Published online Apr 28, 2007. doi: 10.3748/wjg.v13.i16.2363
Revised: February 28, 2007
Accepted: March 8, 2007
Published online: April 28, 2007
AIM: To investigate the expression of selenoprotein P mRNA (SePmRNA) in tissues of normal liver, liver cirrhosis and hepatocellular carcinoma (HCC), and its relationship with HCC occurrence and development.
METHODS: The expression of SePmRNA in tissues of normal liver, liver cirrhosis and HCC were detected by in situ hybridization using a cDNA probe.
RESULTS: The enzyme digesting products of PBluescript-Human Selenoprotein P were evaluated by electrophoresis. The positive expression of SePmRNA was found in the tissues of normal liver, liver cirrhosis and HCC. The expression of SeP mRNA was found in hepatic interstitial substance, especially in endothelial cells and lymphocytes of vasculature. The positive rate of SePmRNA in normal liver tissue was 84.6% (11/13) and the positive signals appeared in the nucleus and cytoplasm, mostly in the nucleolus, and the staining granules were larger in the nucleolus and around the nucleus. The positive rate of SePmRNA in liver cirrhosis tissue was 45.0% (9/20) and the positive signals were mainly in the nucleolus and cytoplasm, being less around the nucleus and inner nucleus than that in normal liver tissue. The positive rate of SePmRNA in HCC tissue was 30.0% (9/30) and the positive signals were in the cytoplasm, but less in the nucleus.
CONCLUSION: SePmRNA expression in the tissues of normal liver and HCC is significantly different (84.6% vs 30.0%, P = 0.003), suggesting that SeP might play a role in the occurrence and development of HCC.
- Citation: Li CL, Nan KJ, Tian T, Sui CG, Liu YF. Selenoprotein P mRNA expression in human hepatic tissues. World J Gastroenterol 2007; 13(16): 2363-2368
- URL: https://www.wjgnet.com/1007-9327/full/v13/i16/2363.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i16.2363
The trace element selenium (Se) is an essential human nutrient[1]. The relationship between Se and cancers has attracted attention of scholars. There is strong evidence that the Se deficiency can result in certain malignant tumors. It has been proved that the occurrence of some cancers, such as prostate cancer, colon cancer, lung cancer and HCC, can be decreased by administration of selenium as a dietary supplement. Se has been shown to prevent cancers directly[2,3], especially liver and stomach cancers. It can resist and relieve the carcinogenesis of the mutagen and carcinogen. Se also can block the precancerous lesions. Se plays its biological role as the form of selenoproteins. Se incorporates into protein in the forms of the rare amino acids selenocysteine (Sec) and selenomethionine (SeMet)
in vivo. Sec is an active form of Se in selenoproteins and has important biological functions. The term “selenoprotein” is restricted to the proteins containing Sec. Selenoprotein P (SeP) is unique in its highest Se content, it contains 8 or 10 Sec residues in its polypeptide chain[4]. Almost all the tissues contain SeP. In recent years, many studies have confirmed that SeP is related to various malignant tumors. Eighteen kinds of selenoproteins including SeP have been found to have biological functions. The relationship between SeP and future risk of cancers has become the hot spot of studies in recent years. However, the expression of SeP in HCC tissues is rarely known. For this reason, plasmid PBluescript-Human Selenoprotein P from Georgia University in America was set into the SeP cDNA probe to detect the expression of SePmRNA in the tissues of normal liver, liver cirrhosis and HCC in this trial, and to investigate the relationship between SeP and the occurrence and the development of HCC so that to provide new ideas for the treatment and prevention of HCC.
Thirty biopsy tissues of HCC patients (18 men and 12 women, with a mean age of 56.9 years) including 16 with chronic viral hepatitis and 20 biopsy tissues of liver cirrhosis patients (12 men and 8 women, with a mean age of 53.6 years), including 13 with chronic viral hepatitis, were collected from The First Affiliated Hospital of the School of Medicine, Xi’an Jiaotong University from January 1999 to January 2001. All the patients were pathologically confirmed and none of them had the history of chemotherapy or radiotherapy. Thirteen liver tissues of healthy men (with a mean age 49.8 years) who died accidentally were collected as controls at the same period. All the specimens were fixed in 40 g/L formaldehyde, embedded in paraffin, and cut into 3 μm serial sections. HE staining was done to confirm the pathological diagnosis once more.
