: To characterize the bifidobacterial microbiota of the colonic mucosa in patients with colon cancer, inflammatory bowel disease or diverticulitis.
METHODS: A sample of the distal colonic mucosa was taken during surgery from a total of 34 patients, twenty-one with diagnosed colorectal cancer, nine with diverticulitis and four with inflammatory bowel disease, requiring surgery for their condition. Bacterial DNA was extracted from the resected mucosal samples and bifidobacterial mucosa-associated microbiota was qualitatively and quantitatively determined by means of qualitative and quantitative PCR.
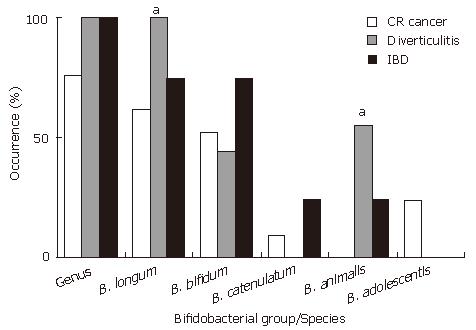
RESULTS: Bifidobacteria were found in 100% of the samples from patients with diverticulitis or IBD and a 76% of those suffering colon cancer. The species B. longum and B. bifidum were the most widely found, followed by B. animalis, B. catenulatum and B. adolescentis. B. breve, B. dentium and B. angulatum were not detected in any sample. A significantly higher occurrence of B. longum was observed in patients with diverticulitis than in those with colon cancer or IBD (100%, 62% and 75%, respectively, P < 0.05). Similar results were obtained for B. animalis (56%, 0% and 25%, P < 0.05), while B. adolescentis was only found in the mucosa from patients with colon cancer (5 out of 21, 24%). At the quantitative level, patients with colon cancer or IBD showed lower counts of total Bifidobacterium (4.94 and 5.91 vs 6.96 log Cells/sample, respectively, P < 0.05) and of the species B. longum (4.05 and 4.79 vs 6.76, P < 0.05) than those with diverticulitis.
CONCLUSION: Aberrancies in mucosa associated microbiota are present in different intestinal diseases. This may indicate a role of the microbiota in the pathogenesis of these diseases.
- Citation: Gueimonde M, Ouwehand A, Huhtinen H, Salminen E, Salminen S. Qualitative and quantitative analyses of the bifidobacterial microbiota in the colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease. World J Gastroenterol 2007; 13(29): 3985-3989
- URL: https://www.wjgnet.com/1007-9327/full/v13/i29/3985.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i29.3985
The microbiota present in the human gastrointestinal tract has a significant influence on our health and well-being. Different bacterial groups and levels are found throughout the gastrointestinal tract from mouth to colon. The stomach and the upper bowel are sparsely populated regions (103-104 CFU/g contents) while the colon is heavily populated (1011-1012 CFU/g contents). In the large intestine, genera such as Bacteriodes, Bifidobacterium, Eubacterium, Clostridium, Fusobacterium and Ruminococcus are usually found. This intestinal microbiota provides the most important contact with the environment for the host and a barrier against harmful food components and pathogenic bacteria[1,2]. The microbiota has also been shown to have a direct impact on the morphology of the gut[3] and it plays an important role in the intestinal immuno-physiological regulation. A healthy microbiota can thus be defined as the normal individual microbiota which both preserves and promotes well-being and absence of disease, especially in the gastrointestinal tract, but also beyond[4].
Major dysfunctions of the gastrointestinal tract are thought to be related to disturbances or aberrancies of the intestinal microbiota[5]. Such aberrancies have been identified in atopic disease[6], rheumatoid arthritis[7], juvenile chronic arthritis[8], ankylosing spondylitis[9], irritable bowel syndrome (IBS)[10,11], inflammatory bowel disease (IBD)[12,13], colon cancer[14] and diverticulitis patients[15]. Specifically, aberrancies in bifidobacterial microbiota, including decreased numbers or an atypical composition have been identified in some of these diseases including atopic disease[6,16], IBS[11] or IBD[12,17,18]. Many diseases and their prevention can thus be linked to the microbiota in the gut. Demonstration of this has stimulated research on the composition and function of the microbiota.
