Published online Apr 28, 2007. doi: 10.3748/wjg.v13.i16.2324
Revised: February 3, 2007
Accepted: March 28, 2007
Published online: April 28, 2007
AIM: To observe the inhibition of hepatitis B virus (HBV) replication and expression by combination of siRNA and lamivudine in HepG2.2.15 cells.
METHODS: Recombinant plasmid psil-HBV was constructed and transfected into HepG2.2.15 cells. The transfected cells were cultured in lamivudine-containing medium (0.05 μmol/L) and harvested at 48, 72 and 96 h. The concentration of HBeAg and HBsAg was determined using ELISA. HBV DNA replication was examined by real-time PCR and the level of HBV mRNA was measured by RT-PCR.
RESULTS: In HepG2.2.15 cells treated with combination of siRNA and lamivudine, the secretion of HBeAg and HBsAg into the supernatant was found to be inhibited by 91.80% and 82.40% (2.89 ± 0.48 vs 11.73 ± 0.38, P < 0.05; 4.59 ± 0.57 vs 16.25 ± 0.48, P < 0.05) at 96 h, respectively; the number of HBV DNA copies within culture medium was also significantly decreased at 96 h (1.04 ± 0.26 vs 8.35 ± 0.33, P < 0.05). Moreover, mRNA concentration in HepG2.2.15 cells treated with combination of siRNA and lamivudine was obviously lower compared to those treated either with siRNA or lamivudine (19.44 ± 0.17 vs 33.27 ± 0.21 or 79.9 ± 0.13, P < 0.05).
CONCLUSION: Combination of siRNA and lamivudine is more effective in inhibiting HBV replication as compared to the single use of siRNA or lamivudine in HepG2.2.15 cells.
- Citation: Li GQ, Xu WZ, Wang JX, Deng WW, Li D, Gu HX. Combination of small interfering RNA and lamivudine on inhibition of human B virus replication in HepG2.2.15 cells. World J Gastroenterol 2007; 13(16): 2324-2327
- URL: https://www.wjgnet.com/1007-9327/full/v13/i16/2324.htm
- DOI: https://dx.doi.org/10.3748/wjg.v13.i16.2324
Chronic HBV infection is one of the most severe infectious diseases in humans. More than 400 million people worldwide suffer from chronic HBV infection and the infected individuals are at high risk of developing liver cirrhosis and hepatocellular carcinoma[1]. Current efforts to treat HBV infection have only limited success. Only 20% of the patients benefit from anti-HBV therapy with recombinant interferon and nucleotide analogues such as lamivudine[2-5]. Thus, there is an urgent need to develop more effective strategies to combat HBV infection.
RNA interference (RNAi) is a cellular progress in which double-stranded RNA of 21-23 nt induces the post-transcriptional gene silencing of homologous mRNA[6-10]. Based on the important role on the reverse transcription step in HBV replication[11-15], several researches have demonstrated that RNAi could suppress the replication of HBV through silencing the expression of the viral genes and provide a new method against chronic HBV infection[16-19]. Considering that lamivudine has been a leading drug which is highly effective in inhibiting HBV DNA synthesis[20] and RNAi is a newly discovered antiviral mechanism[6,7], we hypothesize that combination of siRNA and lamivudine may exhibit greater inhibitory effect on HBV replication. To address this question, we systemically measured the anti-HBV effect of siRNA combined with lamivudine on HepG2.2.15 cells at HBeAg, HBsAg, DNA and mRNA levels. Our findings suggested that combination of siRNA and lamivudine exerted a pronounced inhibition of HBV replication.
Dulbecco’s modified Eagle’s medium (DMEM) and G418 were purchased from GIBCO BRL (USA). HepG2.2.15 cells were maintained in our laboratory. Plasmid psi/U6 was supplied by Wuhan JS Biotech (China). All PCR primers were synthesized by Shanghai Boya Biological Company (China). Trizol, M-MLV reverse transcriptase, Lipofection 2000 reagents were purchased from Invitrogen Company (USA). Restriction enzymes were purchased from Bejing New England Biolab (China).