Plasmid named PBluescript-Human Selenoprotein P was kindly given as a present by Georgia University in the USA. It has 6400 base pairs and was resistant to ampicillin. Its host bacteria JM109 were from Huamei Shengwu Engineering Company. A small amount of plasmid extraction kit and gel extraction kit were obtained from Huashun Shengwu Engineering Company. The restriction endonuclease SacI and XhoI were purchased from Takara. Digoxin probe labelling and detection kits were products of Boehringer Mannheim Company in Germany.
PBluescript-Human Selenoprotein P was eluted from the membrane with 20 μL pH8.0 Tris-HCl buffer for 30 min. The extraction solution was added into the JM109 competent cells, mixed slightly, ice bathed for 30 min, then heat shocked (42°C) for 90 s, quickly transferred onto ice for 2 min, and 800 μL LB culture medium were added, incubated at 37°C for 45 min, and centrifugated at 5000 r/min for 5 min. The deposit and 100 μL supernatant were mixed absolutely, and spread on the LB agarose plate containing ampicillin (100 mg/L) at 37°C overnight. One monoclonal colony was randomly chosen, put into 5 mL of 10 g/L LB culture medium containing ampicillin (100 mg/L), and vigorously shaken (2000 r/min) at 37°C overnight in an air bath to make the absorbance (A) value of the liquid to 280. The cultural product was centrifugated at 12 000 r/min for 30 s three times to harvest the bacteria. The liquid was added to extract the plasmid. The procedure for plasmid extraction was performed according to the instructions of the low-dose plasmid extraction kit from Huashun. The 6 μL plasmid was diluted to 600 μL with sterilized pure water to detect the A value at 260, 280 and 320 nm with pure water as the control and the ratio of A 260 and A 280 so that the quantity of the extracted plasmid could be calculated and the efficiency of the purification could be analyzed.
PBluescript-Human Selenoprotein P was digested with endonuclease and evaluated to ensure this trial could proceed smoothly. The 5 μL plasmid of SeP previously extracted was digested with SacI and XhoI. The reaction system included 5 μL plasmid, 1 μL SacI, 1 μL XhoI, 2 μL 10 × T buffer, 2 μL 10 × BSA buffer and 9 μL sterilized water. The mixture was incubated at 37°C for 3h. Finally 10 μL reaction mixtures were used for 10 g/L agarose gel electrophoresis for 45 min. The plasmid that was not digested served as the control. The standard marker was DL2000 + 15 000 DNA marker. The result was registered by a gel electrophoresis auto-analysis system. Then 500 μL liquid containing bacteria was added to 5 mL LB culture medium, and shaken (160 r/min) overnight in an air bath at 37°C. When the mixture looked like heavy fog, the plasmid was extracted according to the above-mentioned method. SacI and XhoI were used to digest the plasmid. The only difference was that the reaction mixture was enlarged to 50 μL. All the reaction mixtures were used for 10 g/L agarose gel electrophoresis for 45 min. Objective fragments were cut. Finally, the gel extraction was performed according to the instructions of the low-dose gel extraction kit from Huashun. The objective DNA fragments extracted above were further extracted to wipe off impurity with phenol and chloroform, and deposited in ethanol. The deposition was dissolved with 15 μL sterilized water. The procedure was performed according to the methods for molecular biology experiments. The objective DNA fragments were labeled with a digoxin probe labelling kit according to the instructions from Boehringer Mannheim Company in Germany.
