Published online Jun 28, 2007. doi: 10.3748/wjg.v13.i24.3369
Revised: April 3, 2007
Accepted: April 7, 2007
Published online: June 28, 2007
AIM: To identify the difference in gene expression of microphage (Mφ) between normal spleen and portal hypertensive spleen using cDNA microarrays and find new gene functions associated with hypersplenism in portal hypertension.
METHODS: The Biostar-H140s chip containing 14 112 spots of cDNAs were used to investigate the difference of the expression. The total RNA extracted from macrophages isolated from both normal spleen and portal hypertensive spleen was reversely transcribed to cDNA with the incorporation of fluorescent (cy3 and cy5) labeled dCTP to prepare the hybridization probes. After hybridization, the gene chip was scanned for the fluorescent intensity. The differentially expressed genes were screened. That was repeated three times, and only the genes which had differential expression in all three chips were considered to be associated with hypersplenism in portal hypertension.
RESULTS: Eight hundred and ninety-six, 1330 and 898 genes were identified to be differentially expressed in three chips, respectively. One hundred and twenty-one genes (0.86%) were identified to be differentially expressed in all three chips, including 21 up-regulated genes and 73 down-regulated genes. The differentially expressed genes were related to ionic channel and transport protein, cyclin, cytoskeleton, cell receptor, cell signal conduct, metabolism, immune, and so on. These genes might be related to the hypersplenism in portal hypertension.
CONCLUSION: The investigations based on cDNA microarray can screen differentially expressed genes of macrophages between normal spleen and portal hypertensive spleen, thus may provide a new idea in studying the pathogenesis of hypersplenism in portal hypertension.
- Citation: Yan F, Wang XM. Difference in gene expression of macrophage between normal spleen and portal hypertensive spleen idendified by cDNA microarray. World J Gastroenterol 2007; 13(24): 3369-3373
- URL: https://www.wjgnet.com/1007-9327/full/v13/i24/3369.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i24.3369
It is reported that, compared with the macrophage (Mφ) in normal spleen, the Mφ in portal hypertensive spleen has a large amount of acid phosphatase, lysosome and pseudopodium, and can destruct much more erythrocytes and thrombocytes. This proved that the destruction of hemocytes by Mφ of spleen plays an important role in the development of hypersplenism in portal hypertension[1,2].Our previous studies suggested that phagocytosis of Mφ was augmented in hypersplenism in portal hypertension; however, the specific mechanisms are not clear. In this study, cDNA microarrays were used to detect the difference in gene expression of Mφ between normal spleen and portal hypertensive spleen and find new gene functions associated with hypersplenism in portal hypertension in an attempt to explore the pathogenesis of hypersplenism in portal hypertension.
The excised human spleen specimens used in this study were provided with the approval of the hospital authorities. The experimental group included 3 cases of excised human spleen of portal hypertension and hypersplenism (all 3 cases had chronic hepatitis B), and the control group included 2 cases of excised human spleen of traumatic splenic rupture.
Mφ was isolated and purified by adherent culture[3]. Total RNA was extracted from Mφ by the TRIzol method[4].
The Biostar-H140s cDNA microarray provided by Shanghai BioStar Genechip Inc., consists of a total of 14 112 human genes. The cDNA inserts were amplified using the polymerase chain reaction (PCR) with universal primers, and then purified according to standard method. All PCR products were examined by agarose gel electrophoresis to ensure the quality. Then the amplified PCR products were dissolved in a buffer solution. The solution with amplified PCR products were spotted onto silylated slides (TeleChem International, USA) using a Cartesian PixSys 7500 motion control robot (Cartesian Technologies, USA). Glass slides with spotted cDNA were hydrated for 2 h in 700 mL/L humidity, dried for 0.5 h at room temperature, and UV crosslinked (65 mj/cm). They were further processed at room temperature by soaking in 2 g/L sodium dodecyl sulfate (SDS) for 10 min, in distilled H2O for 10 min, and 2 g/L sodium borohydride (NaBH4) for 10 min. The slides were dried again and ready for use.
