Published online May 21, 2007. doi: 10.3748/wjg.v13.i19.2717
Revised: February 20, 2007
Accepted: March 1, 2007
Published online: May 21, 2007
AIM: To investigate the expression patterns of TTYH2 in the human colon cancer and colon cancer cell lines and to evaluate the inhibitory effect of small interfering RNA (siRNA) on the expression of TTYH2 in colon cancer cell lines.
METHODS: We investigated the expression patterns of TTYH2 in colon cancer, adjacent non-tumorous colon mucosa, and cancer cell lines (DLD-1, caco-2, and Lovo) by RT-PCR. Furthermore, a siRNA plasmid expression vector against TTYH2 was constructed and transfected into DLD-1 and Caco-2 with LipofectamineTM 2000. The down regulation of TTYH2 expression was detected by RT-PCR and the role of siRNA in inducing cell proliferation and cell aggregation was evaluated by MTT and aggregation assay.
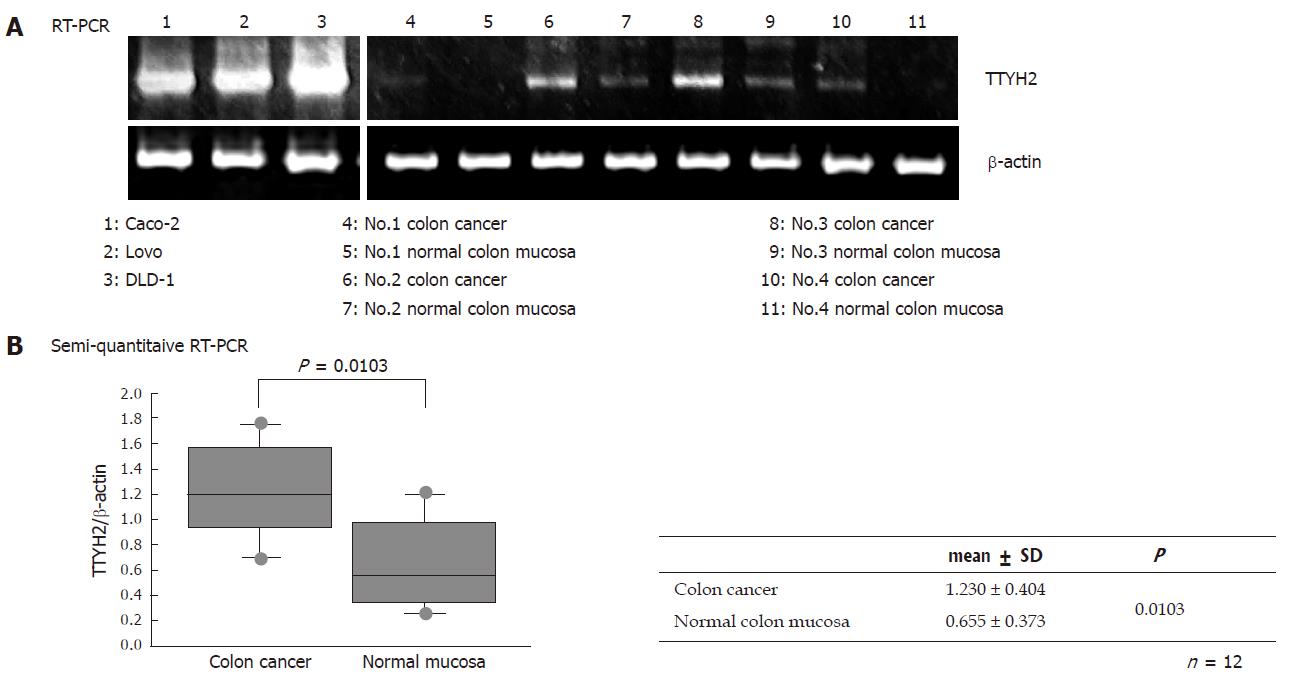
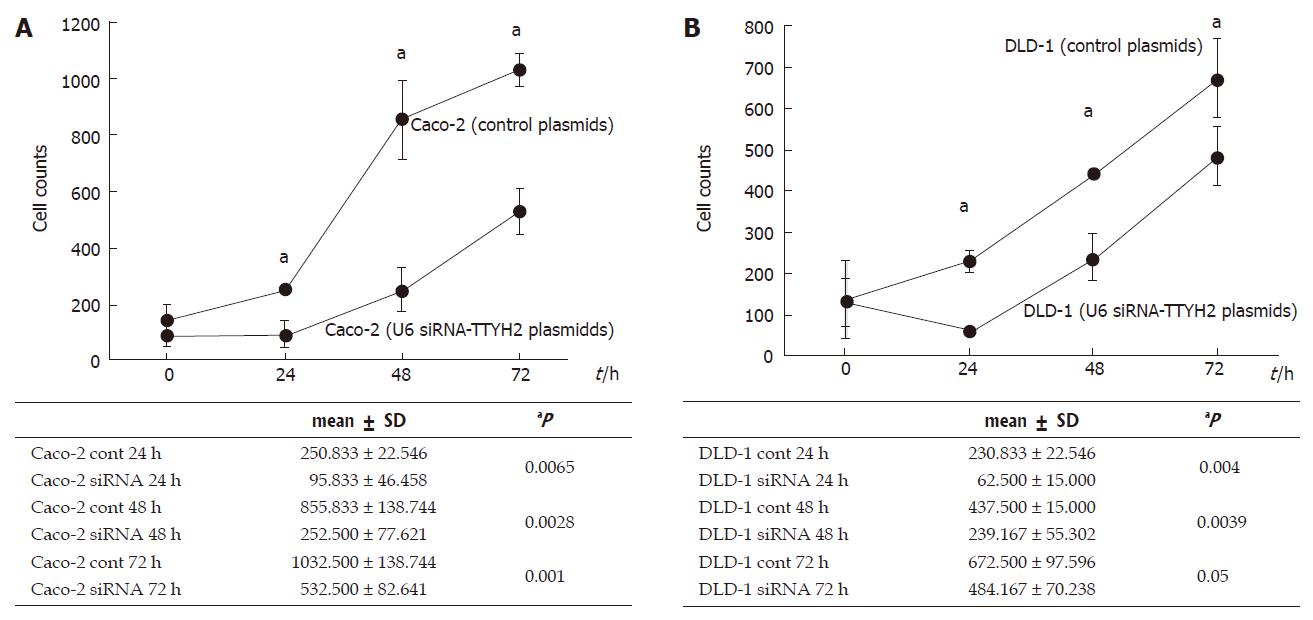
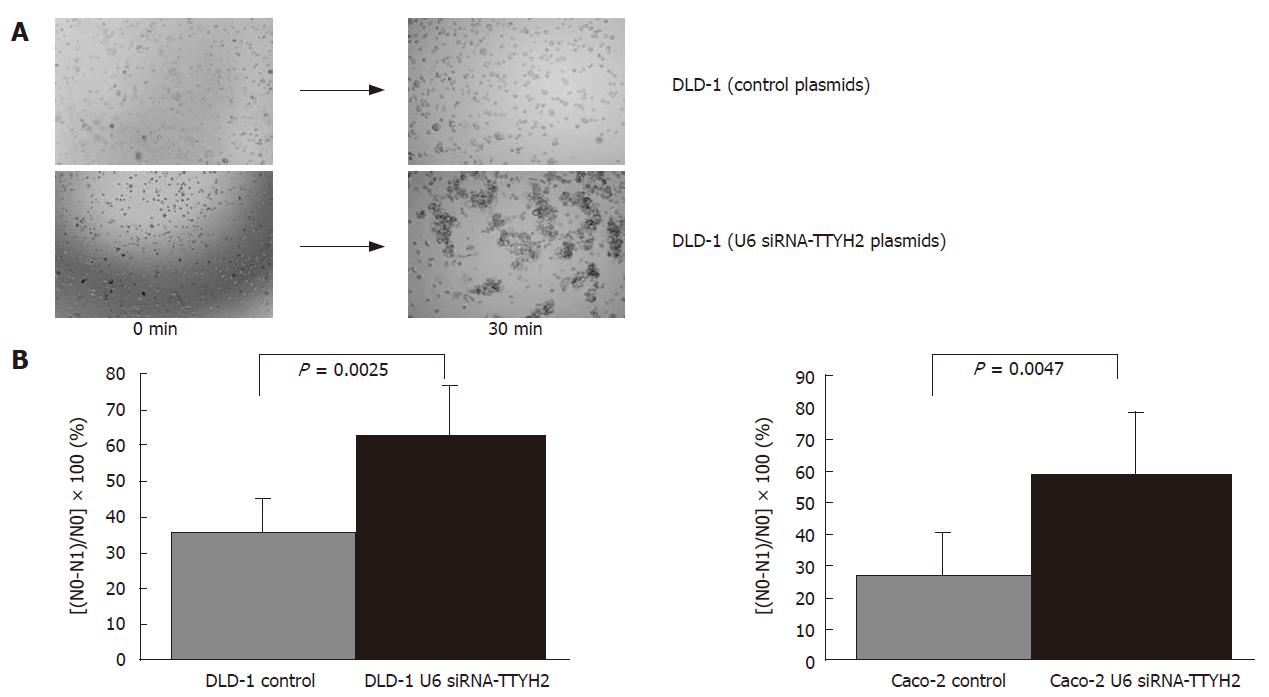
RESULTS: TTYH2 gene expression in colon cancer tissue was significantly up-regulated compared with normal colonic mucosa (1.23 ± 0.404 vs 0.655 ± 0.373, P = 0.0103). Colon cancer derived cell lines including DLD-1, Caco-2, and Lovo also expressed high levels of TTYH2. In contrast, transfection with siRNA-TTYH2 significantly inhibited both proliferation and scattering of these cancer cell lines.