All the specimens including tissues of HCC, liver cirrhosis, and normal liver were sectioned into 3-μm slices serially. The slides were treated with DEPC and polylysine, dewaxed normally, put in water, and treated with 30 mL/L H2O2 at room temperature for 10 min to inactivate the endogenous peroxidase, and with 0.2 mol/L hydrochloric acid to denature the proteins. Fresh diluted protease K of 0.01 mol/L was put on the sections at 37°C and was kept for 5-15 min, washed with distilled water three times, and washed again with 2 g/L glycine for 5 min. After a while, it was washed with PBS for 5 min, fixed with 4 g/L polymethanel and kept for 30 min, again washed with PBS for 5 min, dehydrated with low to high gradient alcohol to the degree of 100%. Then it was washed with DEPC, and the digoxin labeled probe was boiled in 90-100°C water and kept for 10 min to denature the probe. After that, it was taken out and immediately put into shattered ice and kept for 10 min. After the sections became dry in the air, 20 μL in situ hybridization solution containing digoxin-labelling probe was put onto each sections. After that the protective membrane was taken off, and a spare in situ hybridized slide was placed onto the sections, and the hybridization was continued in a wet box at 42°C overnight, the sections were then taken out and washed twice with 2 × SSC in 20-30°C water for 5 min and with 1 × SSC preheated at 37°C for 10 min. Then the blocking liquid was put on the sections and kept at room temperature for 30 min to block out the nonspecific antigen. After that anti-mouse digoxin was put on the sections and kept at 42°C for 2 h, washed with BufferIthree times, each time for 5 min, then washed with Buffer III once for 10 s. NBT was used to show the color. It was dehydrated with low to high gradient alcohol, made transparent with dimethylbenzene and sealed with resin. Finally, the results were observed under an Olympus light microscope. All the equipments and buffers used in in situ hybridization must be treated with DEPC. In the negative control, the hybridization solution was replaced by reserve hybridization solution containing no SeP cDNA probe. Blue granules in the cytoplasm and/or nucleus were considered as positive staining.
The χ2 test and partition of χ2 method were performed to compare the differences among the three groups. All the data were analyzed with statistical software SPSS10.0 and P < 0.05 was considered significant.
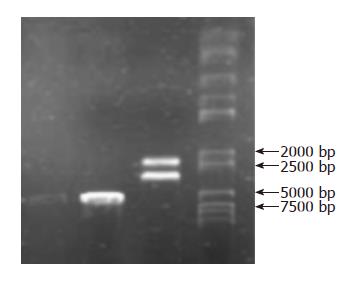
After the low-dose plasmid extraction, the strap of the PBluescript-Human Selenoprotein P was between 5 kb and 7 kb in gel electrophoresis. When digesting with SacI and XhoI, two straps could be found in gel electrophoresis, one was the strap of the SeP segment about 2 kb, the other was the strap of the plasmid carrier about 4.4 kb. All the results were consistent with the expected values (Figure 1).
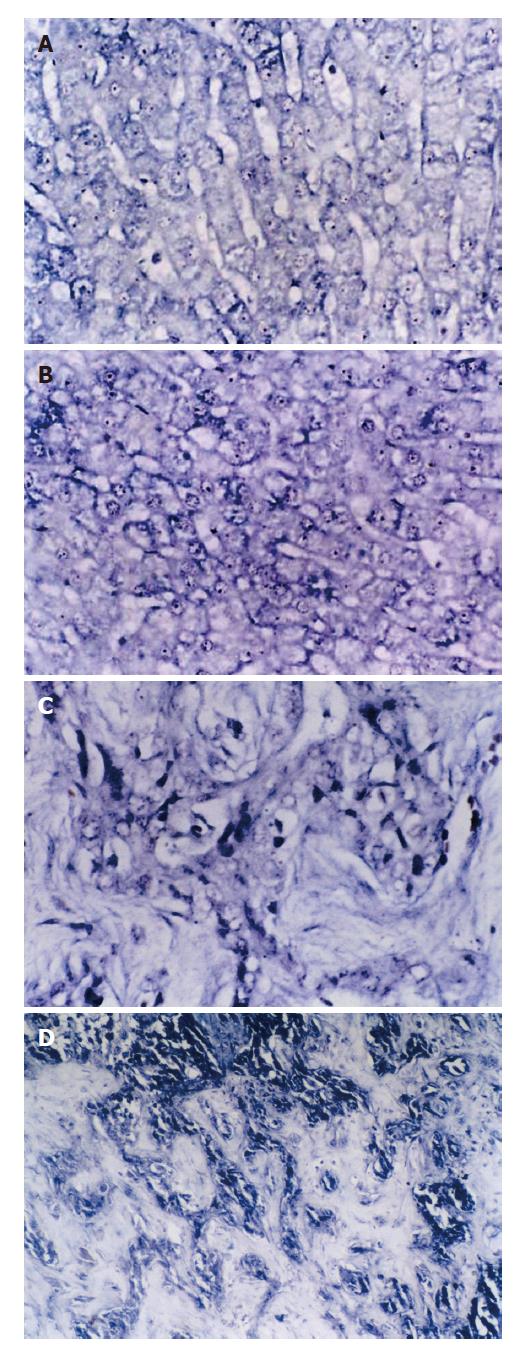
The positive expression of SePmRNA could be found in tissues of normal liver, liver cirrhosis and HCC. In normal liver tissues, the positive signals were found mainly in the nucleus and cytoplasm, most significantly in the nucleolus, and the stained granules were large in the nucleolus and around the nucleus (Figure 2A). In the tissues of liver cirrhosis, the positive signals were found mainly in the nucleolus and cytoplasm, but less around the nucleus and inner nucleus than that in normal liver tissues (Figure 2B). In HCC