Microbiota development and characterization in the human host still rests largely on culture-dependent methods. These conventional methods have low sensitivity and they are time consuming and biased due to the recovery of only the culturable species or strains. These facts can lead to overestimation of some species and underestimation of others. Thus, during the last decade, developments in molecular biology have led to the application of fast and reliable alternative culture-independent methods[19].
Mucosal surfaces are the major sites of contact with external agents including the members of the intestinal microbiota. However, our knowledge on gut microbiota is mainly based in the assessment of faeces and therefore hindered by the fact that the microbes found will mainly represent the microbiota in the lumen of the sigmoid colon, while the composition of the intestinal microbiota may differ between the lumen and the mucosa[20]. For more accurate information on the population elsewhere in the intestine, samples should preferably be taken by endoscopies or during surgery.
Taking into account the possible role of intestinal microbiota in the aetiology of different intestinal diseases, culture-independent studies are needed to elucidate changes in the microbiota of the intestinal mucosa. In this study we characterized the bifidobacterial microbiota of the colonic mucosa in patients with colon cancer, diverticulitis or IBD. The bifidobacterial microbiota was used as an indicator of alterations in the mucosal colonisation pattern. For this purpose the mucosal Bifidobacterium levels and composition were determined by means of quantitative and qualitative PCR.
A total of 34 patients, twenty-one with diagnosed colorectal cancer, nine with diverticulitis and four with IBD requiring surgery for their condition were explained the study and the subjects gave informed consent for a sample taken during surgery for research purposes. The study protocol was approved by the Joint Committee of Ethics for the University of Turku and Turku University Hospital.
The resected mucosal biopsies (area = 0.785 cm2) from the distal colon of patients were cut into small pieces, resuspended in 1.4 mL of buffer ASL (QIAamp DNA stool Mini kit, Qiagen, Hilden, Germany), vortexed thoroughly and used for the DNA extraction by using the QIAamp DNA stool Mini kit following the manufacturer’s instructions for gram-positive bacteria. DNA extracts were stored at -20°C until its use.
For the characterization of the bifidobacterial microbiota, previously described PCR primers targeting total members of the genus[21] and different Bifidobacterium species or groups[22], including B. adolescentis, B. angulatum, B. animalis group (B. animalis ssp. animalis and B. animalis ssp. lactis), B. bifidum, B breve, B. catenulatum group (B. catenulatum and B. pseudocatenulatum), B. dentium and B. longum group (B. longum biotype longum, B. longum biotype infantis) were used as described by Rinne and coworkers[22]. All of the oligonucleotides used were purchased from Thermo Electron Corporation (Thermo Biosciences, Ulm, Germany). Amplification of the DNA was performed using a PCR iCycler apparatus (Bio-Rad, Espoo, Finland). The total volume of each PCR was 50 μL, employing 4 μL of DNA extract as a template. Amplified products were subjected to gel electrophoresis in 1% agarose gels and were visualized by ethidium bromide staining. DNA extracts were analyzed in two independent PCR runs.
Total bifidobacterial levels as well as the levels of the most frequently found species (B. longum group and B. bifidum) were determined by means of quantitative PCR. Previously reported oligonucleotides and methodologies were used to determine the total levels of bifidobacteria[21] and those of B. longum and B. bifidum[23]. For standard curves, different dilutions of microbial cultures from the appropriate bifidobacterial strains (B. infantis and B. longum or B. bifidum; cell numbers ranging from 1 × 104 to 4 × 109 cells/mL) were used for the DNA extraction with the QIAamp DNA stool Mini Kit (Qiagen). Samples (2 μL) were analyzed in 50 μL amplification reactions by using the procedures and conditions previously described[21,23]. The numbers of cells of Bifidobacterium in the fecal samples were determined by comparing the Ct values obtained to the standard curve. DNA extracts from the different samples were analyzed by duplicate in at least two independent PCR runs.