CONCLUSION: The present work demonstrates, for the first time, that the TTYH2 gene expression is significantly up-regulated in colon cancer. The TTYH2 gene may play an important role in regulating both proliferating and metastatic potentials of colorectal cancer.
-
Citation: Toiyama Y, Mizoguchi A, Kimura K, Hiro J, Inoue Y, Tutumi T, Miki C, Kusunoki M. TTYH2, a human homologue of the
Drosophila melanogaster gene tweety, is up-regulated in colon carcinoma and involved in cell proliferation and cell aggregation. World J Gastroenterol 2007; 13(19): 2717-2721 - URL: https://www.wjgnet.com/1007-9327/full/v13/i19/2717.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i19.2717
It is well accepted that colorectal cancer develops in multiple steps with accumulation of genetic aberrations such as activation of oncogenes and inactivation of tumor suppressor genes[1]. However, detailed delineation of the carcinogenesis process has not yet been accomplished and, naturally, a complete cure for patients with advanced colorectal cancer must await new targets and strategies, despite the number of therapeutic approaches that have so far been exploited. As a consequence, colorectal carcinoma is one of the most common causes of cancer-related deaths in the Western world. More desirable therapies require diagnosis of colorectal cancer at early stages and treatments with more efficient drugs. One way to achieve this goal is to identify and characterize the key molecules participating in colon tumorigenesis[2].
Rae et al[3] demonstrated that a novel gene was up-regulated in renal cell carcinoma using differential display PCR. Characterization of the full-length cDNA and gene revealed that the encoded protein is a human homologue of the Drosophila melanogaster tweety protein, and the novel protein has been termed TTYH2.
TTYH2 is located at 17q24. The encoded human TTYH2 protein has 534 amino acids and, like the other members of the tweety-related protein family, is a putative cell surface protein with five transmembrane regions[3]. Using comparative genomic hybridization, high-level amplification of 17q24-q25, the region containing TTYH2, has been found in adrenocortical tumors[4], brain metastases of solid tumors[5], and muscle-invasive bladder cancer[6]. These findings could suggest that up-regulated TTYH2 gene expression might play an important role in the carcinogenesis of various cancers. Furthermore, TTYH2 functions as a cell surface receptor, and its up-regulation may give a growth advantage or metastatic ability to cancer cells.
The aim of this study was to evaluate whether TTYH2 gene expression is altered in colorectal cancer compared with a normal colon, and furthermore to elucidate the role of this putative transmembrane protein in colon cancer cells by sequence-specific silencing of TTYH2 gene expression.
A total of twelve colorectal primary cancer tissues and corresponding normal colonic epithelium specimens were obtained from surgical resections. All patients with tumors were diagnosed at an advanced stage, and all normal tissues were histopathologically confirmed to be free of cancer. A piece of each tissue sample was immediately frozen in liquid nitrogen on resection and stored at -80°C until use. All patients were investigated after obtaining their informed consent to participate in this study. Investigations were performed in accordance with the Helsinki Declaration, and were approved by the Institutional Review Board.
Three human colon cancer cell lines, DLD-1, Caco-2 and Lovo were obtained from the RIKEN Cell Bank (Ibaragi, Japan). These cells were grown in monolayer culture in RPMI 1640 (Sigma-Aldrich Inc. St Louis, MO, USA), supplemented with fetal bovine serum (10% (v/v); Gibco BRL, Tokyo, Japan), glutamine (2 mmol/L), penicillin (100 000 U/L), streptomycin (100 mg/L), and gentamycin (40 mg/L), at 37°C in a 5% CO2 environment. For routine passages, cultures were split 1:10 when they reached 90% confluence, generally every 3 d. For all experiments, cells at the fifth to ninth passage were used. All experiments were performed with exponentially growing cells.