The fluorescent cDNA probes were prepared through reverse transcription and then purified according to the protocol of Schena[5]. The total RNA of Mφ was extracted from 2 cases of normal spleen respectively, and then was mixed as the control group. The total RNA of Mφ was extracted from 3 cases of portal hypertensive spleen respectively, and each case was treated as the experimental group. The probes from the total RNA of control group was labeled with Cy3-dUTP, while those from the total RNA of experimental group were labeled with Cy5-dUTP. The probes were then mixed, precipitated and resolved in a hybridization buffer.
Microarrays were pre-hybridized with hybridization solution containing 0.5 g/L denatured salmon sperm DNA at 42°C for 6 h. Fluorescent probe mixtures were denatured at 95°C for 5 min, and the denatured probe mixtures were applied onto the pre-hybridized chip under a cover glass. Chips were hybridized at 42°C for 16-18 h. The hybridized chips were then washed at 60°C for 10 min each in the mixture of 5 mL/L solution 1 and 20 mL/L solution 2, and 50 mL/L solution 3, then dried at room temperature for scanning (all reagents used in this procedure were contained in the Chip Hybridization Kit provided by Shanghai BioStar Genechip Inc.).
The chips were scanned with a ScanArray 4000 (Packard Biochip Technologies, USA) at two wavelengths to detect emission from both Cy3 and Cy5. The acquired images were analyzed using QuantArray software (Packard Biochip Technologies, USA). Ratios of Cy5 to Cy3 were computed for each location on each microarray. Overall intensities were normalized with a correction coefficient obtained using the ratios of 96 housekeeping genes in each chip. The intensities of each spot at the two wavelengths represent the quantity of Cy3-dUTP and Cy5-dUTP, respectively, hybridized to each spot. Thus, the ratio of each spot represents the ratio of mRNA expression abundance between the gene of Mφ in normal spleen and portal hypertensive spleen. The detection results were described in both scanned microarray images and microarray scatter plots. That was repeated three times, and only the genes that had differential expression in all three chips were considered associated with hypersplenism in portal hypertension.
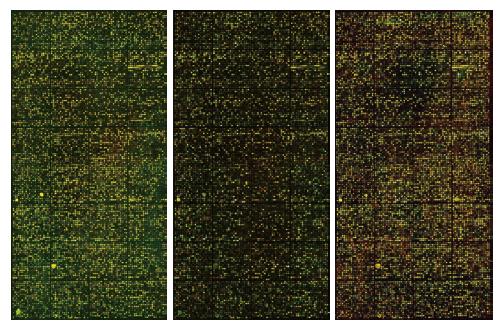
In the scanned microarray images (Figure 1), red points represent the higher expression genes of Mφ in portal hypertensive spleen than those in normal spleen, green points represent the lower expression genes, and yellow points represent the genes that have no change in expression. The hybridization signal of chips is distinct and balanced, indicating that the results are reliable. Compared with the genes of Mφ in normal spleen, a few genes of Mφ in portal hypertensive spleen were highly expressed, some were lowly expressed, however most genes showed no change in expression.
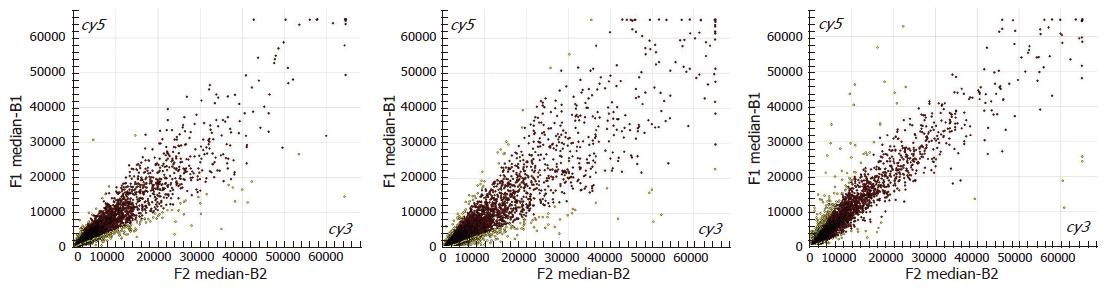
As indicated in the microarray scatter plots (Figure 2), most genes show a concentrated pattern surrounding the diagonal (red points), which means that the ratios range from 0.5 to 2.0, and there is no difference in the gene expression between normal spleen and portal hypertensive spleen. However, the other genes are away from the diagonal (yellow points), indicating that the ratios are beyond the range of 0.5-2.0, and the difference in the expression of those genes is not significant between normal spleen and portal hypertensive spleen.