Statistical analysis was carried out using the SPSS 12.0 software (SPSS Inc., Chicago, IL, USA). χ2 test was used to compare proportions between groups. Data on bacterial levels were subjected to one-way ANOVA using the disease group as factor with three categories; colorectal cancer, diverticulitis and IBD. The least significant difference (LSD) test was used for comparison of means.
The occurrence of the genus Bifidobacterium and the different bifidobacterial species or groups in the mucosal samples analysed are shown in Figure 1. Members of the genus Bifidobacterium were present in all the samples corresponding to patients with diverticulitis or IBD but only in a 76% of the mucosa samples from colorectal cancer patients (n.s. differences).
B. longum group followed by B. bifidum were the species most widely found while B. breve, B. dentium and B. angulatum were not detected in any sample. A significantly higher occurrence of B. longum was observed in patients with diverticulitis (Figure 1) than in those suffering a different disease (100% vs 62% in colorectal cancer and 75% in IBD patients, P < 0.05). Interestingly, a significantly higher occurrence of B. animalis was also found in these patients (56%) than in those suffering from colorectal cancer (0%) or IBD (25%) (P < 0.05). B. adolescentis was only found in colorectal cancer patients (5 out of 21; 24%). The occurrence of B. bifidum was slightly higher in the mucosa of IBD patients (75%) than in those with colorectal cancer (52%) or diverticulitis (44%) although the differences were not statistically significant.
The levels of total bifidobacteria as well as those of the most widely found Bifidobacterium species or groups (B. longum and B. bifidum) are shown in Table 1.
The levels of bifidobacteria in the mucosa samples of patients with colorectal cancer or IBD were significantly lower (2.02 and 1.05 log Cells/sample, respectively) than those observed in diverticulitis patients. The same was observed for B. longum while no differences among the three groups were found for the other Bifidobacterium species tested (B. bifidum). A significantly higher ratio between B. longum and total bifidobacteria was also observed in samples of patients with diverticulitis than in those with colorectal cancer or IBD (Table 1).
Here we studied the bifidobacterial mucosa-associated bifidobacteria in patients with colorectal cancer, diverticulitis or IBD. The qualitative bifidobacterial composition of the distal colon mucosa was characterized and the levels of the predominant Bifidobacterium species determined, showing a different microbiota composition in the different disease groups tested. The findings obtained using culture-independent techniques support the hypothesis that colorectal bacteria may play a role in the pathogenesis of several intestinal diseases.
Traditionally faecal samples have been used to assess the intestinal microbiota but the representativness of these to processes in the colon can be criticized. Mucosa-associated bacteria may be more reflective of the long term milieu in the colon wall and therefore prove to be a better indicator of disease-related microbiota aberrancies, as these microorganisms may have a stronger interaction with the host cells.
Only a few studies with mucosal samples have been conducted in both healthy individuals and patients[24,25]. In this area, the mucosal microbiota of patients suffering inflammatory conditions such as IBS and IBD is the most widely characterized.
When mucosal samples from patients with IBD were analysed and compared with those of healthy controls, lower levels of bifidobacteria and higher of Clostridium and Escherichia coli were observed in the former group[18]. In addition, reduced levels of bifidobacteria in the colorectal mucosa of patients with ulcerative colitis (a manifestation of IBD) have also been reported[26]. In general, our results on the occurrence and levels of total Bifidobacterium are in the range of those reported by Poxton and co-workers[25] in patients with ulcerative colitis. However, our study extends these observations to the level of Bifidobacterium species and differences in the species composition which may more accurately reflect the intestinal microbiota activity. Although the number of patients included in this study is too low to establish firm conclusions, our results indicate differences in the presence of B. longum and B. animalis among the distal colonic mucosa of patients with different diseases. Also the levels of B. longum and its ratio with regard to the total bifidobacterial levels were found to be different.