All mucosa were dissected from the underlying tissue and homogenized in Physcotron (NITI-ON, Japan) for the isolation of total RNA. Total RNA was extracted with Sepasol-RNAI(Nacalai Tesque, Tokyo, Japan), and the quality of the total RNA was judged from the ratio between 28S and 18S RNase after 1% hormarine gel electrophoresis. Contaminated genomic DNA template was removed by DNase1 treatment (Takara 2215A).
Oligo(dT)-primed cDNA was prepared from this RNA (2 μg) by reverse transcription using an Omniscript RT kit (QIAGEN). RT-PCR was performed using specific primers for TTYH2 and β-actin. Primer sequences were as follows: β-actin, 5'-ACAGAGCCTCGCCTTTGC -3' (sense primer) and 5'-GCGGCGATATCATCATCC-3' (antisense primer); and TTYH2, 5'-CGGGATCCGCCATGGGCACGCGTCTGCCGCTCGTCCTG-3’ (sense primer) and 5’-CCGCTCGAGGAACAAGCCTTTAACTTGTTCTGTTTCGG-3’ (antisense primer).
The PCR reactions included 2 × GC buffer, 2.5 mmol/L dNTP mix, 80 ng of each primer, and LA taq DNA polymerase. Optimum cycling parameters, in the linear range of amplification, consisted of 30 sec of denaturation at 94°C, 30 sec of annealing at 58°C, and 2 min of elongation at 72°C, and 30 cycles were performed for the selected gene. A control PCR was also done for β-actin, which served as a standard for sample normalization for 25 cycles. Amplified products were separated electrophoretically, visualized, and photographed under UV light after ethidium bromide staining. Gel density was calculated using Sicon Image (downloaded from the web site: http://www.siconcorp.com), and gene expression was quantified.
Construction of siRNA expression plasmids was based on the U6 siRNA expression vector (Takara, Mie, Japan), which includes a human U6 promoter, a puromycin-resistance gene, and two BspMI sites. The set of sense and antisense oligonucleotides were annealed and ligated into the vector. Sense oligo: CACCGCTTGAATCTCGTCTTCCTACGTGTGCTGTCCGTAGGAAGATGAGGTTCAGGCTTTTT; antisense oligo: GCATAAAAAGCCTGAACCTCATCTTCCTACGGACAGCACACGTAGGAAGACGAGATTCAAGC. Approximately 1.5 × 105 cells/well were plated in a 12-well plate in media containing 10% FCS to give 80%-90% confluence, and transfection of U6 siRNA-TTYH2 plasmids was performed using Lipofectoamine 2000 (4 μL/well; Life Technologies Inc.) to result in a final RNA concentration of 100 nmol/L. After 48 h, the cells were lysed, and RNA was isolated as described above. Expression of the gene was evaluated by RT-PCR, and siRNA expression vectors without inserts were used as controls (Takara).
Cell proliferation assay was evaluated using a Cell Counting Kit (Dojindo Laboratories, Japan) according to the manufacturer’s instructions. Cells 24 h after transfection and control cells (1 × 102 cells/well) were seeded into 96-well cell plates (Becton Dickinson Labware, NJ, USA) in 100 μL of culture medium. After incubation for 0 h, 24 h, 48 h, and 72 h, the medium was discarded and replaced with 90 μL of fresh medium, followed by the addition of 10 μL WST-8 reagent solution and incubation for 4 h at 37°C in the incubator. Cell viability was determined according to a colorimetric comparison by reading optical density values from a microplate reader (SoftMax, Molecular Devices Corporation, CA, USA) at an absorption wavelength of 450 nm.
Cells were grown to confluence in 60-mm tissue culture dishes. Cultures were trypsinized (0.04% trypsin in PBS containing 1 mmol/L Ca2+) to prepare single-cell suspensions and were re-suspended in complete medium. Samples of 2 mL containing 2 × 106 single cells were incubated at 37°C with constant shaking at 90 r/min for a total of 30 min. At 0 min and 30 min, the number of single cells was counted using a hemocytometer. The rate of aggregation was calculated as the percentage of the number of single cells in a microscope field using the formula [(N0-N1)/N0] × 100, where N0 is the total number of cells and N1 is the number of single cells detected in the cultures at the different incubation times.