There were 896, 1330 and 898 genes identified to be differentially expressed in three chips, respectively; 121 genes (0.86%) were differentially expressed in all three chips, including 95 genes which could be found in the GenBank, the other 26 genes were not reported and probably were unidentified novel genes. Among 95 known genes, 1 gene (GenBank No: NM_012218) was related to hepatitis B, and the other 94 genes might be those that were differentially expressed between the Mφ in normal spleen and the Mφ in portal hypertensive spleen, including 21 up-regulated known genes and 73 down-regulated known genes. Ten differentially expressed genes that were up-regulated in macrophges of portal hypertensive spleen are listed in Table 1 and 18 differentially expressed genes that were down-regulated are demonstrated in Table 2.
| GenBank No. | Gene name and description | Average ratio |
| AF189009 | UBQLN2, ubiquilin 2 | 2.117 |
| BX537509 | NET1, neuroepithelial cell transforming gene 1 | 2.270 |
| NM_004830 | CRSP3, cofactor required for Sp1 transcriptional activation | 2.334 |
| AF025771 | ZNF189, zinc finger protein 189 | 2.453 |
| AK090727 | Homo sapiens cDNA FLJ33408 fis, clone BRACE2010550 | 2.511 |
| BX537955 | TRIM37, tripartite motif-containing 37 | 2.553 |
| NM_006716 | ASK, activator of S phase kinase | 2.620 |
| NM_181523 | PIK3R1, phosphoinositide-3-kinase, regulatory subunit 1 | 2.755 |
| XM_376537 | Homo sapiens BCL2-associated transcription factor 1 (BCLAF1), mRNA | 2.993 |
| NM_004416 | DTX1, deltex homolog 1 (Drosophila) | 3.553 |
| GenBank No. | Gene name and description | Average ratio |
| BC068441 | IL1RN, interleukin 1 receptor antagonist | 0.179 |
| NM_014909 | KIAA1036 | 0.268 |
| BU732296 | Homo sapiens cDNA clone UI-E-CI1-afo-e-04-0-UI 3', mRNA sequence | 0.275 |
| AK092248 | Homo sapiens cDNA FLJ34929 fis, clone NT2RP7004728 | 0.293 |
| NM_006254 | PRKCD, protein kinase C, delta | 0.298 |
| BX648172 | OAZ2, ornithine decarboxylase antizyme 2 | 0.302 |
| BM542499 | Homo sapiens cDNA clone IMAGE:5521023 5', mRNA sequence | 0.311 |
| BF240734 | Homo sapiens cDNA clone IMAGE:4091885 5', mRNA sequence | 0.324 |
| NM_003902 | FUBP1, far upstream element (FUSE) binding protein 1 | 0.338 |
| BM993772 | Homo sapiens cDNA clone IMAGE:5869020 3', mRNA sequence | 0.344 |
| NM_005373 | MPL, myeloproliferative leukemia virus oncogene | 0.359 |
| BX647757 | SCML1, sex comb on midleg-like 1 (Drosophila) | 0.370 |
| NM_005781 | ACK1, activated Cdc42-associated kinase 1 | 0.381 |
| AL832249 | Homo sapiens mRNA; cDNA DKFZp686P1077 | 0.390 |
| AK092130 | LOC285378, hypothetical protein LOC285378 | 0.406 |
| AF466367 | Homo sapiens clone KU011197 unknown mRNA | 0.434 |
| NM_001067 | TOP2A, topoisomerase (DNA) II alpha | 0.437 |
| AF051151 | TLR5, toll-like receptor 5 | 0.452 |
Since the microarray analysis was first reported by Schena[6] in 1995, gene chips have been widely used in studying the functions of genes. The results of this study proved that gene chips can successfully profile changes in gene expression on a genomic scale with low consuming, high sensitivity and high-flux. In this study, cDNA microarrays were used to detect the difference in gene expression of Mφ between normal spleen and portal hypertensive spleen and find new gene functions associated with hypersplenism in portal hypertension in an attempt to explore the pathogenesis of hypersplenism in portal hypertension. No similar study has been reported until now.