tissues the positive signals were in the cytoplasm, but less in the nucleus (Figure 2C). SePmRNA expression could be observed in all 3 kinds of liver tissues, mainly in vascular endothelial cells and lymphocytes of vasculature (Figure 2D). Blue granules in the cytoplasm and/or nucleus were regarded as positive. Negative controls showed negative expression, and there was no blue granule in either the cytoplasm or nucleus. The positive rate of SePmRNA expression in tissues of normal liver, liver cirrhosis and HCC were 84.6% (11/13), 45.0% (9/20) and 30.0% (9/30) respectively. SePmRNA expression in the tissues above had significant difference (corrected χ2 = 10.903, P = 0.004; Table1). Further analysis showed that SePmRNA expression in the tissues of normal liver and HCC had significant difference (84.6% vs 30.0%, corrected χ2 = 8.790, P = 0.003; Table 1). There was no significant difference in SePmRNA expression in the tissues between normal liver and liver cirrhosis (84.6% vs 45.0%, corrected χ2 = 3.653, P = 0.056; Table 1), and significant difference between HCC and liver cirrhosis (30.0% vs 45.0%, corrected χ2 = 0.611, P = 0.434; Table 1).
| Liver tissue | n | SePmRNA expression | Positive rate (%) | |
| (+) | (-) | |||
| Normal liver | 13 | 11 | 2 | 84.6 |
| Liver cirrhosis | 20 | 9 | 11 | 45 |
| HCC | 30 | 9 | 21 | 30.0a |
Liver cancer is one of the most common cancers in China[5]. It seriously endangers the people’s health. There have been a number of studies on liver cancer, but there is no great breakthrough. The molecular mechanism of the occurrence and development of liver cancer is still unknown. A number of epidemiological studies, animal model systems and interventional trials have found that a low dietary intake of Se is related to an increased risk of liver cancer. There was inverse association between plasma Se levels and the risk of HCC. Buljevac et al found that the mean Se levels were significantly lower in the HCC and liver cirrhosis cases than in the controls (P < 0.01). Another study examined the association between plasma Se levels and the risk of HCC among chronic carriers of hepatitis B and/or C virus in a cohort of 7342 men in Taiwan. Mean Se levels were significantly lower in HCC cases than in the HBsAg-positive controls (P = 0.01). Adjusted odds ratios of HCC for subjects in increasing quintiles of plasma Se were 1.00, 0.52, 0.32, 0.19, and 0.62, respectively[6]. Many scholars analyzed the plasma Se levels of people in Qidong County, Jiangsu Province of China, where the incidence of HCC is highest in China. They found the inverse relation between plasma Se levels and the epidemic of HCC. A dietary supplement of Se could reduce the incidence of HBV infection by 77.2% and the precancerous lesions by 75.8%. An interventional trail on 130 471 people with Se refined salt was made. After a follow-up of 8 years, the incidence of HCC was found reduced by 35.1% in the treatment group. But after stopping the dietary supplement of Se, the incidence would increase to the level of the placebo group. Young et al[7] compared 226 kinds of epidemiological risk factors of cancers, and concluded that Se supplement might reduce the incidence of HCC[7]. Basic research indicated that Se could selectively restrain and kill the HCC cells in vitro, but had no influence on normal hepatocytes. Recent research has proven in human supplementation trials that Se should be safe as SeMet supplementation but not as selenite[8].