The results are expressed as mean ± SD. The Mann-Whitney U test was used for comparisons among unpaired groups. Results of two-sided statistical test in which P values were less than 0.05 were considered to be statistically significant.
Using RT-PCR, we showed that TTYH2 is upregulated in four colon cancers of four paired samples and essentially no expression, or low levels, of TTYH2 expression in the normal colon samples. In addition, RT-PCR performed on the colon cancer-derived cell lines Caco-2, Lovo and DLD-1 indicated that this gene is high expressed in these cell lines (Figure 1A). To confirm the up-regulation of this gene in colon cancer and to examine a larger number of samples, we performed semi-quantitative RT-PCR eight matched colon cancer and normal colon paired samples. As shown in Figure 1B, TTYH2 gene expression in colon cancer tissue were significantly up-regulated compared with normal colonic mucosa (1.230 ± 0.404 vs 0.655 ± 0.373, respectively: P = 0.013).
To determine any changes occurring in the cell growth pattern due to reduced TTYH2 expression, 1.5 × 105 cells/well were plated in a 12-well plate in media containing 10% FCS to give 80%-90% confluence. We examined whether down-regulation of TTYH2 influenced the growth of Caco-2 and DLD-1. Figure 2A and B show that transfection with U6 siRNA-TTYH2 plasmids obviously inhibited the proliferation of Caco-2 and DLD-1 cells compared with the control plasmid during the course of the study.
Varieties of phenotypic characteristics are required for a cancer cell to successfully complete the metastatic cascade. Among these, acquisition of a motile phenotype is needed for the cell to become metastatically competent. The other factors that may critically affect tumor cell metastasis are the aggregation and adhesive properties of the cells. These properties are usually altered in tumor cells, either because of direct changes in the expression of cell surface adhesion molecules or because of changes in other membrane proteins that can potentially disrupt the interaction between these adhesion molecules[7,8,12]. Therefore, we sought to determine whether reduced expression of TTYH2 would influence the aggregation of DLD-1 and Caco-2 cells. Down-regulations of TTYH2 in DLD-1and Caco-2 by transfection with U6 siRNA-TTYH2 plasmids resulted in significantly enhanced aggregation compared with control cells (P = 0.0025, P = 0.0047). The rates of aggregation potential for transfected DLD-1 cells and control DLD-1 cells were 62.3% ± 9.1% and 35.2% ± 5.9% respectively. The rates of aggregation potential for transfected Caco-2 cells and control caco-2 cells showed the same pattern of DLD-1 cells (59.3% ± 12.4% and 27.1% ± 7.9%, respectively) (Figure 3A and B).
This study is the first to compare TTYH2 gene expression patterns between paired colon cancer and histologically normal colonic mucosa. Using semi-quantitative RT-PCR, TTYH2 gene expression was significantly up-regulated in colon cancer tissue compared with normal mucosa. TTYH2 gene expression in colon cancer cell lines (Caco-2, Lovo, and DLD-1) was also high compared with normal colonic mucosa. It is therefore conceivable that up-regulation of TTYH2 is an event in the malignant transformation of the colonic mucosa.
A motif scan showed that TTYH2 contains five predicted transmembrane domains. Furthermore, a recent study[9] demonstrated that TTYH2 was expressed as a Ca2+-dependent inward rectifier chloride channel in transfected Chinese hamster ovary cells. Its up-regulation may give a growth advantage or metastatic ability to colon cancer. To further elucidate the role of this putative transmembrane protein in colon cancer, we investigated whether the silencing of TTYH2 expression results in altered tumor cell behavior (e.g. growth pattern and metastasis) in colon cancer cell lines.
Normal cell growth is maintained and modulated by positive (cell proliferative) and negative (apoptotic) signals. Loss of regulatory mechanisms as a consequence of either loss of tumor suppressor gene function or deregulated tumor progressor gene function may contribute to the oncogenic process. The present study shows that Caco-2 and DLD-1 transfected with a silencing-TTYH2 vector exhibited suppressed tumor cell growth with low TTYH2 expression. Therefore, the data indicates that TTYH2 possesses a tumor progressor function in colon cancer.
In vivo metastasis of tumor cells is an extremely complicated process that involves several consecutive events (i.e. detachment of tumor cells from primary site, intravasation into bloodstream, evasion of immune surveillance, adherence to vascular endothelial cells of distant organs, and finally extravasation into such tissues)[10,11]. During the initial process of tumor cell metastasis, weakening of cell-matrix and cell-cell interactions is imperative for the detachment of tumor cells from primary sites.