In order to obtain enough amounts of total RNA and eliminate individual variation, the total RNA of Mφ extracted from 2 cases of normal spleen respectively was mixed as the control group, and then was matched with that of 3 cases of the experimental group to 3 match-pairs for the cDNA microarray analysis. The differentially expressed genes were found to be related to ionic channel and transport protein, cyclin, cytoskeleton, cell receptor, cell signal conduct, metabolism, immune, and so on.
PRKCD encoding protein kinase C delta[7] was found down-regulated in the Mφ of portal hypertensive spleen. Protein kinase C delta plays an important role in regulating IL-13-induced 15-lipoxygenase (15-LO) expression in human monocytes and subsequently modulates the inflammatory responses mediated by 15-LO products[8]. Besides, protein kinase C delta is related to monocytic differentiation[9,10]. Those findings indicate that PRKCD has a close relationship with the function of human monocytes (including Mφ). The mature dendritic cell (DC) is considered to be the most potent antigen-presenting cell. Regulation of the DC, particularly its survival, is therefore critical. Bertho et al[11] found that MHC classII-mediated apoptosis of mature DC is produced by activation of the protein kinase C delta isoenzyme. Thrombin can stimulate the production of vascular adhesion molecule-1 (VCAM-1) in endothelial cells, however, it is found to be mediated by the signaling pathways invoved with protein kinase C delta[12]. These findings indicate that PRKCD plays an important role in inducing apoptosis and producing cytokines. However, the effects of down-regulated PRKCD on the Mφ of portal hypertensive spleen remain to be further investigated. IL-1 is an important mediator of inflammation and tissue damage in multiple organs in both experimental animal models and humans[13-15]. The balance between IL-1 and IL-1Ra (interleukin 1 receptor antagonist, IL-1Ra) in local tissues plays an important role in the susceptibility to and severity of many diseases[16,17]. Treatment of rheumatoid arthritis (RA) with daily subcutaneous injections of recombinant IL-1Ra protein has been shown to be efficacious. Gene therapy with IL-1Ra is being evaluated for the treatment of RA and other human diseases[18]. IL1RN encoding IL-1Ra was found down-regulated significantly (the average ratio was 0.179) in the Mφ of portal hypertensive spleen. This leads to the imbalance between IL-1 and IL-1Ra, and it might be related to the pathogenesis of hypersplenism in portal hypertension, but the specific mechanisms need to be further studied.