HBV siRNA expression plasmid was generated by inserting a 21-nt specific sequence encoding corresponding siRNA into BamHI-HindIII sites of plasmid psil/U6 as previously described[21] and further analyzed by BLAST to ensure that it does not has sequence homology with human genome sequence. The oligonucleotide pairs used to coding for the same strand of siRNA were as follows: 5’-GATCCGCACTTCCGGAAACTGTTTCAAGACGACAGTAGTTTCCGGAAGTGTTTTTTGTCGACA-3’ (sense) and 3’-GCGTGAAGGCCTTTGATGACAAAGTTCTGCTGTCATCAAAGGCCTTCACAAAAAACAGCTGTTCGA-5’ (antisense). The recombinant plasmid, named as psil-HBV, was transformed into E. coli strain DH5α and identified by restricted endonuclease digestion and DNA sequence analysis. Construction and identification of an irrelevant control plasmid HK were essentially the same as described above.
HepG2.2.15 cells were cultured in DMEM supplemented with 100 mL/L fetal calf serum (FCS), 100 U/mL penicillin, 100 μg/mL streptomycin and 200 μg/mL G418 and incubated at 37°C in a humidified atmosphere containing 50 mL/L CO2 in air. Cells were cultured in 6-well plates until they were 60%-70% confluence 24 h prior to transfection. Transfection was performed using Lipofection 2000 reagent and psil/HBV complex at 2.5:1 ratio following the manufacturer’s instructions. The transfected cells were cultured in lamivudine-containing medium (0.5 μmol/L) and harvested at 48 h, 72 h and 96 h for further analysis. All experiments were performed in triplicate and divided into five groups.
The levels of HBeAg and HBsAg in culture supernatants were measured using ELISA kits (SA Biotech, Shanghai, China) at 48, 72 and 96 h according to the manufacturer’s instructions.
HBV DNA was extracted from the culture medium and examined by real-time PCR using HBV diagnostic kit (PG Biotech, Shenzhen, China) at 48, 72 and 96 h according to the manufacturer’s protocol.
Total RNA of the transfected cells was isolated with Trizol reagent and reverse transcribed into cDNA with 200 U of M-MLV reverse transcriptase at 48, 72 and 96 h.
Samples were amplified through 30 cycles, each amplification cycle consisting of denaturation at 94°C for 15 s, primer annealing at 52°C for 30 s, and extension at 72°C for 30 s. Cycles were preceded by incubation at 94°C for 3 min to ensure full denaturation of the target gene and were followed by an extra incubation at 72°C for 10 min to ensure full extension of the products. Primers for PCR were as follows: HBV, 5’-ACCTCTGCCTAATCATCTC-3’ (forward) and 5’-GTAAGACAGGAAATGT GAAAC-3’ (reverse); and GAPDH 5’-GTCGGTGTGAACGGATTT-3’ (forward) and 5’-ACTCCACGACGTACTCAGC-3’ (reverse). The PCR amplification product was analyzed by 1.2% agarose gel electrophoresis.
Data were expressed as mean ± SD. Data were compared using F-test and one-way Anova. All statistical analyses were performed using the SPSS 12.0 software. P < 0.05 was considered statistically significant.
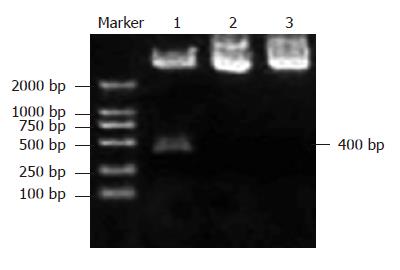
The PstIsite in the plasmid psil-HBV was replaced by SalIsite that we designed in the inserted DNA sequence. The fragment digested by SalIwas about 400 bp in size (Figure 1). The DNA sequence analysis of positive clones confirmed the results as expected.
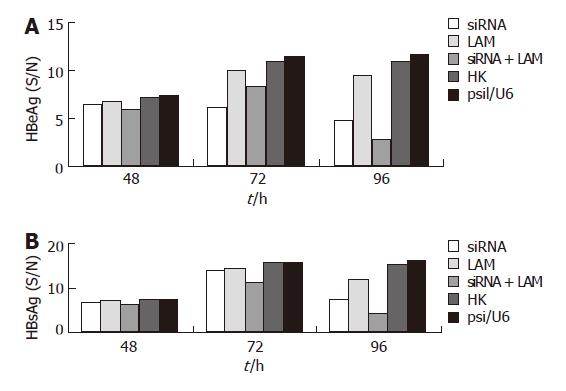
The concentrations of HBeAg and HBsAg in the culture supernatant were measured at 48, 72 and 96 h by ELISA. As is shown in Figure 2 and Table 1, there was no significant inhibition on the expression of HBeAg and HBsAg at 48 and 72 h; an obvious inhibition (P < 0.05) was, however, observed at 96 h, with the reduction in levels of HBsAg and HBeAg by 91.8% and 82.4%, respectively, in the supernatant of cells treated with combined siRNA and lamivudine as compared to the single use of siRNA or lamivudine.