Over expression of cell surface Muc4/SMC has been reported to disrupt integrin-mediated cell adhesion as well as homotypic cell-cell interactions, causing the dissociation of tumor cells in culture[12]. Furthermore, MUC1 has been implicated in determining the adhesion properties of cells, primarily through the modulation of integrin-mediated adhesion in melanoma cell lines[13]. Similarity, TTYH2 contains a RGD motif potentially capable of integrin binding[9]. Consistent with the proposed mechanism, the present data demonstrated enhanced adhesion and cell-cell aggregation in low TTYH2-expressing Caco-2 and DLD-1 cell lines.
In conclusion, we propose that TTYH2 is implicated in tumor growth and metastasis by directly altering tumor cell properties (aggregation). The present work is the first demonstration of a direct association of TTYH2 with the metastatic colon cancer phenotype. Moreover, it provides evidence for the functional role of TTYH2 in tumor cell growth and behavior and suggests a critical role for TTYH2 up-regulation in colon adenocarcinoma.
S- Editor Liu Y L- Editor Li M E- Editor Chen GJ
| 1. | Kountouras J, Boura P, Lygidakis NJ. New concepts of molecular biology for colon carcinogenesis. Hepatogastroenterology. 2000;47:1291-1297. [PubMed] |
| 2. | Kitahara O, Furukawa Y, Tanaka T, Kihara C, Ono K, Yanagawa R, Nita ME, Takagi T, Nakamura Y, Tsunoda T. Alterations of gene expression during colorectal carcinogenesis revealed by cDNA microarrays after laser-capture microdissection of tumor tissues and normal epithelia. Cancer Res. 2001;61:3544-3549. [PubMed] |
| 3. | Rae FK, Hooper JD, Eyre HJ, Sutherland GR, Nicol DL, Clements JA. TTYH2, a human homologue of the Drosophila melanogaster gene tweety, is located on 17q24 and upregulated in renal cell carcinoma. Genomics. 2001;77:200-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 4. | Dohna M, Reincke M, Mincheva A, Allolio B, Solinas-Toldo S, Lichter P. Adrenocortical carcinoma is characterized by a high frequency of chromosomal gains and high-level amplifications. Genes Chromosomes Cancer. 2000;28:145-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 5. | Petersen I, Hidalgo A, Petersen S, Schlüns K, Schewe C, Pacyna-Gengelbach M, Goeze A, Krebber B, Knösel T, Kaufmann O. Chromosomal imbalances in brain metastases of solid tumors. Brain Pathol. 2000;10:395-401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 45] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 6. | Simon R, Burger H, Semjonow A, Hertle L, Terpe HJ, Bocker W. Patterns of chromosomal imbalances in muscle invasive bladder cancer. Int J Oncol. 2000;17:1025-1029. [PubMed] |
| 7. | Sommers CL. The role of cadherin-mediated adhesion in breast cancer. J Mammary Gland Biol Neoplasia. 1996;1:219-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 8. | Truant S, Bruyneel E, Gouyer V, De Wever O, Pruvot FR, Mareel M, Huet G. Requirement of both mucins and proteoglycans in cell-cell dissociation and invasiveness of colon carcinoma HT-29 cells. Int J Cancer. 2003;104:683-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 9. | Suzuki M, Mizuno A. A novel human Cl(-) channel family related to Drosophila flightless locus. J Biol Chem. 2004;279:22461-22468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 113] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 10. | Fidler IJ. Critical factors in the biology of human cancer metastasis: twenty-eighth G.H.A. Clowes memorial award lecture. Cancer Res. 1990;50:6130-6138. [PubMed] |
| 11. | Folkman J. Endothelial cells and angiogenic growth factors in cancer growth and metastasis. Introduction. Cancer Metastasis Rev. 1990;9:171-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 97] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 12. | Komatsu M, Carraway CA, Fregien NL, Carraway KL. Reversible disruption of cell-matrix and cell-cell interactions by overexpression of sialomucin complex. J Biol Chem. 1997;272:33245-33254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 122] [Cited by in RCA: 136] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 13. | Wesseling J, van der Valk SW, Vos HL, Sonnenberg A, Hilkens J. Episialin (MUC1) overexpression inhibits integrin-mediated cell adhesion to extracellular matrix components. J Cell Biol. 1995;129:255-265. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 359] [Cited by in RCA: 373] [Article Influence: 12.4] [Reference Citation Analysis (0)] |