ASK encoding activator of S phase kinase was found up-regulated in the Mφ of portal hypertensive spleen. Cdc7-Dbf4 kinase complexes, conserved widely in eukaryotes, play essential roles in initiation and progression of the S phase. Cdc7 kinase activity fluctuates during cell cycle, and this is mainly the result of oscillation of expression of the Dbf4 subunit. Yamada et al[19] had isolated and characterized the promoter region of the human ASK gene encoding Dbf4-related regulatory subunit for human Cdc7 kinase, and identified one ASK promoter segment, which was sufficient for mediating growth stimulation. In the Mφ of portal hypertensive spleen, the up-regulation of ASK may lead to the activity enhancement (including phagocytosis) of Mφ, resulting in the pathogenesis of hypersplenism in portal hypertension. Phosphatidylinositol 3-kinase (PIK3) is a key step in the metabolic actions of insulin. One 85 KDa regulatory subunit of PIK3 is encoded by PIK3R1 (phosphoinositide-3-kinase, regulatory subunit 1). It was proved that the expression of PIK3R1 was associated with alterations in glucose/insulin homeostasis[20]. In our study, PIK3R1 was found up-regulated significantly in the Mφ of portal hypertensive spleen, indicating that more insulin existed and the glycometabolism was enhanced in Mφ. Furthermore, enhancement of glycometabolism is regarded as an index of enhanced cell functions, therefore we presume that the up-regulation of PIK3R1 may cause the functional enhancement of Mφ in spleen. However, the possible molecular mechanisms remain undiscovered.
Many differentially expressed genes of Mφ between normal spleen and portal hypertensive spleen have been successfully screened by cDNA microarrays, providing clues and target genes in studying the molecular mechanisms of pathogenesis of hypersplenism in portal hypertension. However, the implication of the gene expression needs to be further investigated.
The destruction of hemocytes by macrophage of spleen plays an important role in the development of hypersplenism in portal hypertension. The authors have proved that phagocytosis of Mφ is augmented in hypersplenism in portal hypertension; however, the specific mechanisms are not clear.
The functions of macrophage are focused in the investigation of pathogenesis of hypersplenism in portal hypertension.
This study based on cDNA microarray has screened differentially expressed genes of macrophages between normal spleen and portal hypertensive spleen, which may provide a new idea in studying the pathogenesis of hypersplenism in portal hypertension.
Small interfering RNAs or other techniques may alter the expression of the differentially expressed genes, and may be used in treatment of hypersplenism in portal hypertension.
The authors identified the difference in gene expression of Mφ between normal spleen and portal hypertensive spleen using cDNA microarrays, found that 121 genes were differentially expressed in all three chips, including 21 up-regulated known genes and 73 down-regulated known genes. The differentially expressed genes were related to the ionic channel and transport protein, cyclin, cytoskeleton, cell receptor, cell signal conduct, metabolism, immune, and so on. These genes might be also related to the hypersplenism in portal hypertension. These differentially expressed genes may provide some clues for further studies of hypersplenism in portal hypertension.
S- Editor Liu Y L- Editor Ma JY E- Editor Lu W
| 1. | Yongxiang W, Zongfang L, Guowei L, Zongzheng J, Xi C, Tao W. Effects of splenomegaly and splenic macrophage activity in hypersplenism due to cirrhosis. Am J Med. 2002;113:428-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 2. | Li ZF, Zhang Y, Gao J, Zhang PJ, Wang JX, Liu XG. Expression and significance of Toll-like receptor 4 of splenic macrophage in patients with hypersplenism due to portal hypertension. Zhonghua YiXue ZaZhi. 2004;84:1088-1091. [PubMed] |
| 3. | Yan F, Li ZF, Zhang S, Yang JH, Li AM, Liu XG. Isolation and purification of macrophages from human spleen. Xi'an Jiaotong Daxue Xuebao. 2004;25:452-455. |
| 4. | Yan F, Li ZF, Su QH, Ma SHY, Cao G, Zhang S. Extraction and productivity of total RNA in macrophages isolated from human spleen. Zhonghua Shiyan Waike Zazhi. 2004;22:176-177. |
| 5. | Zhong WD, He HC, Bi XC, Ou RB, Jiang SA, Liu LS. cDNA macroarray for analysis of gene expression profiles in prostate cancer. Chin Med J (Engl). 2006;119:570-573. [PubMed] |