Unfortunately, samples from healthy controls were not available for comparison but our results indicate that the intestinal mucosal microbiota is altered in different diseases. Patients with colorectal cancer or IBD harbor lower levels of total bifidobacteria and especially B. longum showing a reduced occurrence of B. longum and also B. animalis in their colonic mucosa when compared to diverticulitis patients. This may reflect the long time frame involved with colon cancer development. It may also suggest that microbiota screening procedures could be developed to identify potential aberrancies that may be correlated to the colorectal cancer risk of the subject.
A thorough knowledge of the gut microbiota composition will offer a basis for future interventional studies to improve colonic health. To this regard, probiotic or prebiotic intervention has been suggested to be beneficial in the prevention and treatment of colon cancer by different mechanisms[26]. A specific probiotic has been found to modulate the expression of genes involved in cell growth and differentiation in the intestinal mucosa of human volunteers[27]. Probiotics have also been successfully applied in patients with IBD[28,29] or diverticulitis[30].
Several studies indicate that aberrancies in faecal microbiota are present in different diseases, and our results strengthen those studies with colonic mucosa data. These results indicate that the mucosa-associated microbiota is disturbed in different diseases, as indicated by the observed alterations in the bifidobacterial microbiota. This may imply a role of the microbiota in the pathogenesis of different diseases requiring further characterization in both patients and subjects at risk of developing specific diseases. Characterizing the differences in mucosa-associated bacteria more accurately will increase our understanding of their role in health and disease and it may provide targets for intervention and further development in the disease risk reduction area.
We wish to thank Nina Kainulainen for her excellent technical assistance during this study.
The microbiota present in the human gastrointestinal tract has a significant influence on our health and well-being. Major dysfunctions of the gastrointestinal tract are thought to be related to disturbances or aberrancies of the intestinal microbiota. Many diseases and their prevention can thus be linked to the microbiota in the gut. Demonstration of this has stimulated research on the composition and function of the microbiota. A thorough knowledge of the gut microbiota composition will offer a basis for future interventional studies to improve colonic health.
Major dysfunctions of the gastrointestinal tract are thought to be related to disturbances or aberrancies of the intestinal microbiota. Such aberrancies have been identified in atopic disease, rheumatoid arthritis, juvenile chronic arthritis, ankylosing spondylitis, irritable bowel syndrome (IBS), inflammatory bowel disease (IBD), colon cancer and diverticulitis patients. Mucosal surfaces are the major sites of contact with external agents including the members of the intestinal microbiota. However, our knowledge on gut microbiota is mainly based on the assessment of faeces and are therefore hindered by the fact that the microbes found will mainly represent the microbiota in the lumen of the sigmoid colon, while the composition of the intestinal microbiota may differ between the lumen and the mucosa. For more accurate information on the population elsewhere in the intestine, samples should preferably be taken by endoscopies or during surgery. Here we studied the bifidobacterial mucosa-associated bifidobacteria in patients with colorectal cancer, diverticulitis or IBD. The qualitative bifidobacterial composition of the distal colon mucosa was characterized and the levels of the predominant Bifidobacterium species determined, showing a different microbiota composition in the different disease groups tested.
The intestinal mucosal microbiota has been reported to be altered in patients with different diseases. Specifically, reduced levels of intestinal bifidobacteria have been observed in some diseases. This study extends these observations to the level of Bifidobacterium species and differences in the species composition which may more accurately reflect the intestinal microbiota activity. The results of this study show that aberrancies in mucosa associated microbiota are present in different intestinal diseases. This may indicate a role of the microbiota in the pathogenesis of these diseases.