Se is present in known human selenoproteins as the amino acid Sec [9]. Sec represents the 21st amino acid and is encoded by the UGA triplet in selenoprotein mRNA[10,11]. SeP is the quantitatively most important selenoprotein in both rat and human plasma as it contains more than 50% of the total amount of plasma Se[4]. So there has been great interest in the relationship between SeP and HCC. SeP was first published in 1977, and many kinds of cells could secrete SeP[12]. The SeP synthesized by the liver is secreted into plasma[13,14]. It is also produced by the astroglia, cerebellum granular cells and striated myocytes. SeP is a kind of extracellular glycoprotein. The gene of SeP has been cloned and sequenced[15]. In human plasma, the SeP concentration is dependent on the dietary Se status, cytokines[16], and is related to the risk of cancers.
Although studies on the mechanism showed that Se suppresses tumors the research is few in cellular and molecular levels. To get a better understanding of the mechanism by which Se suppressed tumors, we chose the newly found selenoprotein SeP to investigate its relationship with HCC and the function of this protein so as to provide new thoughts in the treatment of liver cancer. Our experiment showed that there was positive expression of SePmRNA in tissues of normal liver, liver cirrhosis and HCC with positive rates of 84.6% (11/13), 45.0% (9/20) and 30.0% (9/30), respectively the difference being significant (P < 0.05).
The premorbid level of SeP in plasma from patients with cancer at different sites was compared by Persson-Moschos et al[17] with that from control subjects in a nested case-control study. A total of 12 500 middle-aged men was screened from 1974-1982 in Malmo, Sweden, and 400 cancer cases were identified during follow-up until the end of 1988, and 302 plasma samples were available for analysis of SeP. Two living controls per case of the same screening day and age were chosen. The odds ratio for overall cancer risk in the lowest quintile of SeP level was 5.2 compared with that in the highest (P = 0.01). In subgroups of patients with digestive tract cancers, the odds ratios for cancer risk in the lowest group was 3.4 compared with the highest (P = 0.02). The selenoprotein levels in all cancer cases were higher than that in controls in this study. Mork et al[18] have analyzed the mRNA and protein expression, and enzyme activity of SeP of a matched pairs of biopsies of colorectal adenoma and adjacent normal mucosa from 11 patients. All colorectal adenomas revealed a marked reduction of SeP mRNA, protein and enzyme activity compared with adjacent tissues. Many other studies have proven this result later[19]. Calvo et al[20] found the down-regulation of SeP during the progression of prostate cancer. All these results were consistent with our results.
SePmRNA expression could be observed in all 3 kinds of liver tissues, mainly in vascular endothelial cells and lymphocytes of vasculature. This finding was in agreement with the previous reports that SeP was expressed in the vessel endothelial cells. Previous research using immunohistochemistry proved SeP was mainly in vessel endothelial cells in mouse liver, kidney and brain. As SeP can combine with biomembrane, interstitial substance might be its target. It was presumed that some fragments of SeP might promote its combination with carbohydrate on cell surface and in interstitial substance.
The positive expression of SePmRNA could be found in tissues of HCC, liver cirrhosis and normal liver. In normal liver tissues, the positive signals were found in the nucleus, mostly in the nucleolus and around the nucleus. In liver cirrhosis tissues, the positive signals were mainly in the nucleolus. In HCC tissue, the positive signals were in the cytoplasm. This result suggests that SeP mRNA is secreted into the cytoplasm from the nucleus. The characteristics of SePmRNA expression indicate that SeP may be involved in liver carcinogenesis from normal liver to liver cirrhosis and to HCC. The marked reduction of SePmRNA in tissues of liver cirrhosis and HCC may be attributed to the reduction in synthesizing the SePmRNA after liver cell damage, or to inadequate Se intake to synthesize the SePmRNA. The positive expression rate of SePmRNA in HCC tissue was the lowest, which was mainly in the cytoplasm but rarely in the nucleus. The difference was significant. It is indicated that there might be mutation or deletion of the SeP gene in HCC. Selenium in HCC tissues is found obviously lower than that in perifocal tissues by diamino-naphthalene fluorescence analysis assay. In HCC cells, the activity of SeP promotor can be repressed by IFN-γ, TNF-α, IL-1β and TGF-β1, which lowered the expression level of SeP mRNA and the production of SeP. Therefore, SeP mRNA deletion might be caused by some cytokines in HCC tissues. SeP is a Se-supplier protein accounting for 50% of blood plasma selenium. The low SeP level in HCC tissues may be due to not only lack of the selenium but also SeP gene mutation or deletion. The positive expression signals of SePmRNA in HCC tissues were in the cytoplasm possibly due to the increased reactive or protective synthesis of SePmRNA in the early stage of the carcinomatous change. In this stage, a large amount of SePmRNA secreted to the plasma from cells to confront the carcinomatous change of the cells. This phenomenon indicates that the reduction of SeP expression would be the early event in the carcinomatous change of HCC. And because the cancer is one of the genetic diseases, we can infer that SeP may relieve the degree of the damage or adduction of DNA in the nucleus because the level of drug metabolic enzyme increased to detoxicate the carcinogen in vitro after Se supplementation and that the most important role of SeP is cyto-protection against oxidation[21,22]. SeP might act as a scavenger that diminished oxygen-derived free radicals in vivo to protect DNA in cells against oxidative toxicity so as to escape averting to cancer cells. The deficiency of SeP may enhance the susceptibility of DNA to oxidative damage resulting from the reactive oxygen species (ROS) and the damage from phospholipid peroxidation leading to the deterioration of disease. Furthermore, SeP might improve the immunity of bodies to build up the ability to eliminate malignant cells because the level of IgG of the patients was raised as SeP increased. Also the supplement of Se could build up the immunity of humans.