| HBeAg | HBsAg | |||||
| Group | 48 h | 72 h | 96 h | 48 h | 72 h | 96 h |
| siRNA | 6.54 ± 0.30 (15.43) | 6.23 ± 0.27 (55.78) | 4.90 ± 0.29 (70.92)a | 6.94 ± 0.14 (13.88) | 14.00 ± 0.64 (15.06) | 7.68 ± 0.70 (60.57)a |
| LAM | 6.80 ± 0.22 (10.48) | 10.02 ± 0.45 (15.20) | 9.53 ± 0.44 (22.85)a | 7.10 ± 0.19 (11.03) | 14.39 ± 0.45 (12.38) | 11.93 ± 0.36(30.53)a |
| siRNA+LAM | 6.01 ± 0.42 (25.52) | 8.40 ± 0.20(32.55) | 2.89 ± 0.48 (91.8)ac | 6.47 ± 0.27 (22.24) | 11.26 ± 0.77 (34.62) | 4.59 ± 0.57(82.40)ac |
| HK | 7.25 ± 0.27 (0) | 11.06 ± 0.21 (0) | 10.92 ± 0.33 (0) | 7.52 ± 0.18 (0) | 15.93 ± 0.44(0) | 15.43 ± 0.47 (0) |
| psi/U6 | 7.35 ± 0.34 | 11.44 ± 0.17 | 11.73 ± 0.38 | 7.72 ± 0.22 | 16.11 ± 0.41 | 16.25 ± 0.48 |
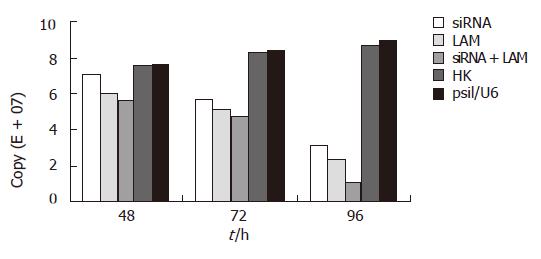
The level of HBV DNA was examined by real-time PCR. This approach can detect HBV DNA in the range of 104 to 108 copies. As shown in Figure 3 and Table 2, the copies of viral DNA in cells treated with siRNA combined with lamivudine reduced by 88.42% at 96 h (P < 0.05) compared to controls. The inhibitory effect was much higher than that obtained with single use of siRNA or lamivudine.
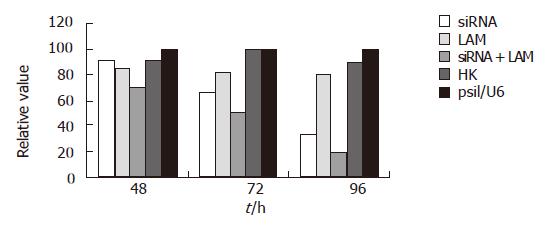
The level of HBV mRNA was examined by RT-PCR. The results revealed that the use of siRNA combined with lamivudine yielded the greatest reduction by 80.56% at 96 h compared to either siRNA or lamivudine alone (Figure 4).
RNAi regulates gene expression via a unique mechanism by degradation of target mRNA in a sequence-specific manner[6,9]. There has been considerable reports on the use of RANi to inhibit viral replication, such as HCV, HIV and poliovirus, etc[22-24]. Although synthetic or vector-based siRNA has been shown to efficiently suppress HBV replication[16-19], the possibility of the combination of siRNA and lamivudine to inhibit HBV replication has not been systemically studied. In this paper, we showed that the use of siRNA combined with lamivudine markedly reduced RNA level in HepG2.2.15 cells. Furthermore, the results indicated that combination of siRNA and lamivudine exhibited a stronger anti-HBV effect than the single use siRNA or Lamivudine.
During HBV replication, HBV genome shows a rapid mutation due to the high rate of replication and the lack of proofreading by reverse transcription[3]. Thus, drug resistant mutations appeared upon completing of antiviral therapy with lamivudine, which concentrated on the inhibition of reverse transcriptase activity[4]. During RNAi process, siRNA first recognizes and degrades the corresponding sequence of viral RNA, consequently, depleting template for reverse transcription. Decreased synthesis of HBV DNA then further influences the life-cycle of viral genome, especially in transcription and translation, thereby resulting in the overall inhibition of viral replication. In addition, the numerous potential targets for RNAi along HBV genome make it possible to target conserved regions, limiting the viral ability to create escape mutations. The ability of siRNAs to reduce the level of viral transcripts and proteins even in the absence of active viral replication makes it a good candidate as an assistant therapy to lamivudine, which acts only on the replication-competent HBV.