| 6. | Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467-470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6477] [Cited by in RCA: 5103] [Article Influence: 170.1] [Reference Citation Analysis (0)] |
| 7. | Huppi K, Siwarski D, Goodnight J, Mischak H. Assignment of the protein kinase C delta polypeptide gene (PRKCD) to human chromosome 3 and mouse chromosome 14. Genomics. 1994;19:161-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 8. | Xu B, Bhattacharjee A, Roy B, Feldman GM, Cathcart MK. Role of protein kinase C isoforms in the regulation of interleukin-13-induced 15-lipoxygenase gene expression in human monocytes. J Biol Chem. 2004;279:15954-15960. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 9. | Liu H, Keefer JR, Wang QF, Friedman AD. Reciprocal effects of C/EBPalpha and PKCdelta on JunB expression and monocytic differentiation depend upon the C/EBPalpha basic region. Blood. 2003;101:3885-3892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 10. | Suh KS, Tatunchak TT, Crutchley JM, Edwards LE, Marin KG, Yuspa SH. Genomic structure and promoter analysis of PKC-delta. Genomics. 2003;82:57-67. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 11. | Bertho N, Blancheteau VM, Setterblad N, Laupeze B, Lord JM, Drénou B, Amiot L, Charron DJ, Fauchet R, Mooney N. MHC class II-mediated apoptosis of mature dendritic cells proceeds by activation of the protein kinase C-delta isoenzyme. Int Immunol. 2002;14:935-942. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 12. | Minami T, Abid MR, Zhang J, King G, Kodama T, Aird WC. Thrombin stimulation of vascular adhesion molecule-1 in endothelial cells is mediated by protein kinase C (PKC)-delta-NF-kappa B and PKC-zeta-GATA signaling pathways. J Biol Chem. 2003;278:6976-6984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 97] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 13. | Firestein GS, Berger AE, Tracey DE, Chosay JG, Chapman DL, Paine MM, Yu C, Zvaifler NJ. IL-1 receptor antagonist protein production and gene expression in rheumatoid arthritis and osteoarthritis synovium. J Immunol. 1992;149:1054-1062. [PubMed] |
| 14. | Koch AE, Kunkel SL, Chensue SW, Haines GK, Strieter RM. Expression of interleukin-1 and interleukin-1 receptor antagonist by human rheumatoid synovial tissue macrophages. Clin Immunol Immunopathol. 1992;65:23-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 55] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 15. | Malyak M, Swaney RE, Arend WP. Levels of synovial fluid interleukin-1 receptor antagonist in rheumatoid arthritis and other arthropathies. Potential contribution from synovial fluid neutrophils. Arthritis Rheum. 1993;36:781-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 63] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Beaulieu AD, McColl SR. Differential expression of two major cytokines produced by neutrophils, interleukin-8 and the interleukin-1 receptor antagonist, in neutrophils isolated from the synovial fluid and peripheral blood of patients with rheumatoid arthritis. Arthritis Rheum. 1994;37:855-859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 32] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 17. | Roux-Lombard P, Modoux C, Vischer T, Grassi J, Dayer JM. Inhibitors of interleukin 1 activity in synovial fluids and in cultured synovial fluid mononuclear cells. J Rheumatol. 1992;19:517-523. [PubMed] |
| 18. | Arend WP. The balance between IL-1 and IL-1Ra in disease. Cytokine Growth Factor Rev. 2002;13:323-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 534] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 19. | Yamada M, Sato N, Taniyama C, Ohtani K, Arai K, Masai H. A 63-base pair DNA segment containing an Sp1 site but not a canonical E2F site can confer growth-dependent and E2F-mediated transcriptional stimulation of the human ASK gene encoding the regulatory subunit for human Cdc7-related kinase. J Biol Chem. 2002;277:27668-27681. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 20. | Almind K, Delahaye L, Hansen T, Van Obberghen E, Pedersen O, Kahn CR. Characterization of the Met326Ile variant of phosphatidylinositol 3-kinase p85alpha. Proc Natl Acad Sci USA. 2002;99:2124-2128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 27] [Article Influence: 1.2] [Reference Citation Analysis (0)] |