On the whole, until now there has been no certain conclusion about the relation between SeP and cancers. Though our results have preliminarily indicated SeP may play an important role in the occurrence and development of cancers, further studies are needed to explore the detained mechanism in the future.
We wish to thank our colleagues of the First Affiliated Hospital of the School of Medicine, Xi’an Jiaotong University and the Fourth Military Medical University for their help in accomplishing the study.
Se deficiency can result in certain malignant tumors. It has been proved that the occurrence of some cancers can be decreased by administration of selenium as a dietary supplement. Se plays its biological role as the form of selenoproteins. SeP is the quantitatively most important selenoprotein. In recent years, many studies have confirmed that SeP is related to various malignant tumors. However, the expression of SeP in HCC tissues is rarely known. This paper tries to address the expression of SePmRNA in the tissues of normal liver, liver cirrhosis and HCC, and to investigate the relationship between SeP and the occurrence and the development of HCC.
A number of epidemiological studies, animal model systems and interventional trials have found that a low dietary intake of Se is related to an increased risk of liver cancer. There is inverse association between plasma Se levels and the risk of HCC. Se is present in known human selenoproteins as the amino acid Sec. SeP contains more than 50% of the total amount of plasma Se. The relationship between SeP and future risk of cancers has become the hot spot of studies in recent years.
Many scholars have analyzed the plasma Se levels of people in Qidong County, Jiangsu Province of China, where the incidence of HCC is highest in China. They found the inverse relation between plasma Se levels and the epidemic of HCC. Dietary supplement of Se could reduce the incidence of HBV infection by 77.2% and the precancerous lesions by 75.8%. This is the only study to compare the expression of SePmRNA in the tissues of normal liver, liver cirrhosis and HCC.
Se plays its biological role as the form of selenoproteins, which has been proved to be safe as SeMet supplementa-tion but not as selenite. The study of SeP mRNA expression helps better understand the mechanism by which Se suppressed tumors and the function of this protein,and provide new thoughts in the treatment of liver cancer.
The trace element selenium (Se) is an essential human nutrient. The term “selenoprotein” is restricted to the proteins containing selenocysteine. Selenoprotein P (SeP) is one of the 18 selenoproteins which have been found to have biological functions.
This manuscript addresses an important issue in development of hepatic diseases. The study focuses on the expression of selenoprotein in normal liver, liver cirrhosis and hepatocellular carcinoma. The human tissue samples were used to determine the mRNA of selenoprotein P using in situ hybridization.