Although the development of a more effective anti-HBV therapy is still a challenge, our findings provide solid evidence for the feasibility of a cocktail treatment, which may enhance anti-HBV efficacy and overcome the shortcomings of current therapy. Therefore, this new method which combined gene therapy and chemotherapy opens a new avenue for treating chronic HBV infection. The precise mechanism by which combination of siRNA and lamivudine exerts more inhibitory efficacy on HBV replication needs to be clarified in further studies.
S- Editor Liu Y L- Editor Kumar M E- Editor Lu W
| 1. | Kao JH, Chen DS. Global control of hepatitis B virus infection. Lancet Infect Dis. 2002;2:395-403. |
| 2. | Ganem D, Prince AM. Hepatitis B virus infection--natural history and clinical consequences. N Engl J Med. 2004;350:1118-1129. |
| 3. | Papatheodoridis GV, Dimou E, Papadimitropoulos V. Nucleoside analogues for chronic hepatitis B: antiviral efficacy and viral resistance. Am J Gastroenterol. 2002;97:1618-1628. |
| 4. | Ayres A, Bartholomeusz A, Lau G, Lam KC, Lee JY, Locarnini S. Lamivudine and Famciclovir resistant hepatitis B virus associated with fatal hepatic failure. J Clin Virol. 2003;27:111-116. |
| 5. | Jonas MM, Mizerski J, Badia IB, Areias JA, Schwarz KB, Little NR, Greensmith MJ, Gardner SD, Bell MS, Sokal EM. Clinical trial of lamivudine in children with chronic hepatitis B. N Engl J Med. 2002;346:1706-1713. |
| 6. | Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293-296. |
| 7. | Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550-553. |
| 8. | Chuang CF, Meyerowitz EM. Specific and heritable genetic interference by double-stranded RNA in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2000;97:4985-4990. |
| 10. | Geiss G, Jin G, Guo J, Bumgarner R, Katze MG, Sen GC. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J Biol Chem. 2001;276:30178-30182. |
| 11. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. |
| 12. | Zhang XN, Xiong W, Wang JD, Hu YW, Xiang L, Yuan ZH. siRNA-mediated inhibition of HBV replication and expression. World J Gastroenterol. 2004;10:2967-2971. |
| 13. | Shlomai A, Shaul Y. Inhibition of hepatitis B virus expression and replication by RNA interference. Hepatology. 2003;37:764-770. |
| 14. | Paddison PJ, Caudy AA, Hannon GJ. Stable suppression of gene expression by RNAi in mammalian cells. Proc Natl Acad Sci USA. 2002;99:1443-1448. |
| 15. | Yu JY, DeRuiter SL, Turner DL. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA. 2002;99:6047-6052. |
| 16. | Tang N, Huang AL, Zhang BQ, Yan G, He TC. [Potent and specific inhibition of hepatitis B virus antigen expression by RNA interference]. Zhonghua Yixue Zazhi. 2003;83:1309-1312. |
| 17. | Uprichard SL, Boyd B, Althage A, Chisari FV. Clearance of hepatitis B virus from the liver of transgenic mice by short hairpin RNAs. Proc Natl Acad Sci USA. 2005;102:773-778. |
| 18. | Wu HL, Huang LR, Huang CC, Lai HL, Liu CJ, Huang YT, Hsu YW, Lu CY, Chen DS, Chen PJ. RNA interference-mediated control of hepatitis B virus and emergence of resistant mutant. Gastroenterology. 2005;128:708-716. |
| 19. | Wu KL, Zhang X, Zhang J, Yang Y, Mu YX, Liu M, Lu L, Li Y, Zhu Y, Wu J. Inhibition of Hepatitis B virus gene expression by single and dual small interfering RNA treatment. Virus Res. 2005;112:100-107. |
| 20. | Lau DT, Khokhar MF, Doo E, Ghany MG, Herion D, Park Y, Kleiner DE, Schmid P, Condreay LD, Gauthier J. Long-term therapy of chronic hepatitis B with lamivudine. Hepatology. 2000;32:828-834. |
| 21. | Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA. 2002;99:5515-5520. |
| 22. | Kapadia SB, Brideau-Andersen A, Chisari FV. Interference of hepatitis C virus RNA replication by short interfering RNAs. Proc Natl Acad Sci USA. 2003;100:2014-2018. |