The results of this study may indicate a role of the microbiota in the pathogenesis of different diseases requiring further characterization in both patients and subjects at risk of developing specific diseases. Characterizing the differences in mucosa-associated bacteria more accurately will increase our understanding of their role in health and disease and it may provide targets for intervention and further development in the disease risk reduction area.
We think the terminology used in this article will be easily understandable for the readers.
The authors qualitatively and quantitatively analyzed the bifidobacterial microbiota in colonic mucosa of patients with colorectal cancer, diverticulitis and inflammatory bowel disease and provide an interesting finding that the component and level of microbiota differed among these three groups. The study provides some relevant information regarding the quantitative and qualitative presence of bifidobacterial microbiota in the colon of patients with colon cancer, diverticulitis and inflammatory bowel disease.
S- Editor Liu Y L- Editor Kremer M E- Editor Lu W
| 1. | Grönlund MM, Arvilommi H, Kero P, Lehtonen OP, Isolauri E. Importance of intestinal colonisation in the maturation of humoral immunity in early infancy: a prospective follow up study of healthy infants aged 0-6 months. Arch Dis Child Fetal Neonatal Ed. 2000;83:F186-F192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 168] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 2. | Kirjavainen PV, Apostolou E, Arvola T, Salminen SJ, Gibson GR, Isolauri E. Characterizing the composition of intestinal microflora as a prospective treatment target in infant allergic disease. FEMS Immunol Med Microbiol. 2001;32:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 98] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 3. | Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol Mol Biol Rev. 1998;62:1157-1170. [PubMed] |
| 4. | Salminen SJ, Gueimonde M, Isolauri E. Probiotics that modify disease risk. J Nutr. 2005;135:1294-1298. [PubMed] |
| 5. | Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361:512-519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2194] [Cited by in RCA: 2114] [Article Influence: 96.1] [Reference Citation Analysis (0)] |
| 6. | Kalliomäki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol. 2001;107:129-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 894] [Cited by in RCA: 811] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 7. | Benno P, Alam M, Henriksson K, Norin E, Uribe A, Midtvedt T. Abnormal colonic microbial function in patients with rheumatoid arthritis. Scand J Rheumatol. 1994;23:311-315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 8. | Malin M, Verronen P, Mykkänen H, Salminen S, Isolauri E. Increased bacterial urease activity in faeces in juvenile chronic arthritis: evidence of altered intestinal microflora? Br J Rheumatol. 1996;35:689-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 9. | Stebbings S, Munro K, Simon MA, Tannock G, Highton J, Harmsen H, Welling G, Seksik P, Dore J, Grame G. Comparison of the faecal microflora of patients with ankylosing spondylitis and controls using molecular methods of analysis. Rheumatology (Oxford). 2002;41:1395-1401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 110] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 10. | Si JM, Yu YC, Fan YJ, Chen SJ. Intestinal microecology and quality of life in irritable bowel syndrome patients. World J Gastroenterol. 2004;10:1802-1805. [PubMed] |
| 11. | Malinen E, Rinttilä T, Kajander K, Mättö J, Kassinen A, Krogius L, Saarela M, Korpela R, Palva A. Analysis of the fecal microbiota of irritable bowel syndrome patients and healthy controls with real-time PCR. Am J Gastroenterol. 2005;100:373-382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 499] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 12. | Favier C, Neut C, Mizon C, Cortot A, Colombel JF, Mizon J. Fecal beta-D-galactosidase production and Bifidobacteria are decreased in Crohn's disease. Dig Dis Sci. 1997;42:817-822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 146] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 13. | Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P. Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut. 2006;55:205-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1633] [Cited by in RCA: 1677] [Article Influence: 88.3] [Reference Citation Analysis (0)] |