S- Editor Wang J L- Editor Ma JY E- Editor Ma WH
| 1. | Xia Y, Hill KE, Byrne DW, Xu J, Burk RF. Effectiveness of selenium supplements in a low-selenium area of China. Am J Clin Nutr. 2005;81:829-834. |
| 2. | Abdulah R, Miyazaki K, Nakazawa M, Koyama H. Chemical forms of selenium for cancer prevention. J Trace Elem Med Biol. 2005;19:141-150. |
| 4. | Traulsen H, Steinbrenner H, Buchczyk DP, Klotz LO, Sies H. Selenoprotein P protects low-density lipoprotein against oxidation. Free Radic Res. 2004;38:123-128. |
| 5. | Lu JB, Sun XB, Dai DX, Zhu SK, Chang QL, Liu SZ, Duan WJ. Epidemiology of gastroenterologic cancer in Henan Province, China. World J Gastroenterol. 2003;9:2400-2403. |
| 6. | Yu MW, Horng IS, Hsu KH, Chiang YC, Liaw YF, Chen CJ. Plasma selenium levels and risk of hepatocellular carcinoma among men with chronic hepatitis virus infection. Am J Epidemiol. 1999;150:367-374. |
| 7. | Young KJ, Lee PN. Intervention studies on cancer. Eur J Cancer Prev. 1999;8:91-103. |
| 8. | Burk RF, Norsworthy BK, Hill KE, Motley AK, Byrne DW. Effects of chemical form of selenium on plasma biomarkers in a high-dose human supplementation trial. Cancer Epidemiol Biomarkers Prev. 2006;15:804-810. |
| 9. | Burk RF, Hill KE. Selenoprotein P: an extracellular protein with unique physical characteristics and a role in selenium homeostasis. Annu Rev Nutr. 2005;25:215-235. |
| 10. | Stoytcheva Z, Tujebajeva RM, Harney JW, Berry MJ. Efficient incorporation of multiple selenocysteines involves an inefficient decoding step serving as a potential translational checkpoint and ribosome bottleneck. Mol Cell Biol. 2006;26:9177-9184. |
| 11. | Mansur DB, Hao H, Gladyshev VN, Korotkov KV, Hu Y, Moustafa ME, El-Saadani MA, Carlson BA, Hatfield DL, Diamond AM. Multiple levels of regulation of selenoprotein biosynthesis revealed from the analysis of human glioma cell lines. Biochem Pharmacol. 2000;60:489-497. |
| 12. | Yang X, Hill KE, Maguire MJ, Burk RF. Synthesis and secretion of selenoprotein P by cultured rat astrocytes. Biochim Biophys Acta. 2000;1474:390-396. |
| 13. | Burk RF, Hill KE, Motley AK, Austin LM, Norsworthy BK. Deletion of selenoprotein P upregulates urinary selenium excretion and depresses whole-body selenium content. Biochim Biophys Acta. 2006;1760:1789-1793. |
| 15. | Tujebajeva RM, Harney JW, Berry MJ. Selenoprotein P expression, purification, and immunochemical characterization. J Biol Chem. 2000;275:6288-6294. |
| 16. | Mostert V, Dreher I, Kohrle J, Abel J. Transforming growth factor-beta1 inhibits expression of selenoprotein P in cultured human liver cells. FEBS Lett. 1999;460:23-26. |
| 17. | Persson-Moschos ME, Stavenow L, Akesson B, Lindgärde F. Selenoprotein P in plasma in relation to cancer morbidity in middle-aged Swedish men. Nutr Cancer. 2000;36:19-26. |
| 18. | Mörk H, al-Taie OH, Bähr K, Zierer A, Beck C, Scheurlen M, Jakob F, Köhrle J. Inverse mRNA expression of the selenocysteine-containing proteins GI-GPx and SeP in colorectal adenomas compared with adjacent normal mucosa. Nutr Cancer. 2000;37:108-116. |
| 19. | Al-Taie OH, Uceyler N, Eubner U, Jakob F, Mörk H, Scheurlen M, Brigelius-Flohe R, Schöttker K, Abel J, Thalheimer A. Expression profiling and genetic alterations of the selenoproteins GI-GPx and SePP in colorectal carcinogenesis. Nutr Cancer. 2004;48:6-14. |
| 20. | Calvo A, Xiao N, Kang J, Best CJ, Leiva I, Emmert-Buck MR, Jorcyk C, Green JE. Alterations in gene expression profiles during prostate cancer progression: functional correlations to tumorigenicity and down-regulation of selenoprotein-P in mouse and human tumors. Cancer Res. 2002;62:5325-5335. |
| 21. | Steinbrenner H, Bilgic E, Alili L, Sies H, Brenneisen P. Selenoprotein P protects endothelial cells from oxidative damage by stimulation of glutathione peroxidase expression and activity. Free Radic Res. 2006;40:936-943. |
| 22. | Steinbrenner H, Alili L, Bilgic E, Sies H, Brenneisen P. Involvement of selenoprotein P in protection of human astrocytes from oxidative damage. Free Radic Biol Med. 2006;40:1513-1523. |