| 14. | Hope ME, Hold GL, Kain R, El-Omar EM. Sporadic colorectal cancer--role of the commensal microbiota. FEMS Microbiol Lett. 2005;244:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 77] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 15. | Colecchia A, Sandri L, Capodicasa S, Vestito A, Mazzella G, Staniscia T, Roda E, Festi D. Diverticular disease of the colon: new perspectives in symptom development and treatment. World J Gastroenterol. 2003;9:1385-1389. [PubMed] |
| 16. | He F, Ouwehand AC, Isolauri E, Hashimoto H, Benno Y, Salminen S. Comparison of mucosal adhesion and species identification of bifidobacteria isolated from healthy and allergic infants. FEMS Immunol Med Microbiol. 2001;30:43-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 134] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 17. | Seksik P, Rigottier-Gois L, Gramet G, Sutren M, Pochart P, Marteau P, Jian R, Doré J. Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut. 2003;52:237-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 532] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 18. | Mylonaki M, Rayment NB, Rampton DS, Hudspith BN, Brostoff J. Molecular characterization of rectal mucosa-associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis. 2005;11:481-487. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 184] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 19. | Gueimonde M, Salminen S. Methods for analysing gut microbiota. Lactic acid bacteria, Microbiological and Functional aspects 3rd edition. New York: Marcel Dekker 2004; 365-374. |
| 20. | Ouwehand AC, Salminen S, Arvola T, Ruuska T, Isolauri E. Microbiota composition of the intestinal mucosa: association with fecal microbiota? Microbiol Immunol. 2004;48:497-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 21. | Gueimonde M, Tölkkö S, Korpimäki T, Salminen S. New real-time quantitative PCR procedure for quantification of bifidobacteria in human fecal samples. Appl Environ Microbiol. 2004;70:4165-4169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 136] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 22. | Rinne MM, Gueimonde M, Kalliomäki M, Hoppu U, Salminen SJ, Isolauri E. Similar bifidogenic effects of prebiotic-supplemented partially hydrolyzed infant formula and breastfeeding on infant gut microbiota. FEMS Immunol Med Microbiol. 2005;43:59-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 113] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 23. | Gueimonde M, Debor L, Tölkkö S, Jokisalo E, Salminen S. Quantitative assessment of faecal bifidobacterial populations by real-time PCR using lanthanide probes. J Appl Microbiol. 2007;102:1116-1122. [PubMed] |
| 24. | Delgado S, Suárez A, Mayo B. Identification of dominant bacteria in feces and colonic mucosa from healthy Spanish adults by culturing and by 16S rDNA sequence analysis. Dig Dis Sci. 2006;51:744-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 25. | Poxton IR, Brown R, Sawyerr A, Ferguson A. Mucosa-associated bacterial flora of the human colon. J Med Microbiol. 1997;46:85-91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 84] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 26. | Geier MS, Butler RN, Howarth GS. Probiotics, prebiotics and synbiotics: a role in chemoprevention for colorectal cancer? Cancer Biol Ther. 2006;5:1265-1269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 27. | Macfarlane S, Furrie E, Cummings JH, Macfarlane GT. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin Infect Dis. 2004;38:1690-1699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 128] [Cited by in RCA: 118] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 28. | Di Caro S, Tao H, Grillo A, Elia C, Gasbarrini G, Sepulveda AR, Gasbarrini A. Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Dig Liver Dis. 2005;37:320-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 97] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 29. | Guandalini S. Use of Lactobacillus-GG in paediatric Crohn's disease. Dig Liver Dis. 2002;34 Suppl 2:S63-S65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 30. | Gosselink MP, Schouten WR, van Lieshout LM, Hop WC, Laman JD, Ruseler-van Embden JG. Delay of the first onset of pouchitis by oral intake of the probiotic strain Lactobacillus rhamnosus GG. Dis Colon Rectum. 2004;47:876-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 136] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 31. | Giaccari S, Tronci S, Falconieri M, Ferrieri A. Long-term treatment with rifaximin and lactobacilli in post-diverticulitic stenoses of the colon. Riv Eur Sci Med Farmacol. 1993;15:29-34. [PubMed] |